Intensity-Based Camera Setup for Refractometric and Biomolecular Sensing with a Photonic Crystal Microfluidic Chip
Abstract
1. Introduction
2. Materials and Methods
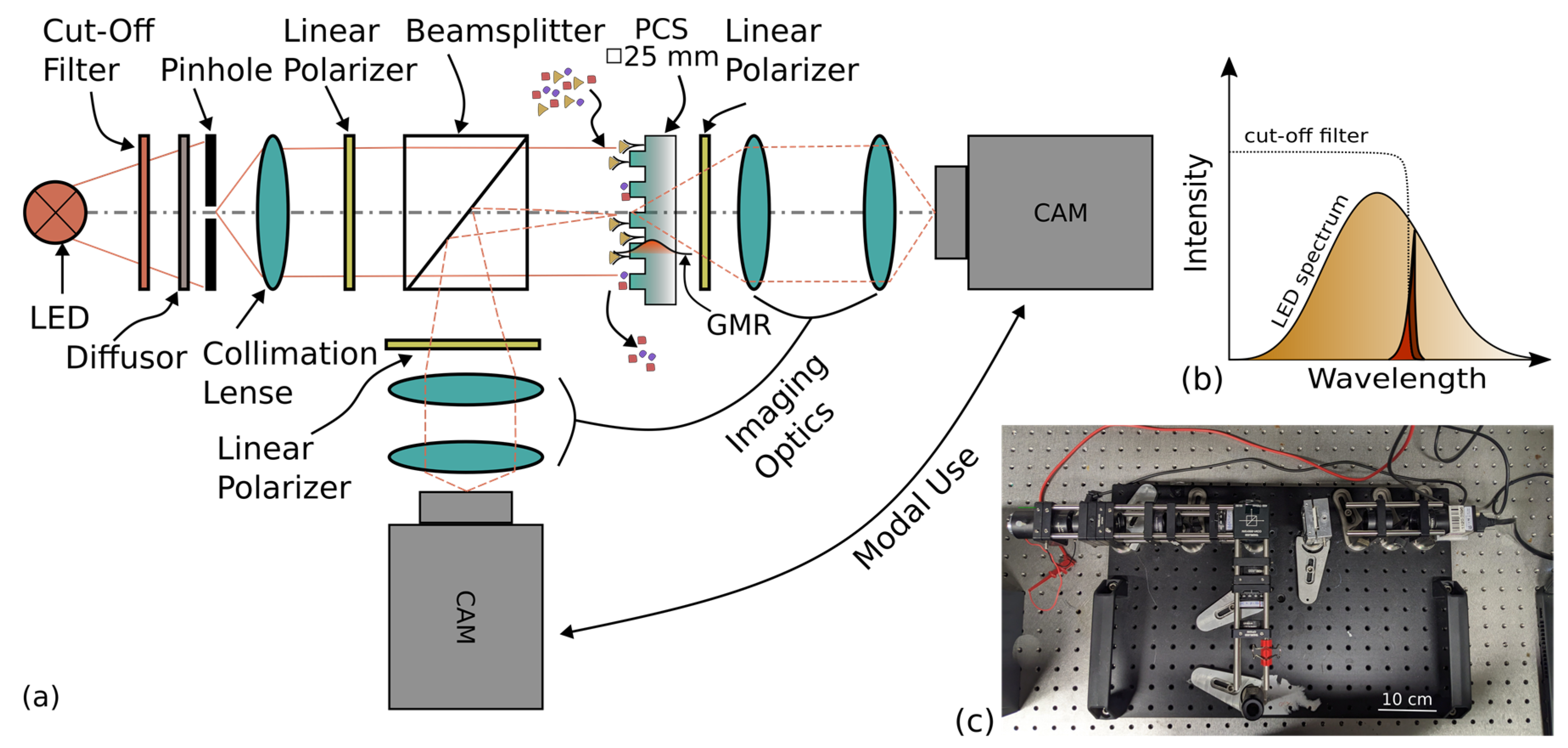
2.1. Intensity-Based Camera Setup (IBCS)
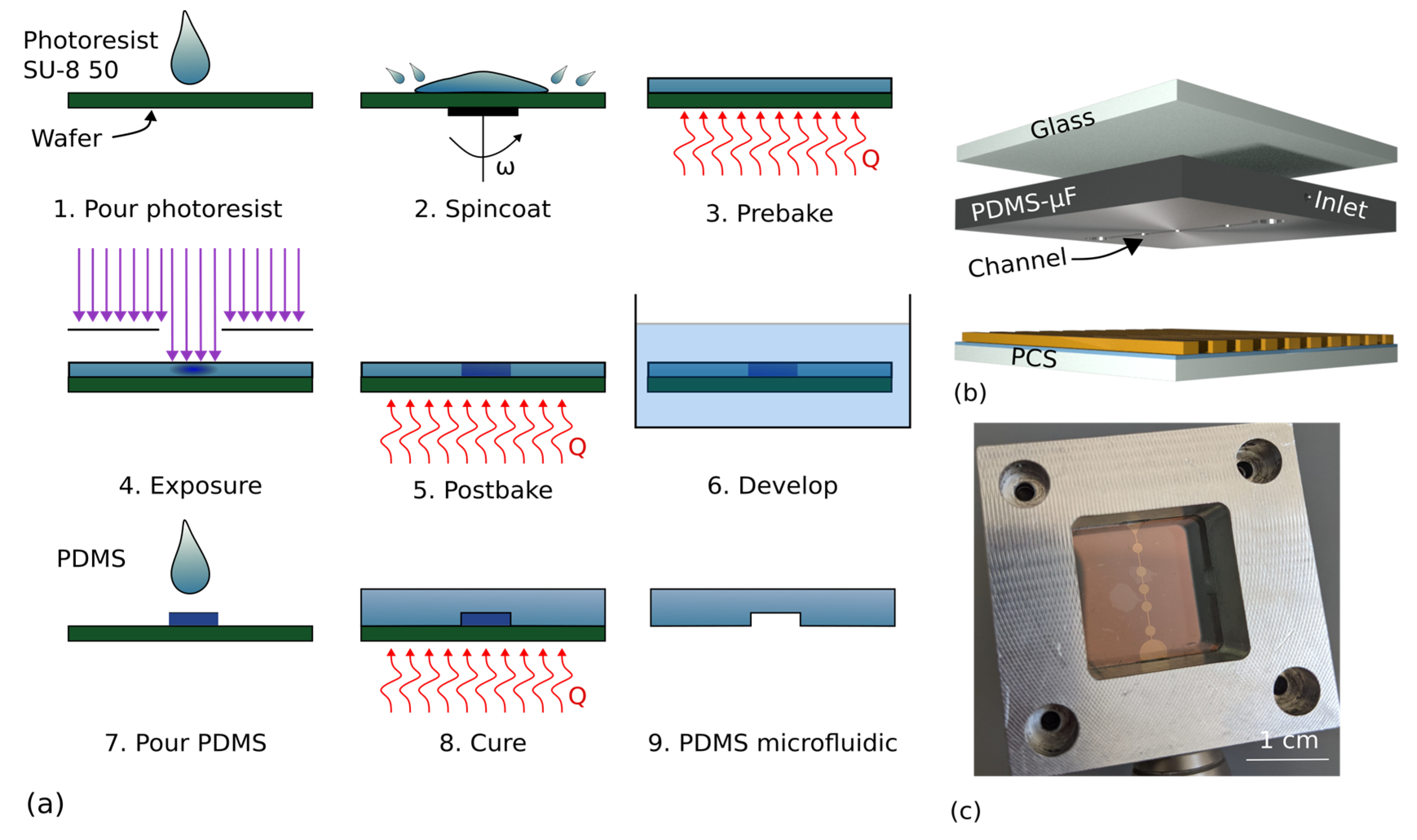
2.2. Fabrication of Photonic Crystal Slabs and Surface Functionalization
2.3. Fabrication of Microfluidics Based on Poly(dimethylsiloxane)
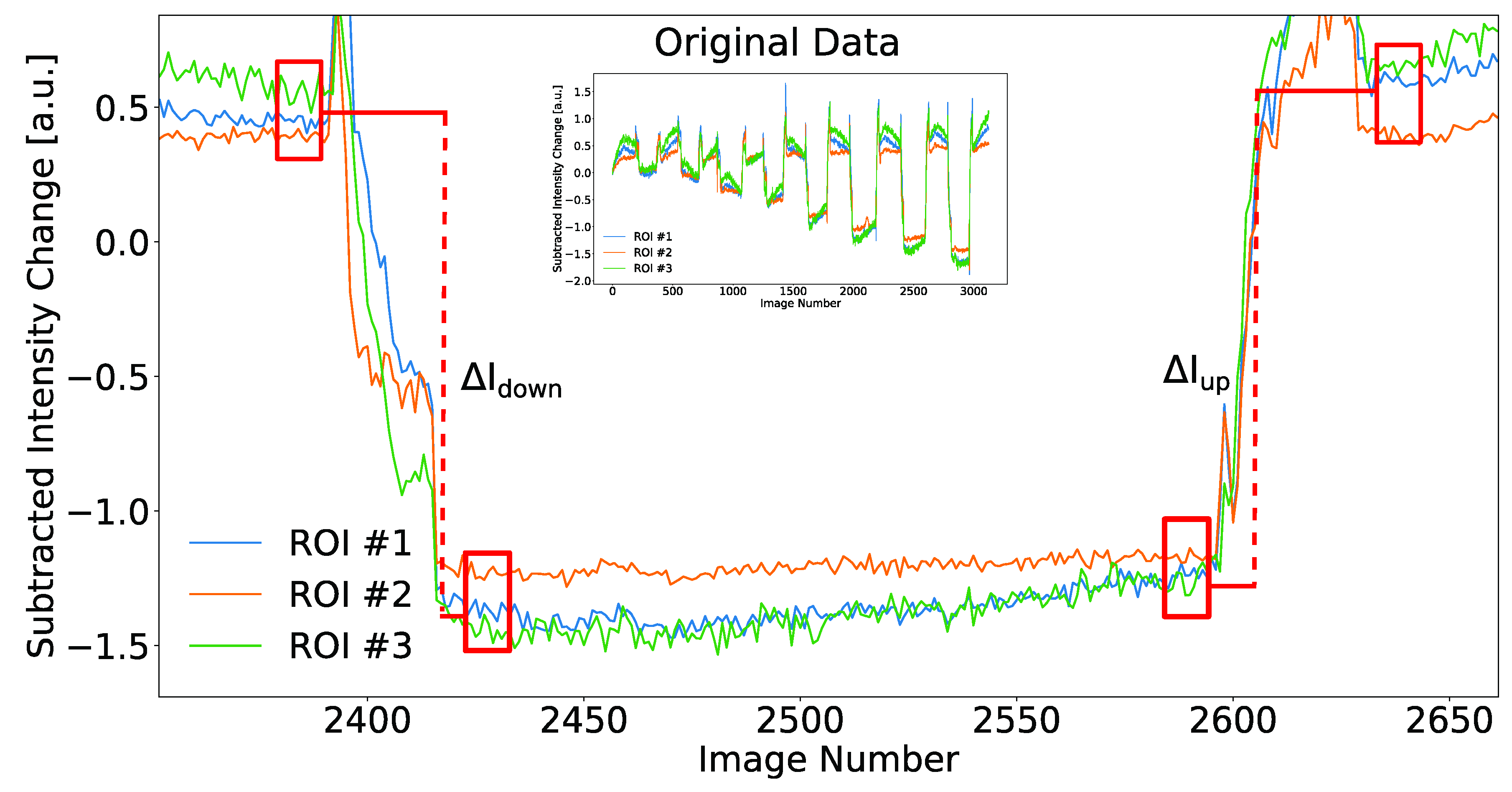
3. Results
3.1. Effects of the Cut-Off Filter on the Limit of Detection
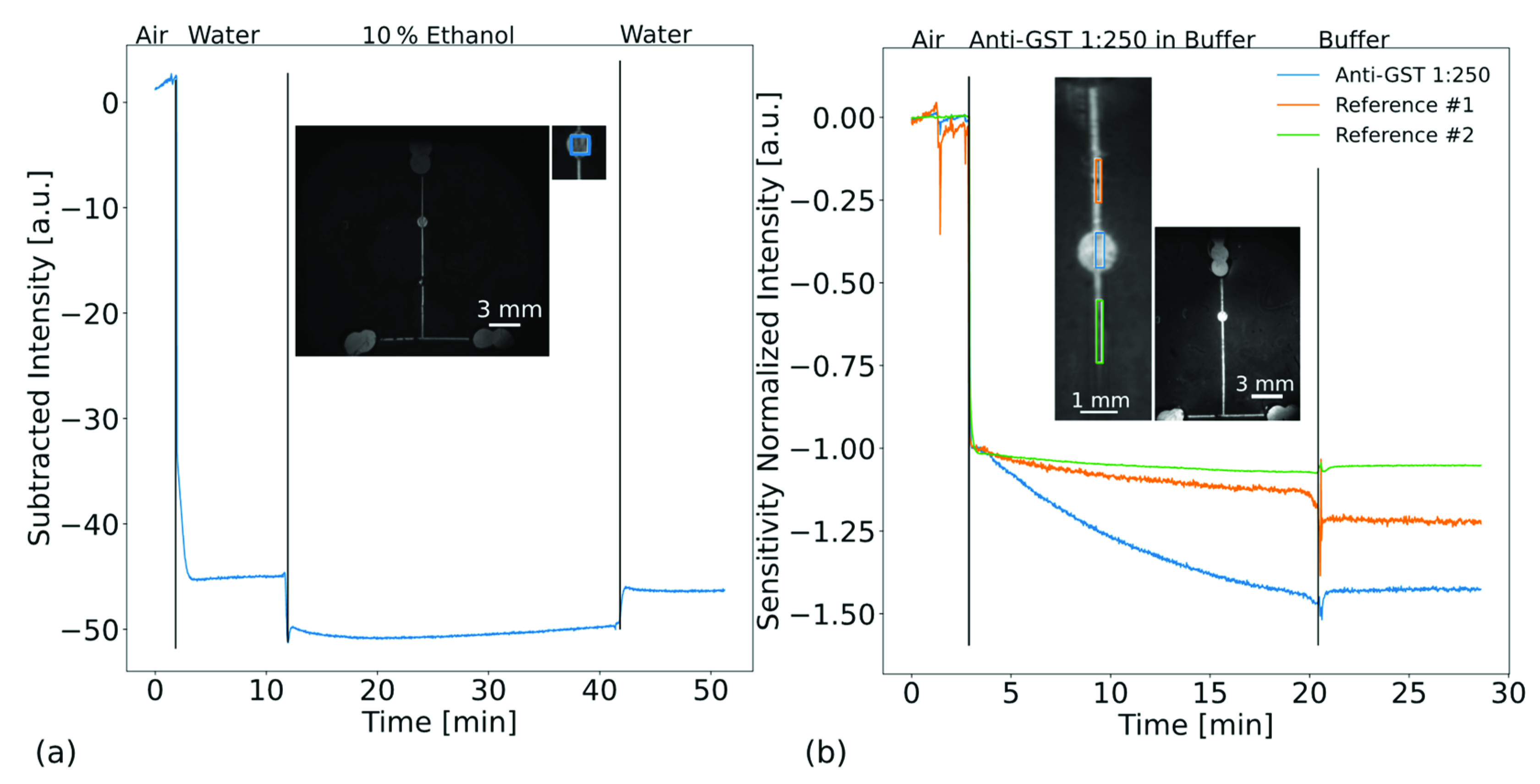
3.2. Refractive Index and Biomolecular Sensing with a Microfluidic Biochip
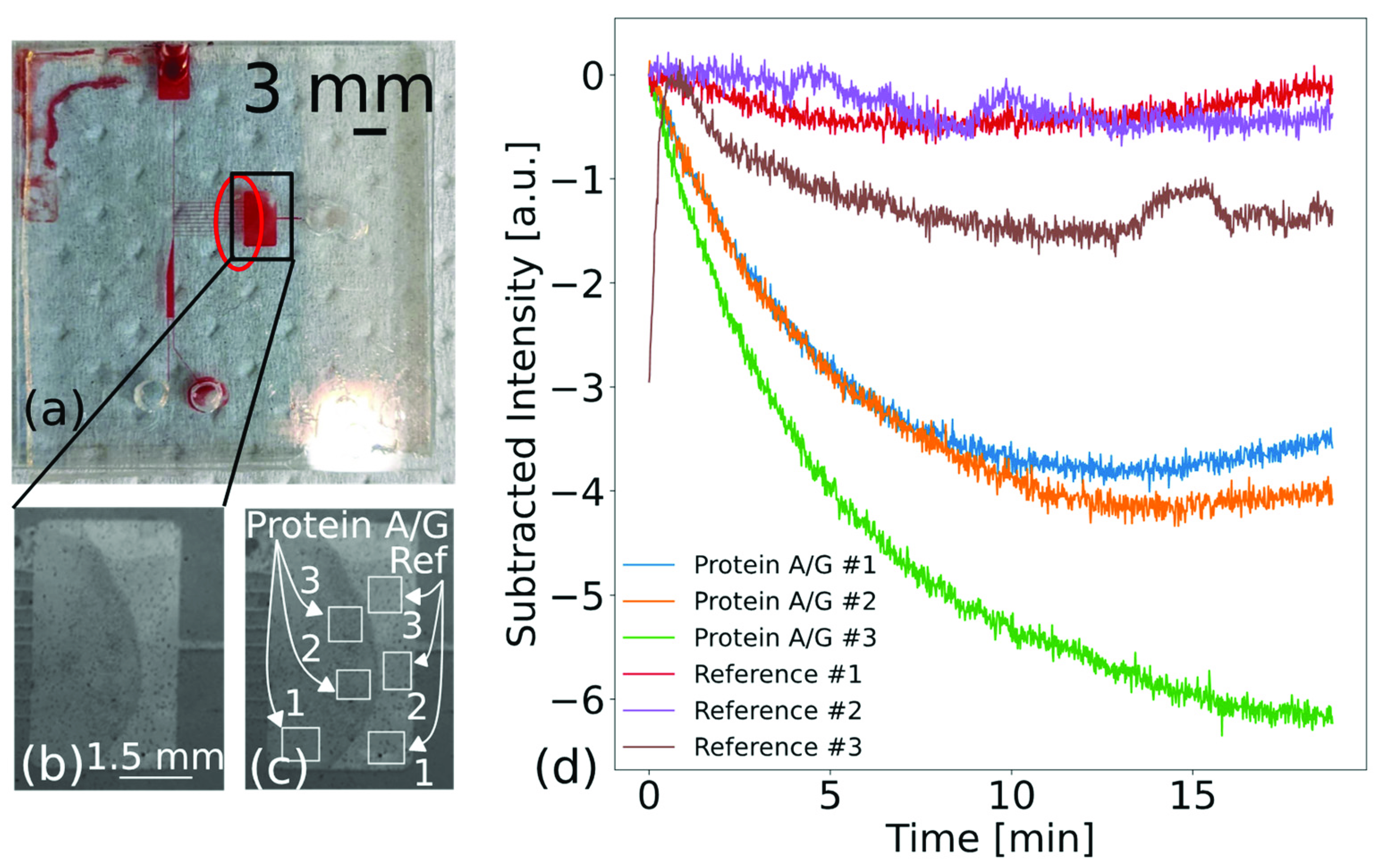
3.3. Biosensing of Immunoglobulins G from Unpurified Whole Blood
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Soler, M.; Huertas, C.S.; Lechuga, L.M. Label-Free Plasmonic Biosensors for Point-of-Care Diagnostics: A Review. Expert Rev. Mol. Diagn. 2019, 19, 71–81. [Google Scholar] [CrossRef]
- Cardenas, J.C. Thrombin Generation Following Severe Trauma: Mechanisms, Modulators, and Implications for Hemostasis and Thrombosis. Shock 2021, 56, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Apple, F.S.; Chung, A.Y.; Kogut, M.E.; Bubany, S.; Murakami, M.M. Decreased Patient Charges Following Implementation of Point-of-Care Cardiac Troponin Monitoring in Acute Coronary Syndrome Patients in a Community Hospital Cardiology Unit. Clin. Chim. Acta 2006, 370, 191–195. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging Applications of Label-Free Optical Biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I.E. Surface Plasmon Resonance Based Immunosensor for the Detection of the Cancer Biomarker Carcinoembryonic Antigen. Talanta 2011, 86, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Ciminelli, C.; Dell’Olio, F.; Conteduca, D.; Campanella, C.M.; Armenise, M.N. High Performance SOI Microring Resonator for Biochemical Sensing. Opt. Laser Technol. 2014, 59, 60–67. [Google Scholar] [CrossRef]
- Cardenosa-Rubio, M.C.; Robison, H.M.; Bailey, R.C. Recent Advances in Environmental and Clinical Analysis Using Microring Resonator–Based Sensors. Curr. Opin. Environ. Sci. Health 2019, 10, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Dell’Olio, F.; Passaro, V.M. Optical Sensing by Optimized Silicon Slot Waveguides. Opt. Express 2007, 15, 4977. [Google Scholar] [CrossRef]
- Claes, T.; Molera, J.G.; De Vos, K.; Schacht, E.; Baets, R.; Bienstman, P. Label-Free Biosensing With a Slot-Waveguide-Based Ring Resonator in Silicon on Insulator. IEEE Photonics J. 2009, 1, 197–204. [Google Scholar] [CrossRef]
- Bläsi, J.; Gerken, M. Multiplex Optical Biosensors Based on Multi-Pinhole Interferometry. Biomed. Opt. Express 2021, 12, 4265–4275. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Fornes, G.; Avella-Oliver, M.; Carrascosa, J.; Fernandez, E.; Brun, E.M.; Maquieira, Á. Disk-Based One-Dimensional Photonic Crystal Slabs for Label-Free Immunosensing. Biosens. Bioelectron. 2019, 126, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, P.K.; Sarkar, S.; Joseph, J. High Sensitivity Guided-Mode-Resonance Optical Sensor Employing Phase Detection. Sci. Rep. 2017, 7, 7607. [Google Scholar] [CrossRef]
- Fan, S.; Joannopoulos, J.D. Analysis of Guided Resonances in Photonic Crystal Slabs. Phys. Rev. B-Condens. Matter Mater. Phys. 2002, 65, 235112. [Google Scholar] [CrossRef]
- Wang, S.S.; Moharam, M.G.; Magnusson, R.; Bagby, J.S. Guided-Mode Resonances in Planar Dielectric-Layer Diffraction Gratings. J. Opt. Soc. Am. A 1990, 7, 1470. [Google Scholar] [CrossRef]
- Wang, S.S.; Magnusson, R. Theory and Applications of Guided-Mode Resonance Filters. Appl. Opt. 1993, 32, 2606. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, M.; Jahns, S.; Gerken, M. Intensity-Based Readout of Resonant-Waveguide Grating Biosensors: Systems and Nanostructures. Photonics Nanostruct.-Fundam. Appl. 2017, 26, 69–79. [Google Scholar] [CrossRef]
- Vörös, J.; Ramsden, J.J.; Csúcs, G.; Szendro, I.; De Paul, S.M.; Textor, M.; Spencer, N.D. Optical Grating Coupler Biosensors. Biomaterials 2002, 23, 3699–3710. [Google Scholar] [CrossRef]
- Kovacs, B.; Kraft, F.A.; Szabo, Z.; Nazirizadeh, Y.; Gerken, M.; Horvath, R. Near Cut-off Wavelength Operation of Resonant Waveguide Grating Biosensors. Sci. Rep. 2021, 11, 13091. [Google Scholar] [CrossRef]
- Quaranta, G.; Basset, G.; Martin, O.J.F.; Gallinet, B. Recent Advances in Resonant Waveguide Gratings. Laser Photonics Rev. 2018, 12, 1800017. [Google Scholar] [CrossRef]
- Paulsen, M.; Neustock, L.T.; Jahns, S.; Adam, J.; Gerken, M. Simulation Methods for Multiperiodic and Aperiodic Nanostructured Dielectric Waveguides. Opt. Quantum Electron. 2017, 49, 107. [Google Scholar] [CrossRef]
- Nazirizadeh, Y.; von Oertzen, F.; Plewa, K.; Barié, N.; Jakobs, P.-J.; Guttmann, M.; Leiste, H.; Gerken, M. Sensitivity Optimization of Injection-Molded Photonic Crystal Slabs for Biosensing Applications. Opt. Mater. Express 2013, 3, 556. [Google Scholar] [CrossRef]
- Kanamori, Y.; Kitani, T.; Hane, K. Control of Guided Resonance in a Photonic Crystal Slab Using Microelectromechanical Actuators. Appl. Phys. Lett. 2007, 90, 031911. [Google Scholar] [CrossRef]
- El Beheiry, M.; Liu, V.; Fan, S.; Levi, O. Sensitivity Enhancement in Photonic Crystal Slab Biosensors. Opt. Express 2010, 18, 22702. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.J.; Block, I.D.; Bole, B.; Dralle, D.; Cunningham, B.T. Label-Free Photonic Crystal Biosensor Integrated Microfluidic Chip for Determination of Kinetic Reaction Rate Constants. IEEE Sens. J. 2009, 9, 1697–1704. [Google Scholar] [CrossRef]
- Kang, C.; Phare, C.T.; Vlasov, Y.A.; Assefa, S.; Weiss, S.M. Photonic Crystal Slab Sensor with Enhanced Surface Area. Opt. Express 2010, 18, 27930. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ye, J.Y.; Divin, C.; Huang, B.; Thomas, T.P.; Baker, J.R.; Norris, T.B. Real-Time Biomolecular Binding Detection Using a Sensitive Photonic Crystal Biosensor. Anal. Chem. 2010, 82, 5211–5218. [Google Scholar] [CrossRef]
- Lin, Y.C.; Hsieh, W.H.; Chau, L.K.; Chang, G.E. Intensity-Detection-Based Guided-Mode-Resonance Optofluidic Biosensing System for Rapid, Low-Cost, Label-Free Detection. Sensors Actuators B Chem. 2017, 250, 659–666. [Google Scholar] [CrossRef]
- Kenaan, A.; Li, K.; Barth, I.; Johnson, S.; Song, J.; Krauss, T.F. Guided Mode Resonance Sensor for the Parallel Detection of Multiple Protein Biomarkers in Human Urine with High Sensitivity. Biosens. Bioelectron. 2020, 153, 112047. [Google Scholar] [CrossRef]
- Jahns, S.; Bräu, M.; Meyer, B.-O.; Karrock, T.; Gutekunst, S.B.; Blohm, L.; Selhuber-Unkel, C.; Buhmann, R.; Nazirizadeh, Y.; Gerken, M. Handheld Imaging Photonic Crystal Biosensor for Multiplexed, Label-Free Protein Detection. Biomed. Opt. Express 2015, 6, 3724–3736. [Google Scholar] [CrossRef] [PubMed]
- Jahns, S. Mobile Biosensorik auf Basis Funktionalisierter, Nanostrukturierter Optischer Wellenleiter. Ph.D. Thesis, Kiel University, Kiel, Germany, 7 November 2016. Available online: https://nbn-resolving.org/urn:nbn:de:gbv:8-diss-199351 (accessed on 4 April 2023).
- Zhou, Y.; Wang, B.; Guo, Z.; Wu, X. Guided Mode Resonance Sensors with Optimized Figure of Merit. Nanomaterials 2019, 9, 837. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Li, Q.; Tian, H.; Wang, Z.; Li, L. An Ultrahigh Q -Factor Dual-Band Terahertz Perfect Absorber with a Dielectric Grating Slit Waveguide for Sensing. J. Phys. D. Appl. Phys. 2020, 53, 235103. [Google Scholar] [CrossRef]
- Beliaev, L.Y.; Stounbjerg, P.G.; Finco, G.; Bunea, A.-I.; Malureanu, R.; Lindvold, L.R.; Takayama, O.; Andersen, P.E.; Lavrinenko, A.V. Pedestal High-Contrast Gratings for Biosensing. Nanomaterials 2022, 12, 1748. [Google Scholar] [CrossRef]
- Sekoguchi, H.; Takahashi, Y.; Asano, T.; Noda, S. Photonic Crystal Nanocavity with a Q-Factor of ~9 Million. Opt. Express 2014, 22, 916. [Google Scholar] [CrossRef]
- Schab, K.; Jelinek, L.; Capek, M.; Ehrenborg, C.; Tayli, D.; Vandenbosch, G.A.E.; Gustafsson, M. Energy Stored by Radiating Systems. IEEE Access 2018, 6, 10553–10568. [Google Scholar] [CrossRef]
- Conteduca, D.; Arruda, G.S.; Barth, I.; Wang, Y.; Krauss, T.F.; Martins, E.R. Beyond Q: The Importance of the Resonance Amplitude for Photonic Sensors. ACS Photonics 2022, 9, 1757–1763. [Google Scholar] [CrossRef]
- Drayton, A.; Li, K.; Simmons, M.; Reardon, C.; Krauss, T. Performance Limitations of Resonant Refractive Index Sensors with Low-Cost Components. Opt. Express 2020, 28, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Gupta, R.; Drayton, A.; Barth, I.; Conteduca, D.; Reardon, C.; Dholakia, K.; Krauss, T.F. Extended Kalman Filtering Projection Method to Reduce the 3σ Noise Value of Optical Biosensors. ACS Sensors 2020, 5, 3474–3482. [Google Scholar] [CrossRef]
- Nazirizadeh, Y.; Bog, U.; Sekula, S.; Mappes, T.; Lemmer, U.; Gerken, M. Low-Cost Label-Free Biosensors Using Photonic Crystals Embedded between Crossed Polarizers. Opt. Express 2010, 18, 19120. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Zhao, D.; Zhou, W.; Sun, Y. High Quality Factor Photonic Crystal Filter at k ≈0 and Its Application for Refractive Index Sensing. Opt. Express 2017, 25, 10536. [Google Scholar] [CrossRef] [PubMed]
- Tarjányi, N.; Turek, I.; Martinček, I. Effect of Mechanical Stress on Optical Properties of Polydimethylsiloxane II—Birefringence. Opt. Mater. 2014, 37, 798–803. [Google Scholar] [CrossRef]
- Lüder, H.; Paulsen, M.; Gerken, M. Photonic Crystal Slab between Orthogonal Polarizers: Details on the Guided Mode Resonance Wavelength. Opt. Quantum Electron. 2020, 52, 180. [Google Scholar] [CrossRef]
- Senior, W.; Gejji, R.; Gai, T.; Slabaugh, C.; Lucht, R. Background Suppression for CARS Thermometry in Highly-Luminous Flames Using an Electro-Optical Shutter. Opt. Lett. 2023, 48, 2010–2013. [Google Scholar] [CrossRef]
- Kehl, F.; Follonier, S. Self-Referenced Waveguide Grating Sensor. Opt. Lett. 2016, 41, 1447. [Google Scholar] [CrossRef]
- Kraft, F.A.; Baur, H.; Bommer, M.; Latz, A.; Fitschen-Oestern, S.; Fuchs, S.; Gerken, M. Label-Free Multiplex Sensing from Buffer and Immunoglobulin G Sensing from Whole Blood with Photonic Crystal Slabs Using Angle-Tuning of an Optical Interference Filter. Biomed. Opt. Express 2023, 14, 2293. [Google Scholar] [CrossRef]
- Bläsi, J.; Gerken, M. Multiplex Microdisk Biosensor Based on Simultaneous Intensity and Phase Detection. Opt. Express 2023, 31, 4319. [Google Scholar] [CrossRef]
- Duffy, J.; Padovani, F.; Brunetti, G.; Noy, P.; Certa, U.; Hegner, M. Towards Personalised Rapid Label Free MiRNA Detection for Cancer and Liver Injury Diagnostics in Cell Lysates and Blood Based Samples. Nanoscale 2018, 10, 12797–12804. [Google Scholar] [CrossRef]
- Petrova, I.; Konopsky, V.; Nabiev, I.; Sukhanova, A. Label-Free Flow Multiplex Biosensing via Photonic Crystal Surface Mode Detection. Sci. Rep. 2019, 9, 8745. [Google Scholar] [CrossRef]
- Hermannsson, P.G.; Sørensen, K.T.; Vannahme, C.; Smith, C.L.C.; Klein, J.J.; Russew, M.-M.; Grützner, G.; Kristensen, A. All-Polymer Photonic Crystal Slab Sensor. Opt. Express 2015, 23, 16529. [Google Scholar] [CrossRef]
- Fang, C.; Dai, B.; Li, Z.; Zahid, A.; Wang, Q.; Sheng, B.; Zhang, D. Tunable Guided-Mode Resonance Filter with a Gradient Grating Period Fabricated by Casting a Stretched PDMS Grating Wedge. Opt. Lett. 2016, 41, 5302. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, N.; Chen, C.; Huang, C. Gradient Waveguide Thickness Guided-Mode. Sensors 2021, 21, 376. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, G.; Liang, S. Several Affinity Tags Commonly Used in Chromatographic Purification. J. Anal. Methods Chem. 2013, 2013, 581093. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.C.; Ohlendorf, D.H.; Wendoloski, J.J.; Salemme, F.R. Structural Origins of High-Affinity Biotin Binding to Streptavidin. Science 1989, 243, 85–88. [Google Scholar] [CrossRef]
- Available online: https://www.thermofisher.com/order/catalog/product/de/en/21186 (accessed on 13 April 2023).
- Xiao, B.; Pradhan, S.K.; Santiago, K.C.; Rutherford, G.N.; Pradhan, A.K. Topographically Engineered Large Scale Nanostructures for Plasmonic Biosensing. Sci. Rep. 2016, 6, 24385. [Google Scholar] [CrossRef] [PubMed]
- Son, J.H.; Lee, S.H.; Hong, S.; Park, S.M.; Lee, J.; Dickey, A.M.; Lee, L.P. Hemolysis-Free Blood Plasma Separation. Lab Chip 2014, 14, 2287–2292. [Google Scholar] [CrossRef]
- Sia, S.K.; Whitesides, G.M. Microfluidic Devices Fabricated in Poly(Dimethylsiloxane) for Biological Studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef]
- Dimov, I.K.; Basabe-Desmonts, L.; Garcia-Cordero, J.L.; Ross, B.M.; Ricco, A.J.; Lee, L.P. Stand-Alone Self-Powered Integrated Microfluidic Blood Analysis System (SIMBAS). Lab Chip 2011, 11, 845–850. [Google Scholar] [CrossRef]
- Yeh, E.; Fu, C.; Hu, L.; Thakur, R.; Feng, J.; Lee, L.P. Self-Powered Integrated Microfluidic Point-of-Care Low-Cost Enabling (SIMPLE) Chip. Sci. Adv. 2017, 3, e1501645. [Google Scholar] [CrossRef] [PubMed]
- Kraft, F.A.; Blaesi, J.; Gerken, M. Regions of Interest and Their Influence on the Limit of Detection in an Intensity-Based Refractive-Index Sensing Setup. In Biophotonics Congress 2021; Optica Publishing Group: Washington, DC, USA, 2021; p. DM1A.3. [Google Scholar] [CrossRef]
- Tie, Y.; Calonder, C.; Van Tassel, P.R. Protein Adsorption: Kinetics and History Dependence. J. Colloid Interface Sci. 2003, 268, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kingshott, P.; Griesser, H.J. Surfaces That Resist Bioadhesion. Curr. Opin. Solid State Mater. Sci. 1999, 4, 403–412. [Google Scholar] [CrossRef]
- Titov, I.; Rutschke, N.; Kraft, F.A.; Köpke, M.; Nebling, E.; Gerken, M. Detection of Fluorescence-Labeled DNA with in-Plane Organic Optoelectronic Devices. Biomed. Opt. Express 2022, 13, 6300. [Google Scholar] [CrossRef]
- Yang, S.; Ündar, A.; Zahn, J.D. A Microfluidic Device for Continuous, Real Time Blood Plasma Separation. Lab Chip 2006, 6, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nunna, B.B.; Talukder, N.; Etienne, E.E.; Lee, E.S. Blood Plasma Self-Separation Technologies during the Self-Driven Flow in Microfluidic Platforms. Bioengineering 2021, 8, 94. [Google Scholar] [CrossRef]
- Fan, R.; Vermesh, O.; Srivastava, A.; Yen, B.K.; Qin, L.; Ahmad, H.; Kwong, G.; Liu, C.-C.; Gould, J.; Hood, L.; et al. Integrated Blood Barcode Chips. Nat. Biotechnol. 2008, 26, 1373–1378. [Google Scholar] [CrossRef]
- Jahns, S.; Gutekunst, S.B.; Selhuber-Unkel, C.; Nazirizadeh, Y.; Gerken, M. Human Blood Microfluidic Test Chip for Imaging, Label-Free Biosensor. Microsyst. Technol. 2016, 22, 1513–1518. [Google Scholar] [CrossRef]
- Jiang, H.; Weng, X.; Li, D. Microfluidic Whole-Blood Immunoassays. Microfluid. Nanofluidics 2011, 10, 941–964. [Google Scholar] [CrossRef]
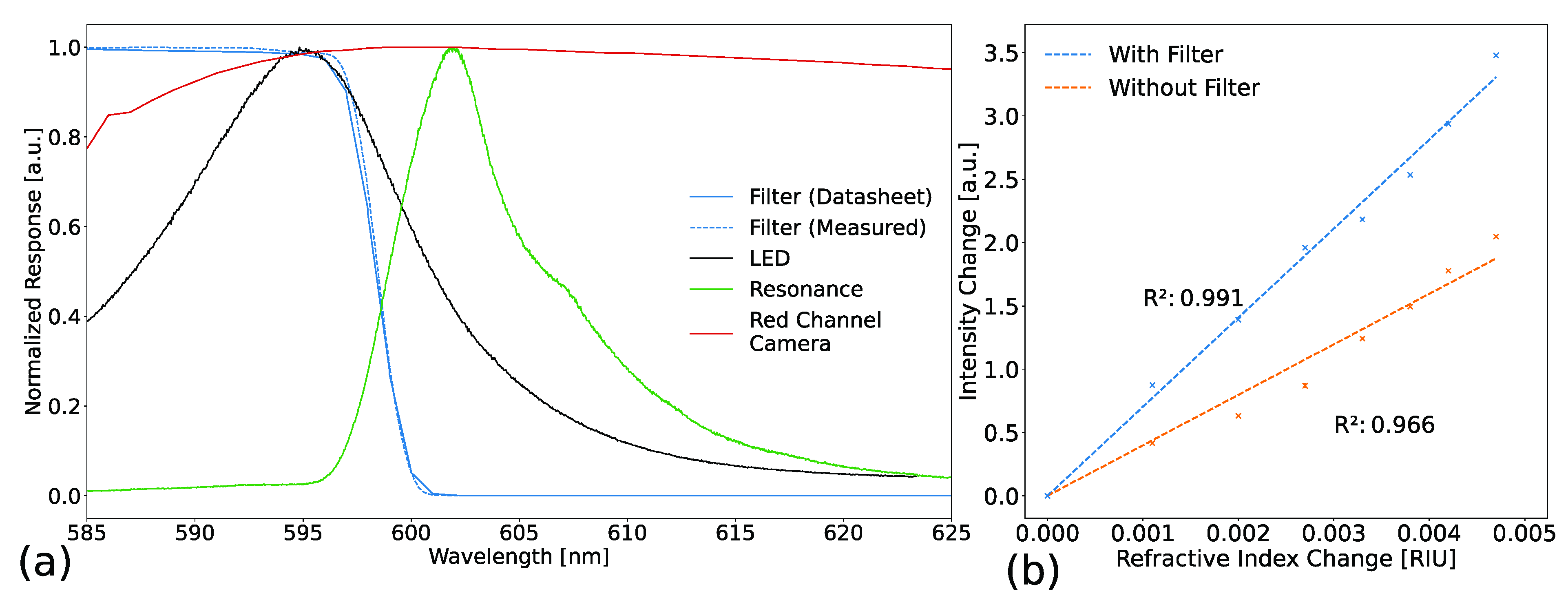

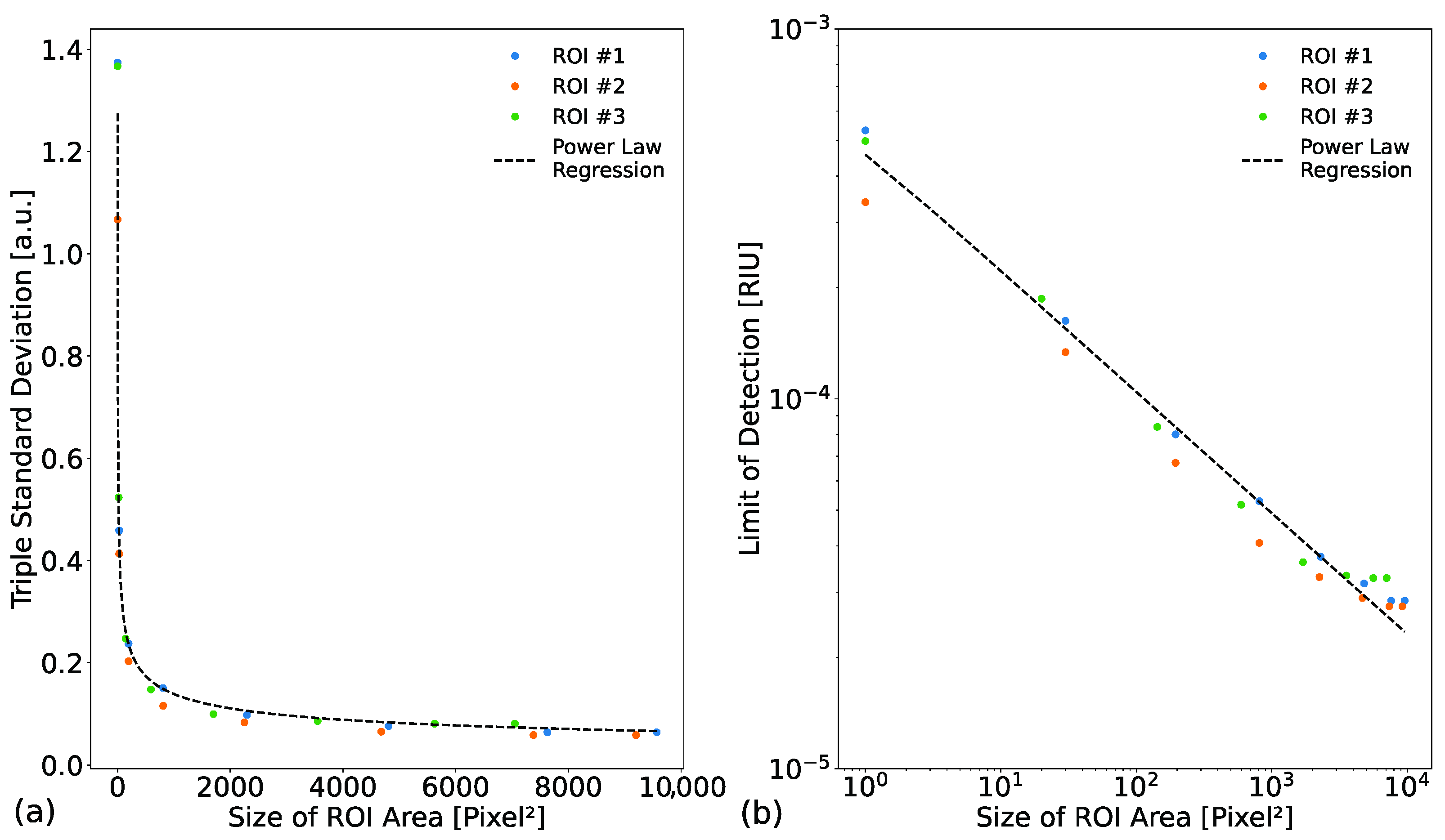

| Cut-Off Filter | Shutter Time [ms] | Triple Standard Deviation [a.u.] | Signal Difference [a.u.] | Sensitivity [a.u./RIU] | LOD [RIU] |
|---|---|---|---|---|---|
| Without filter | 10 | 0.14 ± 0.01 | 1.1 ± 0.2 | 220 ± 50 | (7.1 ± 1.3) × 10−4 |
| With filter | 140 | 0.09 ± 0.01 | 14.7 ± 2.4 | 2940 ± 490 | (3.2 ± 0.7) × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraft, F.A.; Lehmann, S.; Di Maria, C.; Joksch, L.; Fitschen-Östern, S.; Fuchs, S.; Dell’Olio, F.; Gerken, M. Intensity-Based Camera Setup for Refractometric and Biomolecular Sensing with a Photonic Crystal Microfluidic Chip. Biosensors 2023, 13, 687. https://doi.org/10.3390/bios13070687
Kraft FA, Lehmann S, Di Maria C, Joksch L, Fitschen-Östern S, Fuchs S, Dell’Olio F, Gerken M. Intensity-Based Camera Setup for Refractometric and Biomolecular Sensing with a Photonic Crystal Microfluidic Chip. Biosensors. 2023; 13(7):687. https://doi.org/10.3390/bios13070687
Chicago/Turabian StyleKraft, Fabio Aldo, Stefanie Lehmann, Carmela Di Maria, Leonie Joksch, Stefanie Fitschen-Östern, Sabine Fuchs, Francesco Dell’Olio, and Martina Gerken. 2023. "Intensity-Based Camera Setup for Refractometric and Biomolecular Sensing with a Photonic Crystal Microfluidic Chip" Biosensors 13, no. 7: 687. https://doi.org/10.3390/bios13070687
APA StyleKraft, F. A., Lehmann, S., Di Maria, C., Joksch, L., Fitschen-Östern, S., Fuchs, S., Dell’Olio, F., & Gerken, M. (2023). Intensity-Based Camera Setup for Refractometric and Biomolecular Sensing with a Photonic Crystal Microfluidic Chip. Biosensors, 13(7), 687. https://doi.org/10.3390/bios13070687







