Novel Sensitive Electrochemical Immunosensor Development for the Selective Detection of HopQ H. pylori Bacteria Biomarker
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus
2.2. Reagents
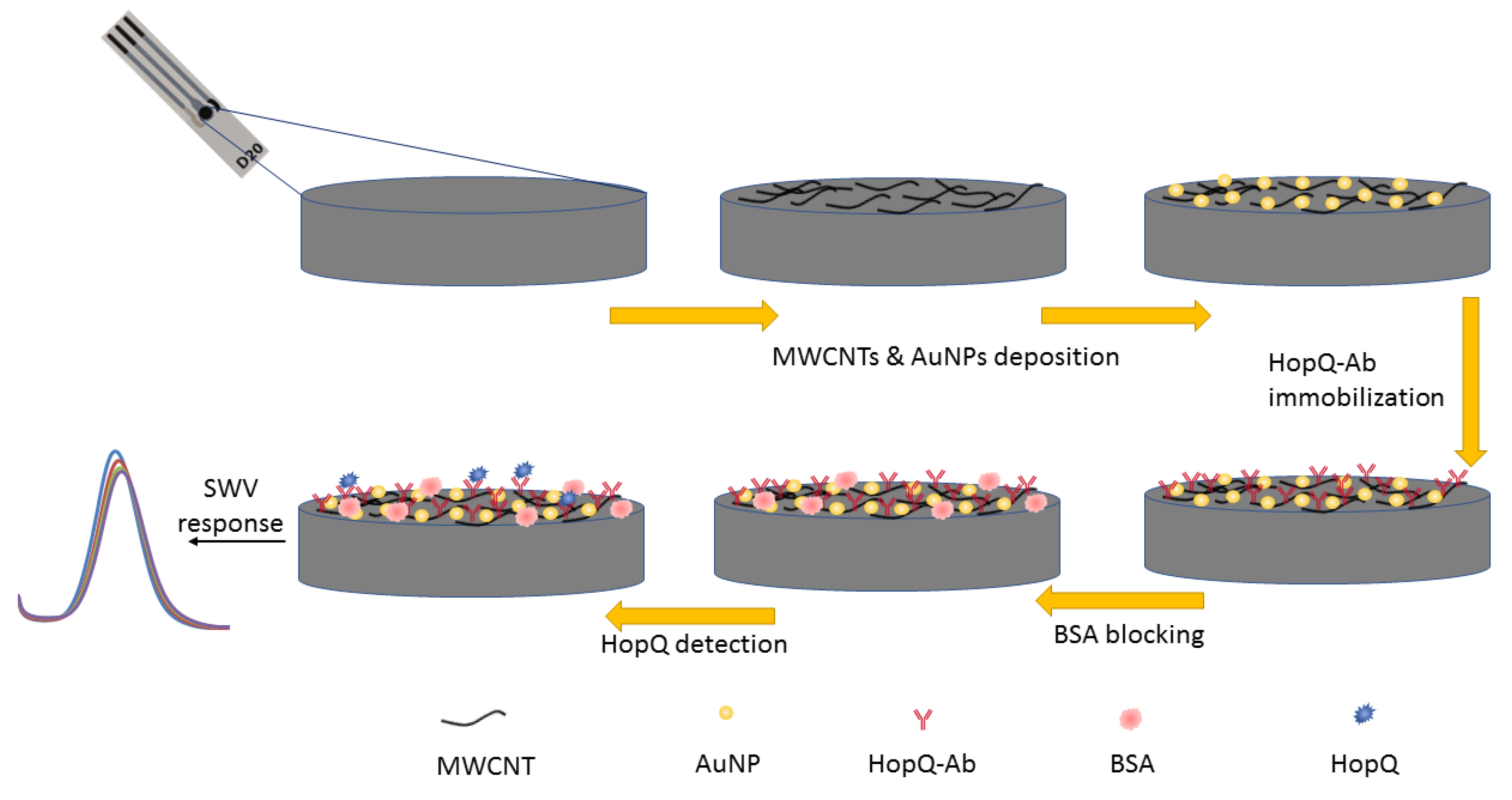
2.3. Immunosensor Preparation
2.3.1. Activation and Pretreatment of SPCE
2.3.2. Nanocomposite Preparation and Surface Modification
2.3.3. WE Preparation and HopQ-Ab Immobilization
2.4. Electrode Characterization
2.4.1. Electrochemical Measurements
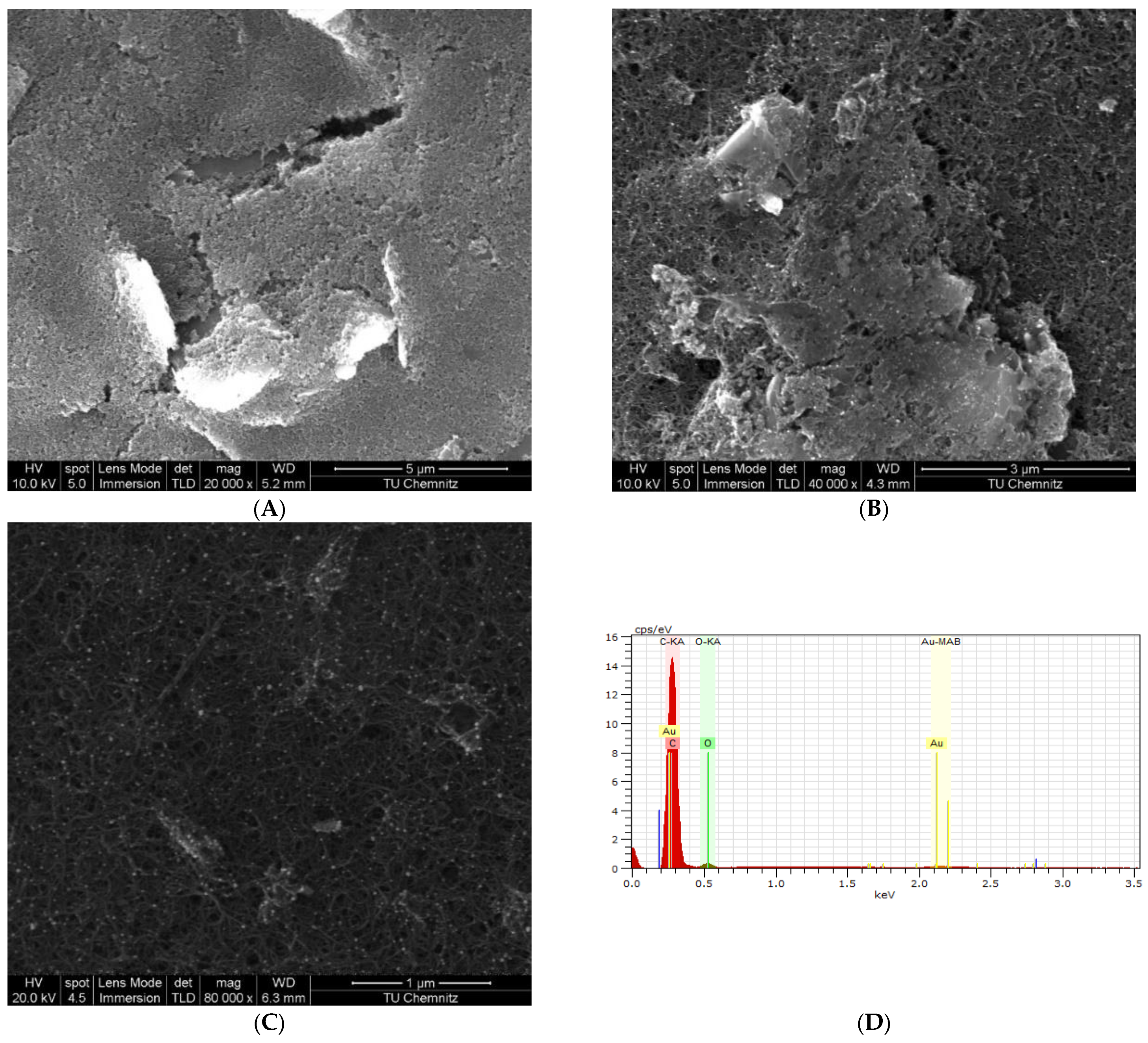
2.4.2. Surface Characterization by Scanning Electron Microscopy
2.5. Analytical Performance and Detection of HopQ
2.5.1. Detection of HopQ and Calibration Curve
2.5.2. Analytical Performance
3. Results
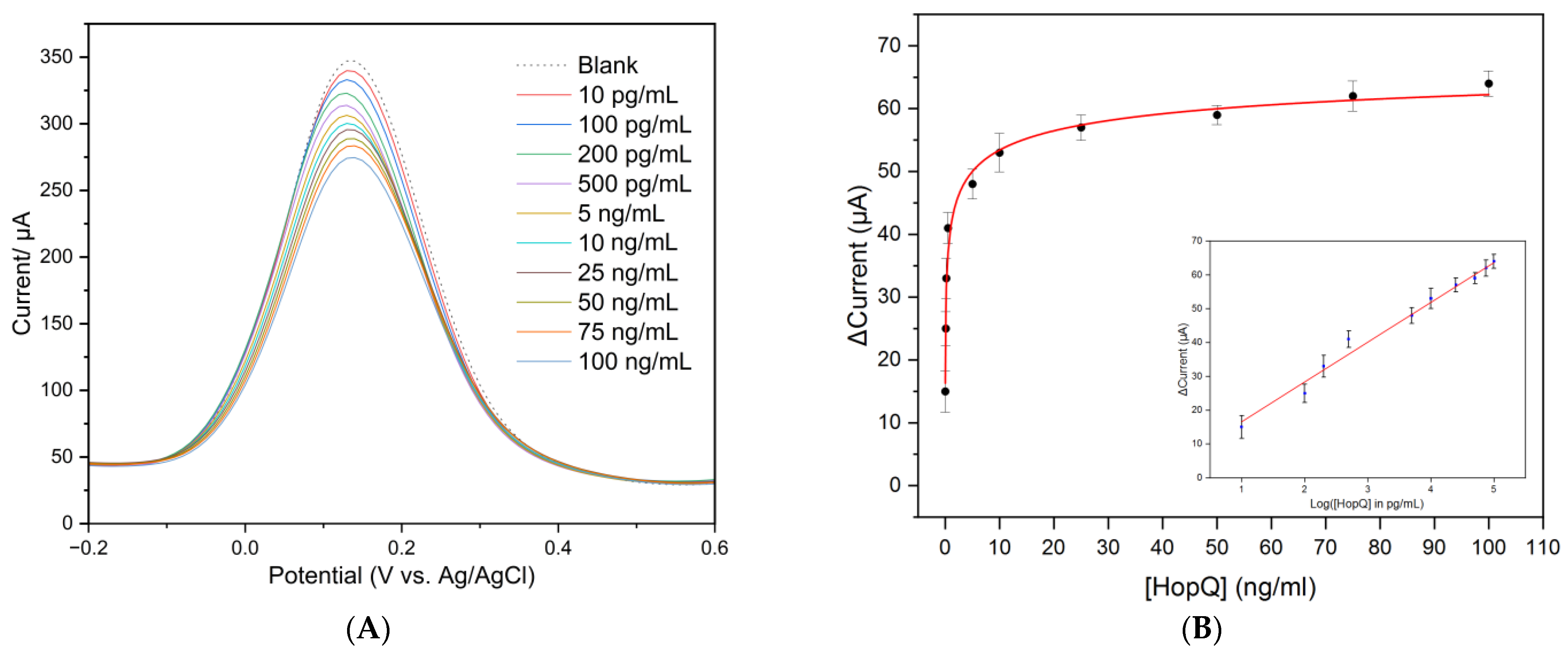
3.1. Electrochemical Measurements and Detection of HopQ
3.1.1. Electrochemical Measurements
3.1.2. Electrochemical Impedance Studies
3.2. Immunosensor Analytical Performance
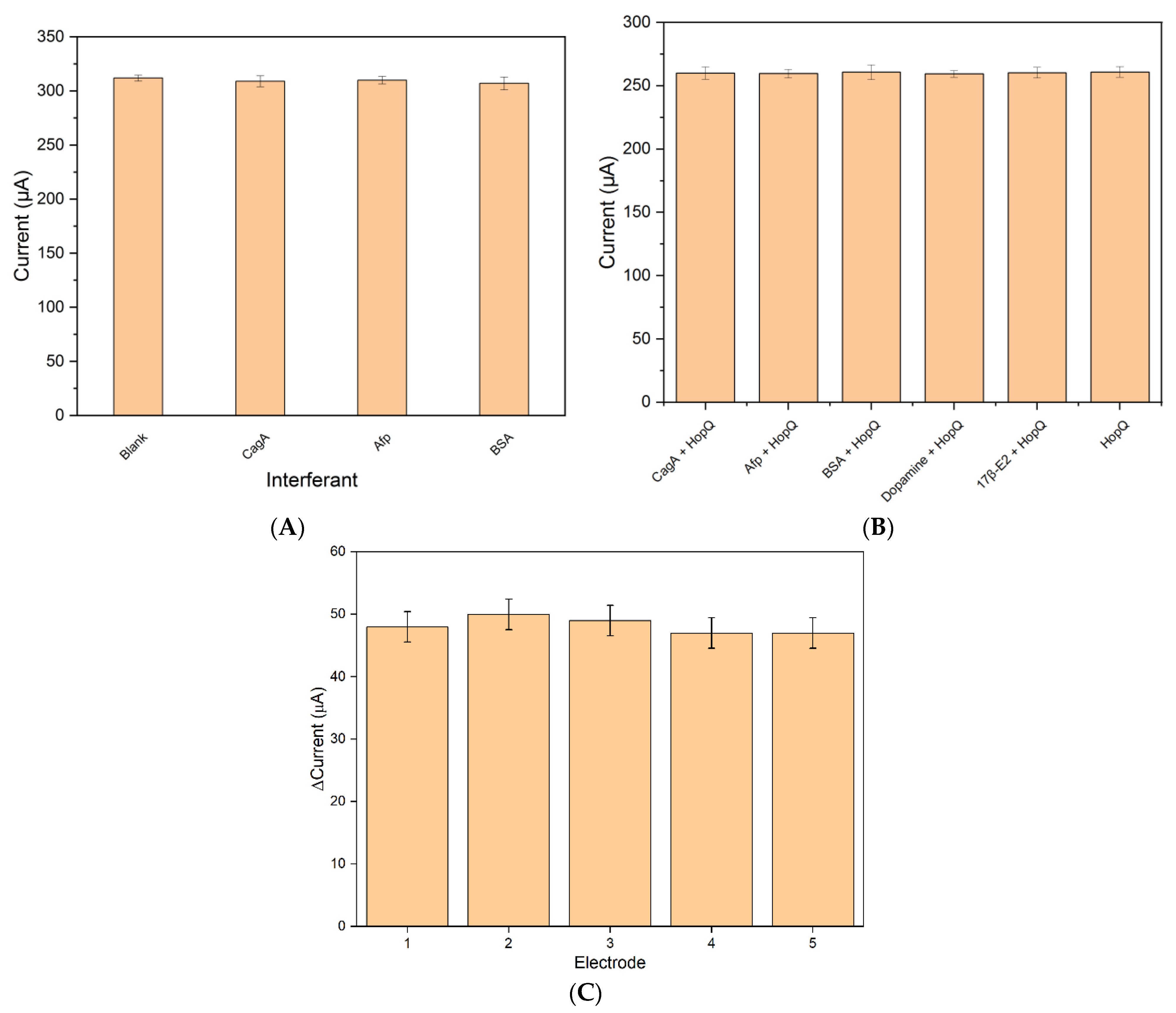
3.3. Selectivity and Cross-Reactivity
3.3.1. Selectivity
3.3.2. Cross-Reactivity
3.4. Reproducibility
3.5. Shelf-Life Studies and Comparison with Other Platforms
3.6. Artificial Saliva Samples
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steiger, C.; Abramson, A.; Nadeau, P.; Chandrakasan, A.P.; Langer, R.; Traverso, G. Ingestible electronics for diagnostics and therapy. Nat. Rev. Mater. 2018, 4, 83–98. [Google Scholar] [CrossRef]
- Suzuki, S.; Esaki, M.; Kusano, C.; Ikehara, H.; Gotoda, T. Development of Helicobacter pylori treatment: How do we manage antimicrobial resistance? World J. Gastroenterol. 2019, 25, 1907–1912. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Kim, J.-B.; Lee, P.; Kim, S.-H. Evodiamine Inhibits Helicobacter pylori Growth and Helicobacter pylori-Induced Inflammation. Int. J. Mol. Sci. 2021, 22, 3385. [Google Scholar] [CrossRef]
- Ahn, H.J.; Lee, D.S. Helicobacter pylori in gastric carcinogenesis. World J. Gastrointest. Oncol. 2015, 7, 455–465. [Google Scholar] [CrossRef]
- IARC. The International Agency for Research on Cancer. Available online: www.iarc.who.int (accessed on 10 March 2023).
- WHO. World Health Organization. August 2022. Available online: https://www.who.int (accessed on 26 February 2023).
- Meégraud, F.; Lehours, P. Helicobacter pylori Detection and Antimicrobial Susceptibility Testing. Clin. Microbiol. Rev. 2007, 20, 280–322. [Google Scholar] [CrossRef] [PubMed]
- Javaheri, A.; Kruse, T.; Moonens, K.; Mejías-Luque, R.; Debraekeleer, A.; Asche, C.I.; Tegtmeyer, N.; Kalali, B.; Bach, N.C.; Sieber, S.A.; et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016, 2, 16189. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, J.; Ma, Q. The relationship between the simultaneity present of cagA and hopQI genes in Helicobacter pylori and the risk of gastric cancer. Cell. Mol. Biol. 2021, 67, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Amieva, M.; Peek, R.M., Jr. Pathobiology of Helicobacter pylori–Induced Gastric Cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.G.; Loke, M.F.; Goh, K.L.; Vadivelu, J.; Ho, B. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol. 2017, 62, 68–76. [Google Scholar] [CrossRef]
- Mladenova, I.; Durazzo, M. Transmission of Helicobacter pylori. Minerva Gastroenterol. Dietol. 2018, 64, 251–254. [Google Scholar] [CrossRef]
- Payão, L.T.R.S.L.M. Helicobacter pylori and its reservoirs: A correlation with the gastric infection. World J. Gastrointest. Pharmacol. Ther. 2016, 7, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Subjakova, V.; Oravczova, V.; Tatarko, M.; Hianik, T. Advances in electrochemical aptasensors and immunosensors for detection of bacterial pathogens in food. Electrochim. Acta 2021, 389, 138724. [Google Scholar] [CrossRef]
- Jaradat, H.; Al-Hamry, A.; Ibbini, M.; Kanoun, O. Early Detection of Helicobacter pylori Bacteria in Complex Samples. In Smart Sensors, Measurement and In-Strumentation; Springer Science and Business Media Deutschland GmbH: Berlin/Heidelberg, Germany, 2021; pp. 165–176. [Google Scholar] [CrossRef]
- Saxena, K.; Chauhan, N.; Jain, U. Advances in diagnosis of Helicobacter pylori through biosensors: Point of care devices. Anal. Biochem. 2021, 630, 114325. [Google Scholar] [CrossRef] [PubMed]
- Nosrati, R.; Golichenari, B.; Nezami, A.; Taghdisi, S.M.; Karimi, B.; Ramezani, M.; Abnous, K.; Shaegh, S.A.M. Helicobacter pylori point-of-care diagnosis: Nano-scale biosensors and microfluidic systems. TrAC Trends Anal. Chem. 2017, 97, 428–444. [Google Scholar] [CrossRef]
- Ricci, C.; Holton, J.; Vaira, D. Diagnosis of Helicobacter pylori: Invasive and non-invasive tests. Best Pract. Res. Clin. Gastroenterol. 2007, 21, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zhu, D.Z.; Gan, H.; Yao, Z.; Fu, Q.; Zhang, X. Prospects and challenges of using electrochemical immunosensors as an alternative detection method for SARS-CoV-2 wastewater-based epidemiology. Sci. Total Environ. 2021, 777, 146239. [Google Scholar] [CrossRef]
- Kim, J.; Park, M. Recent Progress in Electrochemical Immunosensors. Biosensors 2021, 11, 360. [Google Scholar] [CrossRef]
- Mutlaq, S.; Albiss, B.; Al-Nabulsi, A.A.; Osaili, T.; Al-Jaberi, T.; Olaimat, A.N.; Liu, S.-Q.; Ayyash, M.M. Detection of Salmonella Enteritidis in Milk Using Conductometric Immunosensor Coated on Polyaniline/Zinc Oxide Nanocomposite. Foodborne Pathog. Dis. 2023; ahead of print. [Google Scholar] [CrossRef]
- Filik, H.; Avan, A.A. Nanostructures for nonlabeled and labeled electrochemical immunosensors: Simultaneous electrochemical detection of cancer markers: A review. Talanta 2019, 205, 120153. [Google Scholar] [CrossRef]
- Zumpano, R.; Polli, F.; D’agostino, C.; Antiochia, R.; Favero, G.; Mazzei, F. Nanostructure-Based Electrochemical Immunosensors as Diagnostic Tools. Electrochem 2021, 2, 10–28. [Google Scholar] [CrossRef]
- Aydin, M.; Aydin, E.B.; Sezgintürk, M.K. Advances in immunosensor technology. Adv. Clin. Chem. 2021, 102, 1–62. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, J.; Huang, Y.; Zhai, J.; Liao, G.; Wang, Z.; Ning, C. Development of electroactive materials-based immunosensor towards early-stage cancer detection. Coord. Chem. Rev. 2022, 471, 214723. [Google Scholar] [CrossRef]
- Zhang, Z.; Cong, Y.; Huang, Y.; Du, X. Nanomaterials-Based Electrochemical Immunosensors. Micromachines 2019, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Pollap, A.; Kochana, J. Electrochemical Immunosensors for Antibiotic Detection. Biosensors 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Brimo, N.; Serdaroğlu, D. Antibody Immobilization Techniques in Mass Sensitive Immunosensor: Enhanced Sensitivity through Limited Mass Load. Curr. Anal. Chem. 2022, 18, 529–545. [Google Scholar] [CrossRef]
- Benjamin, S.R.; Nascimento, T.D.S.; Roque, C.R.; de Andrade, G.M.; Oriá, R.B. Recent advances in the development of immunosensors for infectious diseases. In Biosensors for Emerging and Re-Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2022; pp. 19–72. [Google Scholar] [CrossRef]
- Xu, C.; Soyfoo, D.M.; Wu, Y.; Xu, S. Virulence of Helicobacter pylori outer membrane proteins: An updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1821–1830. [Google Scholar] [CrossRef]
- Guo, X.; Yin, B.; Wang, C.; Huo, H.; Aziziaram, Z. Risk assessment of gastric cancer in the presence of Helicobacter pylori cagA and hopQII genes. Cell. Mol. Biol. 2021, 67, 299–305. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Dore, M.P.; Graham, D.Y. Diagnosis and Treatment of Helicobacter pylori Infection. Annu. Rev. Med. 2022, 73, 183–195. [Google Scholar] [CrossRef]
- Guevara, B.; Cogdill, A.G. Helicobacter pylori: A Review of Current Diagnostic and Management Strategies. Dig. Dis. Sci. 2020, 65, 1917–1931. [Google Scholar] [CrossRef]
- Mohammadian, T.; Ganji, L. The Diagnostic Tests for Detection of Helicobacter pylori Infection. Monoclon. Antib. Immunodiagn. Immunother. 2019, 38, 1–7. [Google Scholar] [CrossRef]
- Kamboj, A.K.; Cotter, T.G.; Oxentenko, A.S. Helicobacter pylori: The Past, Present, and Future in Management. Mayo Clin. Proc. 2017, 92, 599–604. [Google Scholar] [CrossRef]
- Mao, X.; Jakubovics, N.S.; Bächle, M.; Buchalla, W.; Hiller, K.-A.; Maisch, T.; Hellwig, E.; Kirschneck, C.; Gessner, A.; Al-Ahmad, A.; et al. Colonization of Helicobacter pylori in the oral cavity–An endless controversy? Crit. Rev. Microbiol. 2021, 47, 612–629. [Google Scholar] [CrossRef]
- Bordin, D.S.; Voynovan, I.N.; Andreev, D.N.; Maev, I.V. Current Helicobacter pylori Diagnostics. Diagnostics 2021, 11, 1458. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yang, W.; Gong, W.; Chen, X.; Zhu, Z.; Chen, H. Advanced Sensing Strategies Based on Different Types of Biomarkers toward Early Diagnosis of H. pylori. Crit. Rev. Anal. Chem. 2023, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Bai, H.; Zhang, P.; Zhou, X.; Ying, B. Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int. J. Oral Sci. 2023, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, F.; Vaez, H.; Taheri, R.A.; Sahebkar, A.; Behshod, P.; Khademi, F. Helicobacter pylori in water, vegetables and foods of animal origin: A systematic review and meta-analysis on the prevalence, antibiotic resistance and genotype status in Iran. Gene Rep. 2020, 21, 100913. [Google Scholar] [CrossRef]
- Tyszczuk-Rotko, K.; Kozak, J.; Czech, B. Screen-Printed Voltammetric Sensors—Tools for Environmental Water Monitoring of Painkillers. Sensors 2022, 22, 2437. [Google Scholar] [CrossRef] [PubMed]
- Loock, H.-P.; Wentzell, P.D. Detection limits of chemical sensors: Applications and misapplications. Sens. Actuators B Chem. 2012, 173, 157–163. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.C. Smart Sensors, Measurement and Instrumentation; Volume 38. Available online: http://www.springer.com/series/10617 (accessed on 11 March 2023).
- Gu, Y.; Li, Y.; Ren, D.; Sun, L.; Zhuang, Y.; Yi, L.; Wang, S. Recent advances in nanomaterial-assisted electrochemical sensors for food safety analysis. Food Front. 2022, 3, 453–479. [Google Scholar] [CrossRef]
- Vacek, J.; Hrbac, J. Sensors and microarrays in protein biomarker monitoring: An electrochemical perspective spots. Bioanalysis 2020, 12, 1337–1345. [Google Scholar] [CrossRef]
- Patil, A.V.P.; Chuang, Y.-S.; Li, C.; Wu, C.-C. Recent Advances in Electrochemical Immunosensors with Nanomaterial Assistance for Signal Amplification. Biosensors 2023, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Cheng, N.; Luo, Y.; Lin, Y.; Xu, W.; Du, D. Recent advances in nanomaterials-based electrochemical (bio)sensors for pesticides detection. TrAC Trends Anal. Chem. 2020, 132, 116041. [Google Scholar] [CrossRef]
- Jiang, P.; Wang, Y.; Zhao, L.; Ji, C.; Chen, D.; Nie, L. Applications of Gold Nanoparticles in Non-Optical Biosensors. Nanomaterials 2018, 8, 977. [Google Scholar] [CrossRef]
- Mathew, M.; Radhakrishnan, S.; Vaidyanathan, A.; Chakraborty, B.; Rout, C.S. Flexible and wearable electrochemical biosensors based on two-dimensional materials: Recent developments. Anal. Bioanal. Chem. 2021, 413, 727–762. [Google Scholar] [CrossRef]
- Gupta, R.; Raza, N.; Bhardwaj, S.K.; Vikrant, K.; Kim, K.-H.; Bhardwaj, N. Advances in nanomaterial-based electrochemical biosensors for the detection of microbial toxins, pathogenic bacteria in food matrices. J. Hazard. Mater. 2021, 401, 123379. [Google Scholar] [CrossRef]
- Kour, R.; Arya, S.; Young, S.-J.; Gupta, V.; Bandhoria, P.; Khosla, A. Review—Recent Advances in Carbon Nanomaterials as Electrochemical Biosensors. J. Electrochem. Soc. 2020, 167, 037555. [Google Scholar] [CrossRef]
- Özmen, E.N.; Kartal, E.; Turan, M.B.; Yazıcıoğlu, A.; Niazi, J.H.; Qureshi, A. Graphene and carbon nanotubes interfaced electrochemical nanobiosensors for the detection of SARS-CoV-2 (COVID-19) and other respiratory viral infections: A review. Mater. Sci. Eng. C 2021, 129, 112356. [Google Scholar] [CrossRef]
- Zhou, Y.; Fang, Y.; Ramasamy, R. Non-Covalent Functionalization of Carbon Nanotubes for Electrochemical Biosensor Development. Sensors 2019, 19, 392. [Google Scholar] [CrossRef]
- Alam, A.U.; Deen, M.J. Bisphenol A Electrochemical Sensor Using Graphene Oxide and β-Cyclodextrin-Functionalized Multi-Walled Carbon Nanotubes. Anal. Chem. 2020, 92, 5532–5539. [Google Scholar] [CrossRef]
- Shu, R.; Liu, S.; Huang, L.; Li, Y.; Sun, J.; Zhang, D.; Zhu, M.-Q.; Wang, J. Enzyme-Mimetic nano-immunosensors for amplified detection of food hazards: Recent advances and future trends. Biosens. Bioelectron. 2022, 217, 114577. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, Y.; Liu, S.; Wu, D.; Su, Z.; Chen, G.; Liu, J.; Li, G. Recent advances in enzyme immobilization based on novel porous framework materials and its applications in biosensing. Coord. Chem. Rev. 2022, 459, 214414. [Google Scholar] [CrossRef]
- Ferrier, D.C.; Honeychurch, K.C. Carbon Nanotube (CNT)-Based Biosensors. Biosensors 2021, 11, 486. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, C.; Han, J.; Zhao, W.; Shao, S.; Li, S.; Gao, H.; Xie, H.; Zhang, X. Boosting electrochemical CO2 reduction to formate using SnO2/graphene oxide with amide linkages. J. Mater. Chem. A 2021, 9, 19681–19686. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, Y.; Kianfar, E. Nano Biosensors: Properties, applications and electrochemical techniques. J. Mater. Res. Technol. 2021, 12, 1649–1672. [Google Scholar] [CrossRef]
- Gupta, S.; Tiwari, A.; Jain, U.; Chauhan, N. Synergistic effect of 2D material coated Pt nanoparticles with PEDOT polymer on electrode surface interface for a sensitive label free Helicobacter pylori CagA(Ag-Ab) immunosensing. Mater. Sci. Eng. C 2019, 103, 109733. [Google Scholar] [CrossRef] [PubMed]
- Jain, U.; Gupta, S.; Soni, S.; Khurana, M.P.; Chauhan, N. Triple-nanostructuring-based non-invasive electro-immune sensing of CagA toxin for Helicobacter pylori detection. Helicobacter 2020, 25, e12706. [Google Scholar] [CrossRef]
- Saxena, K.; Kumar, A.; Chauhan, N.; Khanuja, M.; Malhotra, B.D.; Jain, U. Electrochemical Immunosensor for Detection of H. pylori Secretory Protein VacA on g-C3N4/ZnO Nanocomposite-Modified Au Electrode. ACS Omega 2022, 7, 32292–32301. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.; Gupta, S.; Avasthi, D.K.; Adelung, R.; Mishra, Y.K.; Jain, U. Zinc Oxide Tetrapods Based Biohybrid Interface for Voltammetric Sensing of Helicobacter pylori. ACS Appl. Mater. Interfaces 2018, 10, 30631–30639. [Google Scholar] [CrossRef]
- Gupta, S.; Jain, U.; Murti, B.T.; Putri, A.D.; Tiwari, A.; Chauhan, N. Nanohybrid-based immunosensor prepared for Helicobacter pylori BabA antigen detection through immobilized antibody assembly with @ Pdnano/rGO/PEDOT sensing platform. Sci. Rep. 2020, 10, 21217. [Google Scholar] [CrossRef]
- Belova, A.M.; Basmanov, D.V.; Prusakov, K.A.; Lazarev, V.N.; Klinov, D.V. A Microfluidic Platform for the Development of a Biosensor Based on Genetically Modified Helicobacter pylori Single Cells. Biophysics 2018, 63, 735–742. [Google Scholar] [CrossRef]
- Qiu, E.; Jin, S.; Xiao, Z.; Chen, Q.; Wang, Q.; Liu, H.; Xie, C.; Chen, C.; Li, Z.; Han, S. CRISPR-based detection of Helicobacter pylori in stool samples. Helicobacter 2021, 26, e12828. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, H.; Al-Hamry, A.; Wang, Q.; Wang, J.; Zhou, Y.; Song, Y.; Ibbini, M.; Kanoun, O. Performance Investigation of Screen-Printed Carbon Electrodes Activated by MES-Acid. In Proceedings of the 2022 19th International Multi-Conference on Systems, Signals & Devices (SSD), Setif, Algeria, 6–10 May 2022; pp. 410–415. [Google Scholar] [CrossRef]
- O’connell, L.; Marcoux, P.R.; Roupioz, Y. Strategies for Surface Immobilization of Whole Bacteriophages: A Review. ACS Biomater. Sci. Eng. 2021, 7, 1987–2014. [Google Scholar] [CrossRef] [PubMed]
- Sharafeldin, M.; McCaffrey, K.; Rusling, J.F. Influence of antibody immobilization strategy on carbon electrode immunoarrays. Analyst 2019, 144, 5108–5116. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guisán, J.M.; Rocha-Martin, J. Oriented immobilization of antibodies onto sensing platforms—A critical review. Anal. Chim. Acta 2022, 1189, 338907. [Google Scholar] [CrossRef] [PubMed]
- Puertas, S.; Villa, M.D.G.; Mendoza, E.; Jiménez-Jorquera, C.; de la Fuente, J.M.; Fernández-Sánchez, C.; Grazú, V. Improving immunosensor performance through oriented immobilization of antibodies on carbon nanotube composite surfaces. Biosens. Bioelectron. 2013, 43, 274–280. [Google Scholar] [CrossRef]
- Teixeira, P.R.; Santos, M.S.; Silva, A.L.G.; Báo, S.N.; Azevedo, R.B.; Sales, M.J.A.; Paterno, L.G. Photochemically-assisted synthesis of non-toxic and biocompatible gold nanoparticles. Colloids Surf. B Biointerfaces 2016, 148, 317–323. [Google Scholar] [CrossRef]
- Nasraoui, S.; Al-Hamry, A.; Teixeira, P.R.; Ameur, S.; Paterno, L.G.; Ben Ali, M.; Kanoun, O. Electrochemical sensor for nitrite detection in water samples using flexible laser-induced graphene electrodes functionalized by CNT decorated by Au nanoparticles. J. Electroanal. Chem. 2021, 880, 114893. [Google Scholar] [CrossRef]
- Kumar, A.K.S.; Zhang, Y.; Li, D.; Compton, R.G. A mini-review: How reliable is the drop casting technique? Electrochem. Commun. 2020, 121, 106867. [Google Scholar] [CrossRef]
- Yunker, P.J.; Still, T.; Lohr, M.A.; Yodh, A.G. Suppression of the coffee-ring effect by shape-dependent capillary interactions. Nature 2011, 476, 308–311. [Google Scholar] [CrossRef]
- Shakeri, A.; Abu Jarad, N.; Leung, A.; Soleymani, L.; Didar, T.F. Biofunctionalization of Glass- and Paper-Based Microfluidic Devices: A Review. Adv. Mater. Interfaces 2019, 6, 1900940. [Google Scholar] [CrossRef]
- Conde, J.; Dias, J.T.; Grazú, V.; Moros, M.; Baptista, P.V.; de la Fuente, J.M. Revisiting 30 years of biofunctionalization and surface chemistry of inorganic nanoparticles for nanomedicine. Front. Chem. 2014, 2, 48. [Google Scholar] [CrossRef]
- Gao, Y.; Kyratzis, I. Covalent Immobilization of Proteins on Carbon Nanotubes Using the Cross-Linker 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide—A Critical Assessment. Bioconjug. Chem. 2008, 19, 1945–1950. [Google Scholar] [CrossRef] [PubMed]
- Pundir, M.; Prasher, P.; Vasić, K.; Leitgeb, M.; Kumar, A.; Prakash, R.; Knez, Ž.; Pandey, J.K.; Kumar, S. Enzyme modified CNTs for biosensing application: Opportunities and challenges. Colloid Interface Sci. Commun. 2021, 44, 100506. [Google Scholar] [CrossRef]
- Garcia-Mendiola, T.; Bravo, I.; Maria Lopez-Moreno, J.; Pariente, F.; Wannemacher, R.; Weber, K.; Popp, J.; Lorenzo, E. Carbon Nanodots Based Biosensors for Gene Mutation Detection. Sens. Actuators B Chem. 2018, 256, 226–233. [Google Scholar] [CrossRef]
- Qian, L.; Durairaj, S.; Prins, S.; Chen, A. Nanomaterial-based electrochemical sensors and biosensors for the detection of pharmaceutical compounds. Biosens. Bioelectron. 2021, 175, 112836. [Google Scholar] [CrossRef]
- Leva-Bueno, J.; Peyman, S.A.; Millner, P.A. A review on impedimetric immunosensors for pathogen and biomarker detection. Med. Microbiol. Immunol. 2020, 209, 343–362. [Google Scholar] [CrossRef]
- Brodowski, M.; Kowalski, M.; Białobrzeska, W.; Pałka, K.; Walkusz, R.; Roguszczak, J.; Łęga, T.; Sosnowska, M.; Biedulska, M.; Kurzawa, J.K.; et al. Methodology of Selecting the Optimal Receptor to Create an Electrochemical Immunosensor for Equine Arteritis Virus Protein Detection. Chemosensors 2021, 9, 265. [Google Scholar] [CrossRef]
- Mokni, M.; Tlili, A.; Attia, G.; Khaoulani, S.; Zerrouki, C.; Omezzine, A.; Othmane, A.; Bouslama, A.; Fourati, N. Novel sensitive immunosensor for the selective detection of Engrailed 2 urinary prostate cancer biomarker. Biosens. Bioelectron. 2022, 217, 114678. [Google Scholar] [CrossRef]

| Electrode | Rs (Ω) | Rct (Ω) | CPE (F) |
|---|---|---|---|
| SPCE | 192.2 | 2306 | 6.24 × 10−7 |
| AuNP/CNT/SPCE | 165.4 | 416.8 | 3.79 × 10−6 |
| HopQ-Ab/AuNP/CNT/SPCE | 194.05 | 3917.3 | 1.13 × 10−6 |
| BSA/HopQ-Ab/AuNP/CNT/SPCE | 197.7 | 4720 | 9.22 × 10−7 |
| Ref. | Performance of Reported H. pylori Sensors | ||||||
|---|---|---|---|---|---|---|---|
| Interface | Detection Method | Biomarker | LOD (ng/mL) | Linear Range (ng/mL) | Stability at 4 °C (Weeks) | ||
| [64] | CagA-Ab/ZnO*-T/SP-AuE | DPV | CagA | 0.2 | 0.2–50 | 8–9 | Up to 90% |
| [62] | CagA-Ab/TiO2-NPs/c-MWNCT/Pin5COOH/AuE | SWV | CagA | 0.1 | 0.1–8.0 | ~3 | 90% |
| ~6 | 50% | ||||||
| [61] | CagA-Ab/Ptnano/PEDOT/rGO/AuE | EIS | CagA | 0.1 | 0.1–30 | ~8 | 60–70% |
| [65] | BabA-Ab/Pdnano/rGO/PEDOT/AuE | EIS | BabA | 0.2 | 0.2–20 | 8–9 | 70% |
| [63] | VacA-Ab/g-C3N4/ZnO/AuE | DPV | VacA | 0.1 | 0.1–12.8 | ~2 | 94% |
| This work | BSA/HopQ-Ab/AuNP/CNT/SPCE | SWV | HopQ | 0.002 | 0.01–100 | 4 | ~85% |
| 8 | ~60% | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaradat, H.; Al-Hamry, A.; Ibbini, M.; Fourati, N.; Kanoun, O. Novel Sensitive Electrochemical Immunosensor Development for the Selective Detection of HopQ H. pylori Bacteria Biomarker. Biosensors 2023, 13, 527. https://doi.org/10.3390/bios13050527
Jaradat H, Al-Hamry A, Ibbini M, Fourati N, Kanoun O. Novel Sensitive Electrochemical Immunosensor Development for the Selective Detection of HopQ H. pylori Bacteria Biomarker. Biosensors. 2023; 13(5):527. https://doi.org/10.3390/bios13050527
Chicago/Turabian StyleJaradat, Hussamaldeen, Ammar Al-Hamry, Mohammed Ibbini, Najla Fourati, and Olfa Kanoun. 2023. "Novel Sensitive Electrochemical Immunosensor Development for the Selective Detection of HopQ H. pylori Bacteria Biomarker" Biosensors 13, no. 5: 527. https://doi.org/10.3390/bios13050527
APA StyleJaradat, H., Al-Hamry, A., Ibbini, M., Fourati, N., & Kanoun, O. (2023). Novel Sensitive Electrochemical Immunosensor Development for the Selective Detection of HopQ H. pylori Bacteria Biomarker. Biosensors, 13(5), 527. https://doi.org/10.3390/bios13050527







