MoBioS: Modular Platform Technology for High-Throughput Construction and Characterization of Tunable Transcriptional Biological Sensors
Abstract
:1. Introduction
2. Material and Methods
2.1. Strains and Growth Conditions
2.2. Plasmid Construction
2.3. In Vivo Fluorescence Experiments
2.4. Data Processing and Statistical Analysis
3. Results
3.1. Modular Biological Sensor Platform: Plasmid Architecture Overview
3.2. SynSens: Construction and Characterization of Biosensors with an Easily Tunable Synthetic Architecture
3.3. NatSens: Construction and Characterization of Biosensors in Their Natural Architecture
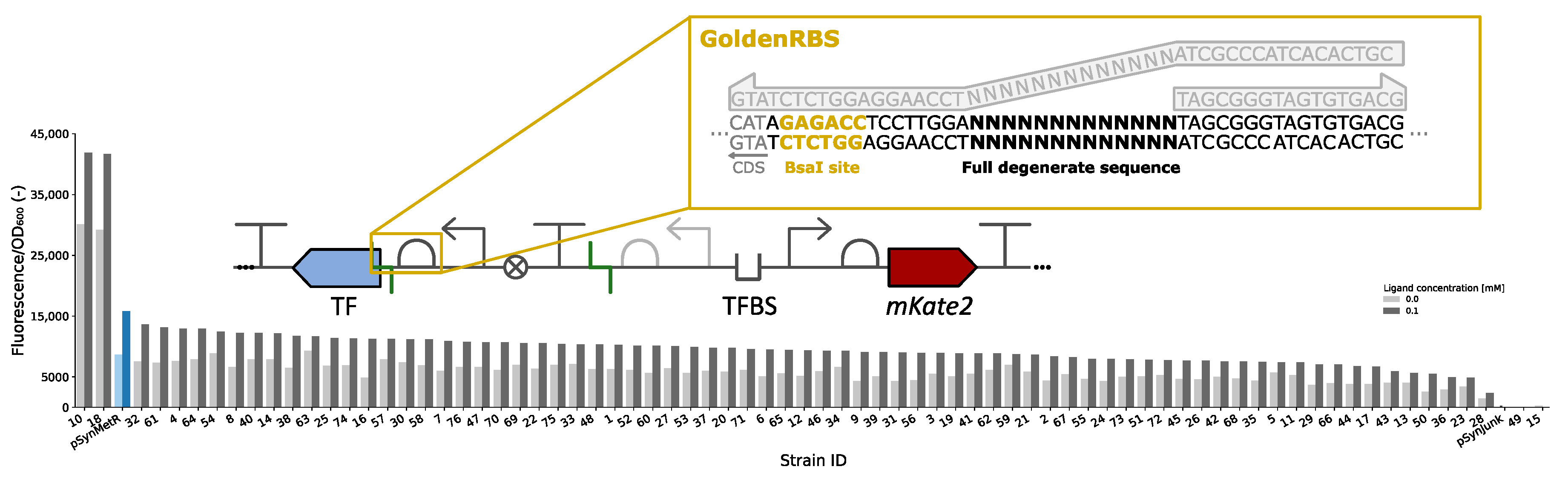
3.4. GoldenRBS Library for Fine-Tuning of Biosensor Response Curve
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Saha, B.C. Commodity Chemicals Production by Fermentation: An Overview. In Fermentation Biotechnology; American Chemical Society: Washington, DC, USA, 2003; Volume 12, pp. 3–17. [Google Scholar]
- Petzold, C.J.; Chan LJ, G.; Nhan, M.; Adams, P.D. Analytics for metabolic engineering. Front. Bioeng. Biotechnol. 2015, 3, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Mahr, R.; Frunzke, J. Transcription factor-based biosensors in biotechnology: Current state and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safaei, M.; Mobini, G.R.; Abiri, A.; Shojaeian, A. Synthetic biology in various cellular and molecular fields: Applications, limitations, and perspective. Mol. Biol. Rep. 2020, 47, 6207–6216. [Google Scholar] [CrossRef]
- Fernandez-López, R.; Ruiz, R.; de la Cruz, F.; Moncalián, G. Transcription factor-based biosensors enlightened by the analyte. Front. Microbiol. 2015, 6, 648. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Jacobsen, F.E.; Giedroc, D.P. Metal Transporters and Metal Sensors: How Coordination Chemistry Controls Bacterial Metal Homeostasis. Chem. Rev. 2009, 109, 4644–4681. [Google Scholar] [CrossRef] [Green Version]
- De Paepe, B.; Peters, G.; Coussement, P.; Maertens, J.; De Mey, M. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J. Ind. Microbiol. Biotechnol. 2017, 44, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Koch, M.; Pandi, A.; Borkowski, O.; Cardoso Batista, A.; Faulon, J.L. Custom-made transcriptional biosensors for metabolic engineering. Curr. Opin. Biotechnol. 2019, 59, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Stewart, A.J.; Plotkin, J.B. Why transcription factor binding sites are ten nucleotides long. Genetics 2012, 192, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Schallmey, M.; Frunzke, J.; Eggeling, L.; Marienhagen, J. Looking for the pick of the bunch: High-throughput screening of producing microorganisms with biosensors. Curr. Opin. Biotechnol. 2014, 26, 148–154. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, J.A.; Prather, K.L.J. Effective use of biosensors for high-throughput library screening for metabolite production. J. Ind. Microbiol. Biotechnol. 2021, 48, 49. [Google Scholar] [CrossRef]
- Boada, Y.; Vignoni, A.; Picó, J.; Carbonell, P. Extended Metabolic Biosensor Design for Dynamic Pathway Regulation of Cell Factories. iScience 2020, 23, 101305. [Google Scholar] [CrossRef]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef] [Green Version]
- Xia, P.-F.; Ling, H.; Foo, J.L.; Chang, M.W. Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 2019, 37, 107393. [Google Scholar] [CrossRef]
- Kim, S.G.; Noh, M.H.; Lim, H.G.; Jang, S.; Jang, S.; Koffas, M.A.G.; Jung, G.Y. Molecular parts and genetic circuits for metabolic engineering of microorganisms. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Min, B.E.; Hwang, H.G.; Lim, H.G.; Jung, G.Y. Optimization of industrial microorganisms: Recent advances in synthetic dynamic regulators. J. Ind. Microbiol. Biotechnol. 2017, 44, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Xu, P.; Ye, C.; Chen, X.; Liu, L. Genetic Circuit-Assisted Smart Microbial Engineering. Trends Microbiol. 2019, 27, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Lee, Y.; Kim, K.; Jang, G.; Yoon, Y. Transcription Factor-Based Biosensors for Detecting Pathogens. Biosensors 2022, 12, 470. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Du, R.; Xie, Z.; He, X.; Huang, K.; Luo, Y.; Xu, W. Using the promoters of MerR family proteins as “rheostats” to engineer whole-cell heavy metal biosensors with adjustable sensitivity. J. Biol. Eng. 2019, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Gallup, O.; Ming, H.; Ellis, T. Ten future challenges for synthetic biology. Eng. Biol. 2021, 5, 51–59. [Google Scholar] [CrossRef]
- Müller, K.M.; Arndt, K.M. Standardization in synthetic biology. Methods Mol. Biol. 2012, 813, 23–43. [Google Scholar] [PubMed]
- Decoene, T.; De Paepe, B.; Maertens, J.; Coussement, P.; Peters, G.; De Maeseneire, S.L.; De Mey, M. Standardization in synthetic biology: An engineering discipline coming of age. Crit. Rev. Biotechnol. 2017, 38, 647–656. [Google Scholar] [CrossRef]
- Quan, J.; Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 2009, 4, e6441. [Google Scholar] [CrossRef] [Green Version]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef] [Green Version]
- De Paepe, B.; Maertens, J.; Vanholme, B.; De Mey, M. Modularization and Response Curve Engineering of a Naringenin-Responsive Transcriptional Biosensor. ACS Synth. Biol. 2018, 7, 1303–1314. [Google Scholar] [CrossRef] [Green Version]
- De Mey, M.; Maertens, J.; Lequeux, G.J.; Soetaert, W.K.; Vandamme, E.J. Construction and model-based analysis of a promoter library for E. coli: An indispensable tool for metabolic engineering. BMC Biotechnol. 2007, 7, 34. [Google Scholar] [CrossRef]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovieva, E.A.; Chepurnykh, T.V.; Shcheglov, A.S.; Verkhusha, V.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Kabir, S.; Tsunedomi, R. Divergent Promoter Organization May Be a Preferred Structure for Gene Control in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2003, 8515, 206–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, C.F.; Warren, R.A. Divergent promoters, a common form of gene organization. Microbiol. Rev. 1988, 52, 318–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Lindquist, S.; Lindberg, F.; Normark, S. Binding of the Citrobacter freundii ampR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J. Bacteriol. 1989, 171, 3746–3753. [Google Scholar] [CrossRef]
- Ding, N.; Zhou, S.; Deng, Y. Transcription-Factor-based Biosensor Engineering for Applications in Synthetic Biology. ACS Synth. Biol. 2021, 10, 911–922. [Google Scholar] [CrossRef]
- Li, C.; Wang, C.; Zhu, J.; Xue, F.; Sun, X.; Gu, Y. Advances and prospects of transcription-factor-based biosensors in high-throughput screening for cell factories construction. Food Bioeng. 2022, 1, 135–147. [Google Scholar] [CrossRef]
- Mitchler, M.M.; Garcia, J.M.; Montero, E.N.; Williams, G.J. Transcription factor-based biosensors: A molecular-guided approach for natural product engineering. Curr. Opin. Biotechnol. 2021, 69, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef] [PubMed]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, I.S.; Juárez, P.; Fernández-Del-Carmen, M.A.; Granell, A.; Orzaez, D. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLoS ONE 2011, 6, e21622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murugesan, K.; Chang, Y.-Y.; Kim, Y.-M.; Jeon, J.-R.; Kim, E.-J. Enhanced transformation of triclosan by laccase in the presence of redox mediators. Water Res. 2010, 44, 298–308. [Google Scholar] [CrossRef]
- Picossi, S.; Belitsky, B.R.; Sonenshein, A.L. Molecular Mechanism of the Regulation of Bacillus subtilis gltAB Expression by GltC. J. Mol. Biol. 2007, 365, 1298–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’oelsnitz, S.; Love, J.D.; Diaz, D.J.; Ellington, A.D. GroovDB: A Database of Ligand-Inducible Transcription Factors. ACS Synth. Biol. 2022, 11, 3534–3537. [Google Scholar] [CrossRef] [PubMed]
- Santos-Zavaleta, A.; Salgado, H.; Gama-Castro, S.; Sánchez-Pérez, M.; Gómez-Romero, L.; Ledezma-Tejeida, D.; García-Sotelo, J.S.; Alquicira-Hernández, K.; Muñiz-Rascado, L.J.; Peña-Loredo, P.; et al. RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019, 47, D212–D220. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Makita, Y.; De hoon, M.; Nakai, K. DBTBS: A database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008, 36, D93–D96. [Google Scholar] [CrossRef] [PubMed]
- Novichkov, P.S.; Kazakov, A.E.; Ravcheev, D.A.; Leyn, S.A.; Kovaleva, G.Y.; Sutormin, R.A.; Kazanov, M.D.; Riehl, W.; Arkin, A.P.; Dubchak, I.; et al. RegPrecise 3.0––A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 2013, 14, 745. [Google Scholar] [CrossRef]
- Dudek, C.-A.; Jahn, D. PRODORIC: State-of-the-art database of prokaryotic gene regulation. Nucleic Acids Res. 2021, 50, D295–D302. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Rao, C.V. The role of configuration and coupling in autoregulatory gene circuits. Mol. Microbiol. 2010, 75, 513–527. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.-Y.; Redfield, B.; Maxon, M.; Weissbach, H.; Brot, N. The effect of homocysteine on metR regulation of metE, metR and metH expression in vitro. Biochem. Biophys. Res. Commun. 1989, 163, 79–83. [Google Scholar] [CrossRef]
- Wu, W.F.; Urbanowski, M.L.; Stauffer, G.V. Characterization of a second MetR-binding site in the metE metR regulatory region of Salmonella typhimurium. J. Bacteriol. 1995, 177, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Glascock, C.B.; Weickert, J.M. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene 1998, 223, 221–231. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demeester, W.; De Baets, J.; Duchi, D.; De Mey, M.; De Paepe, B. MoBioS: Modular Platform Technology for High-Throughput Construction and Characterization of Tunable Transcriptional Biological Sensors. Biosensors 2023, 13, 590. https://doi.org/10.3390/bios13060590
Demeester W, De Baets J, Duchi D, De Mey M, De Paepe B. MoBioS: Modular Platform Technology for High-Throughput Construction and Characterization of Tunable Transcriptional Biological Sensors. Biosensors. 2023; 13(6):590. https://doi.org/10.3390/bios13060590
Chicago/Turabian StyleDemeester, Wouter, Jasmine De Baets, Dries Duchi, Marjan De Mey, and Brecht De Paepe. 2023. "MoBioS: Modular Platform Technology for High-Throughput Construction and Characterization of Tunable Transcriptional Biological Sensors" Biosensors 13, no. 6: 590. https://doi.org/10.3390/bios13060590
APA StyleDemeester, W., De Baets, J., Duchi, D., De Mey, M., & De Paepe, B. (2023). MoBioS: Modular Platform Technology for High-Throughput Construction and Characterization of Tunable Transcriptional Biological Sensors. Biosensors, 13(6), 590. https://doi.org/10.3390/bios13060590




