Recent Progress of Activity-Based Fluorescent Probes for Imaging Leucine Aminopeptidase
Abstract
:1. Introduction
2. Fluorescent/Chemiluminescent Probes for LAP
2.1. Leu Residue Hydrolysis
2.1.1. DCDHF-Based Fluorescence Probes
2.1.2. Fluorescence Probes Based on Rhodamine and Its Derivates
2.1.3. Chemiluminescence LAP Probes
2.1.4. LAP Probes with AIEgens
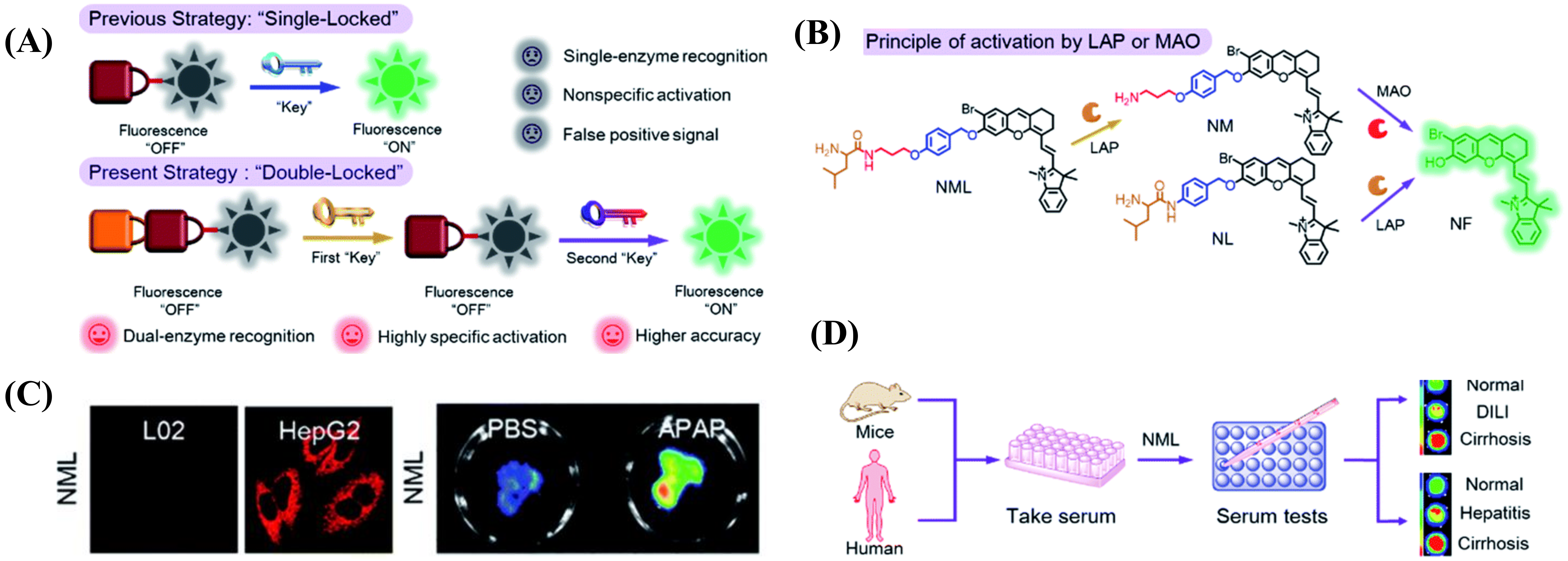
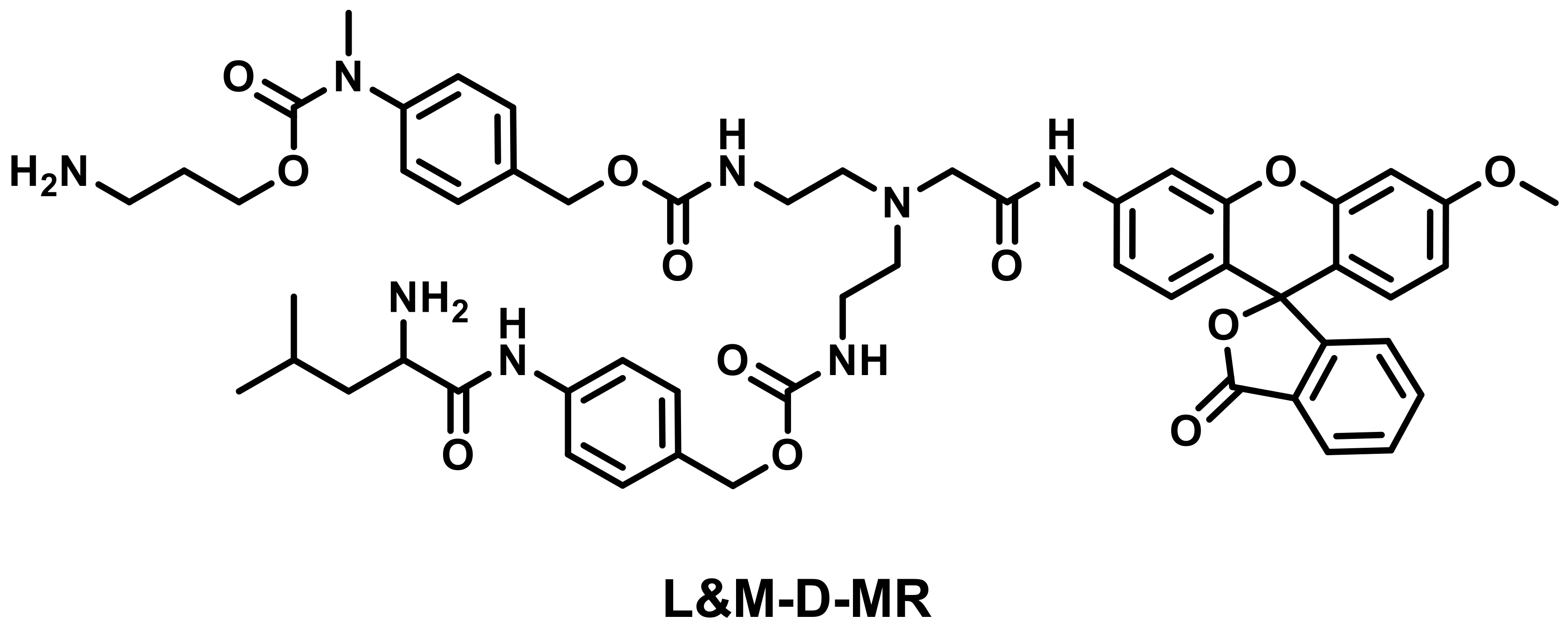
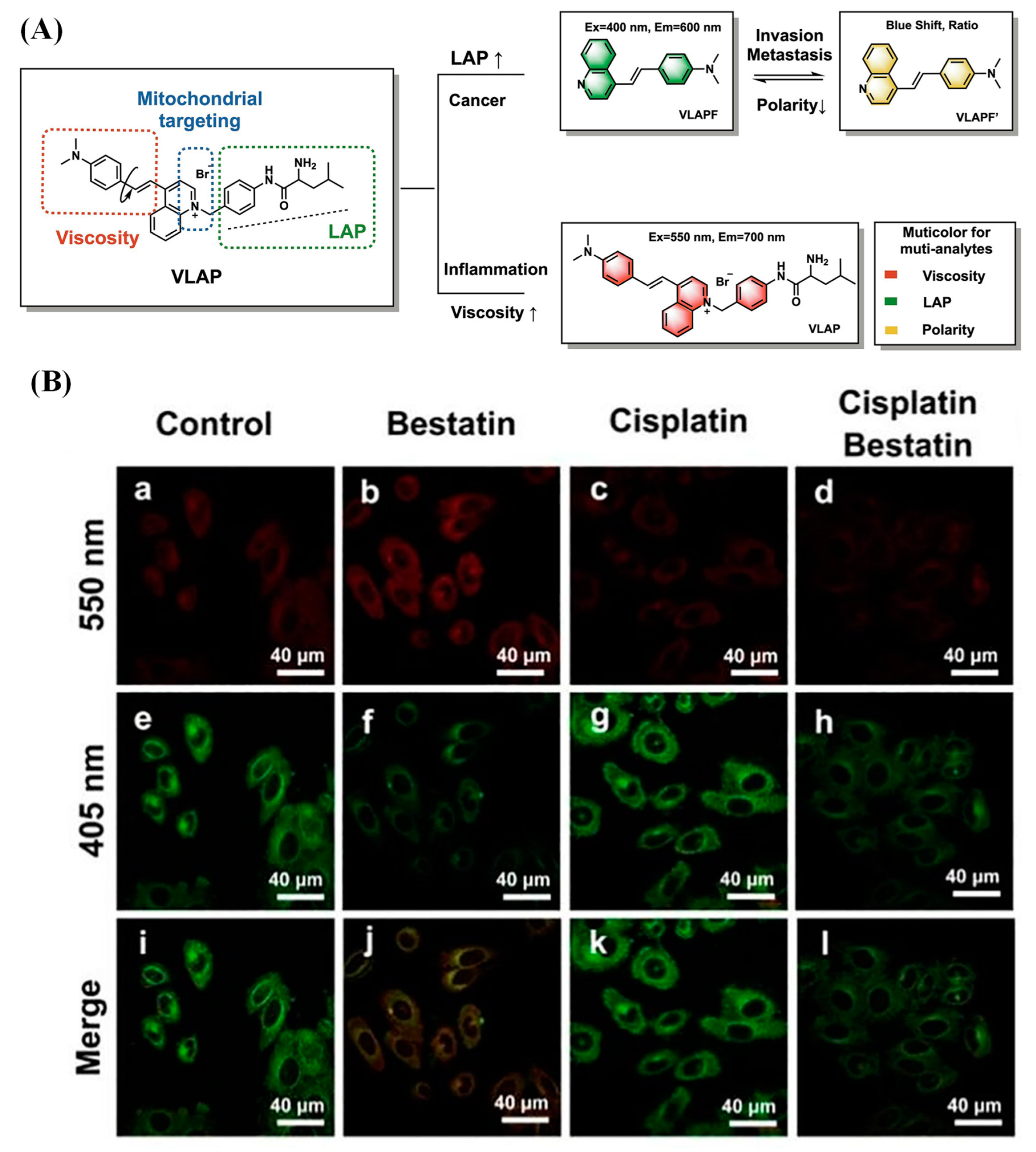
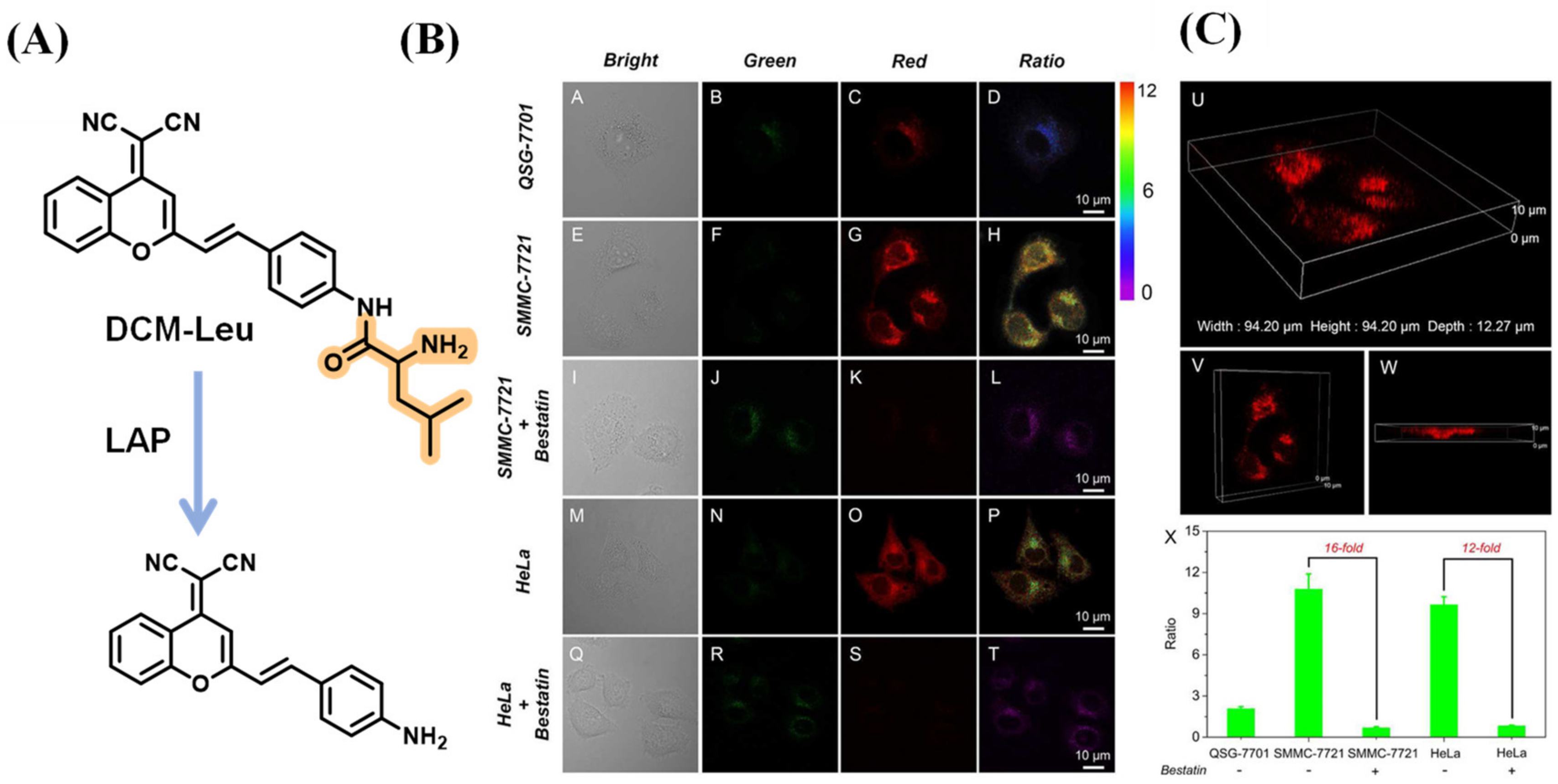
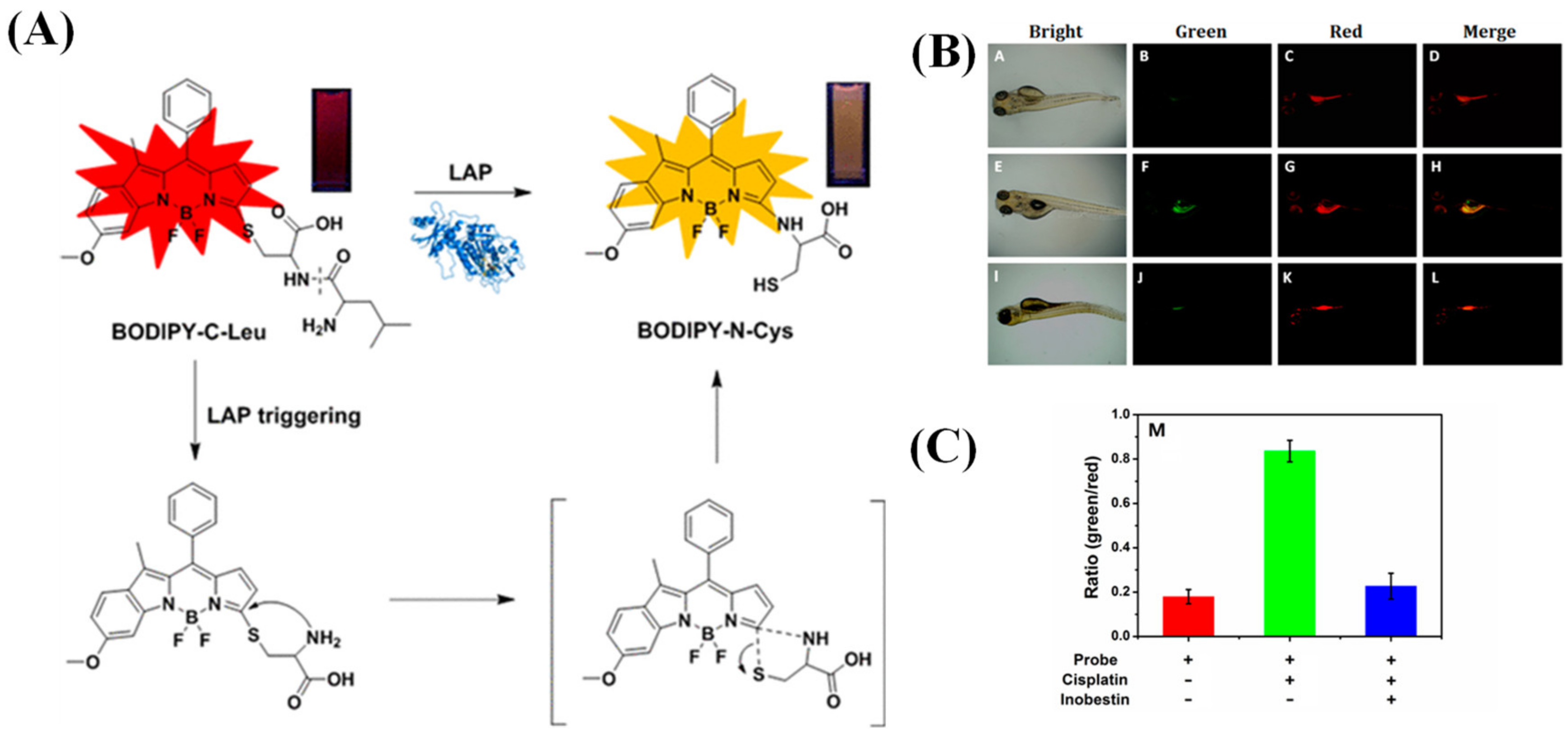
2.1.5. Multi-Detection Probes for Imaging LAP and Diseases Diagnosis
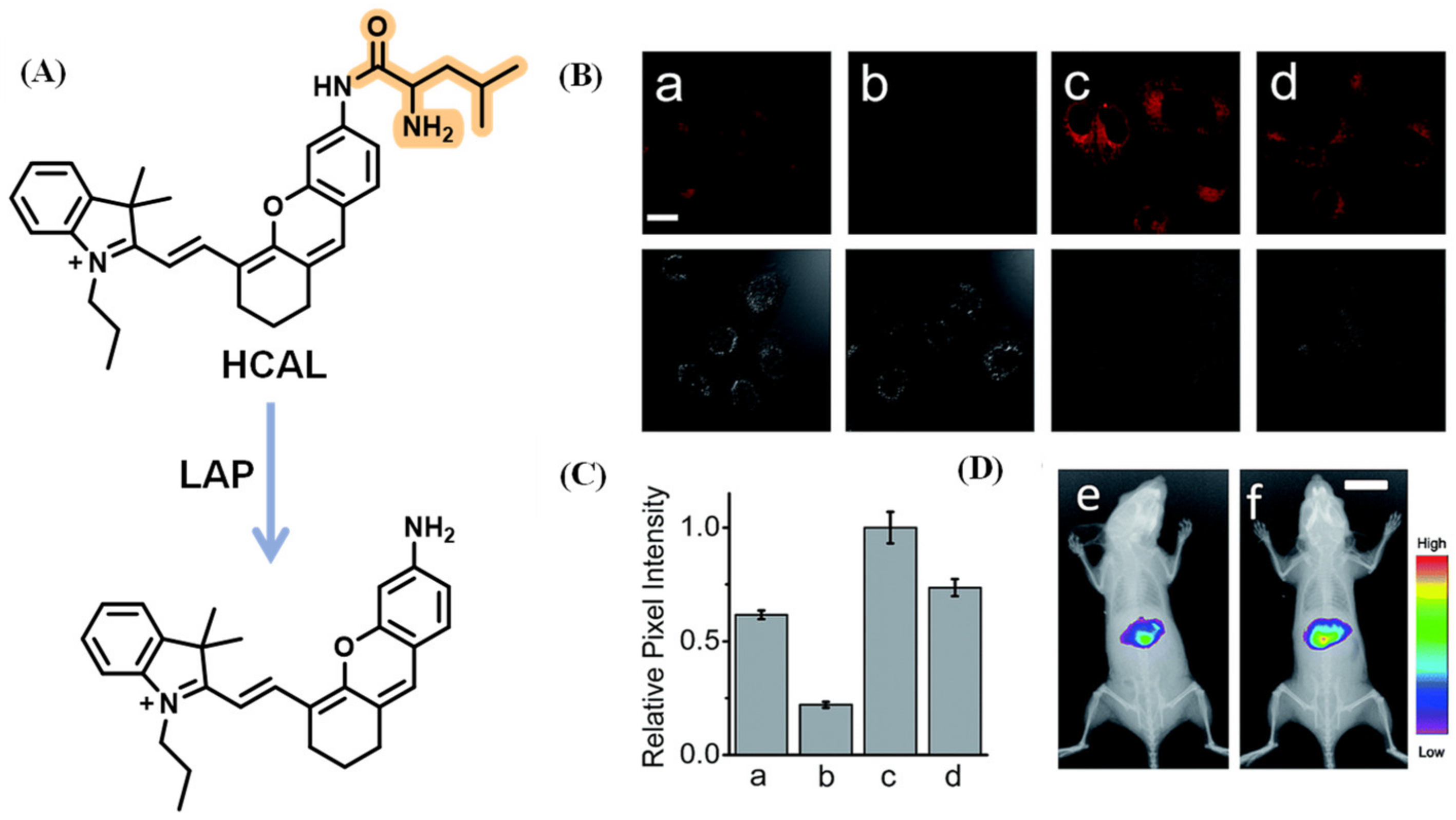
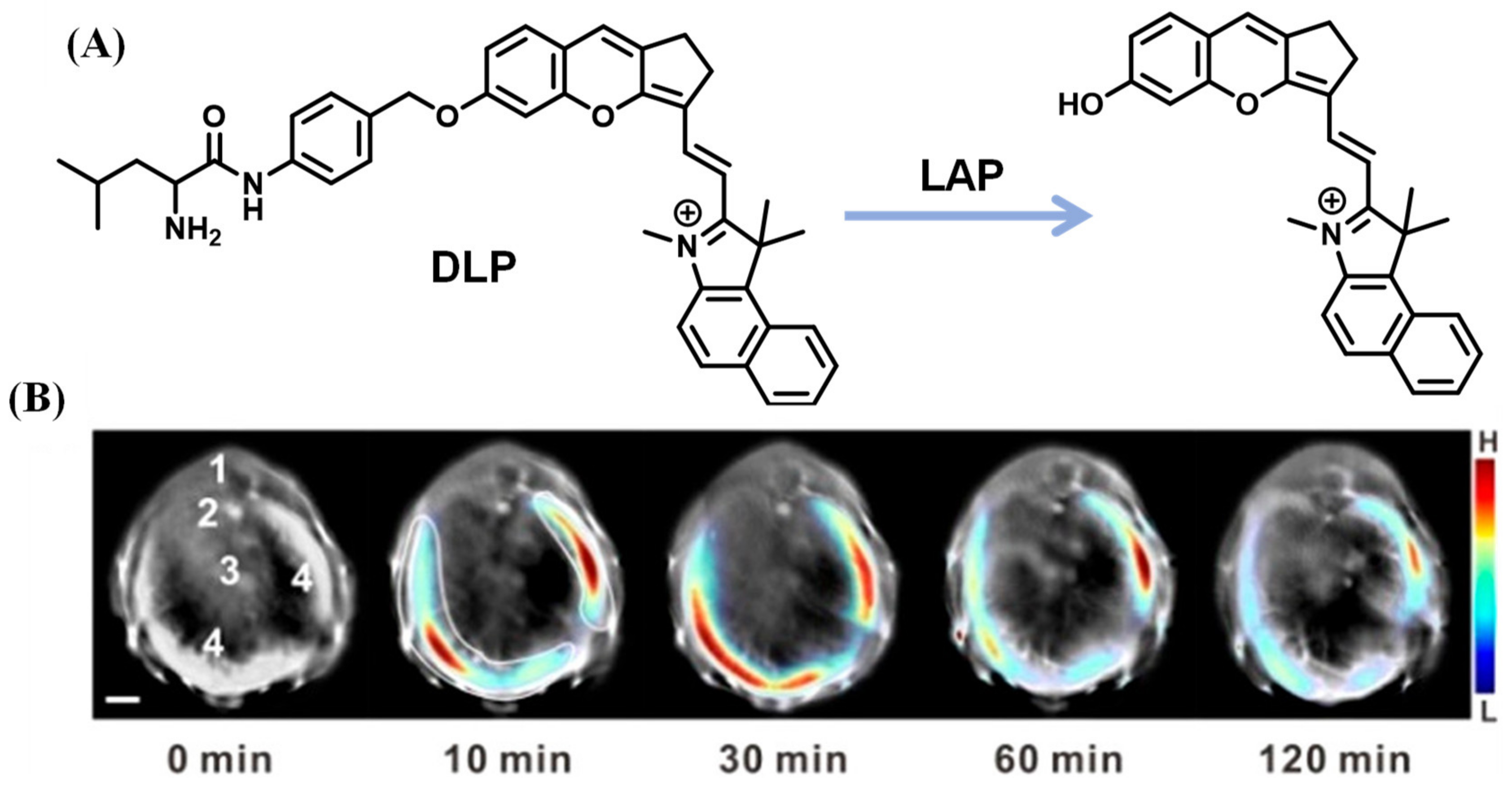
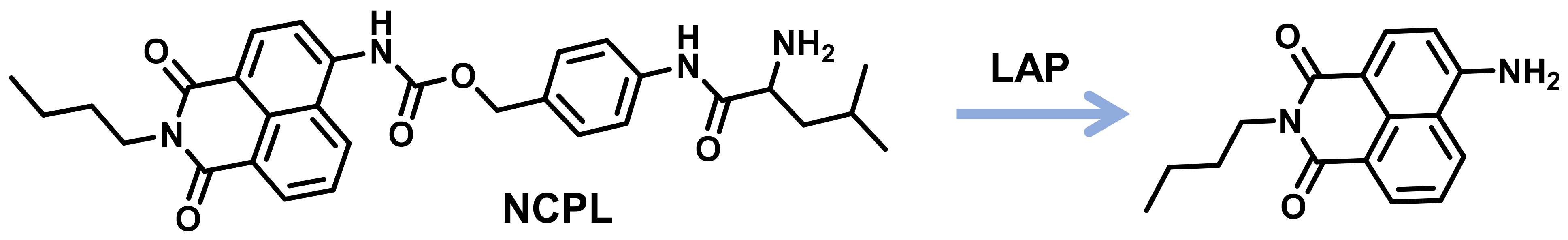
2.1.6. LAP Probes with Targeting Capability
2.1.7. Fluorescent LAP Probes for Integrated Diagnosis and Treatment
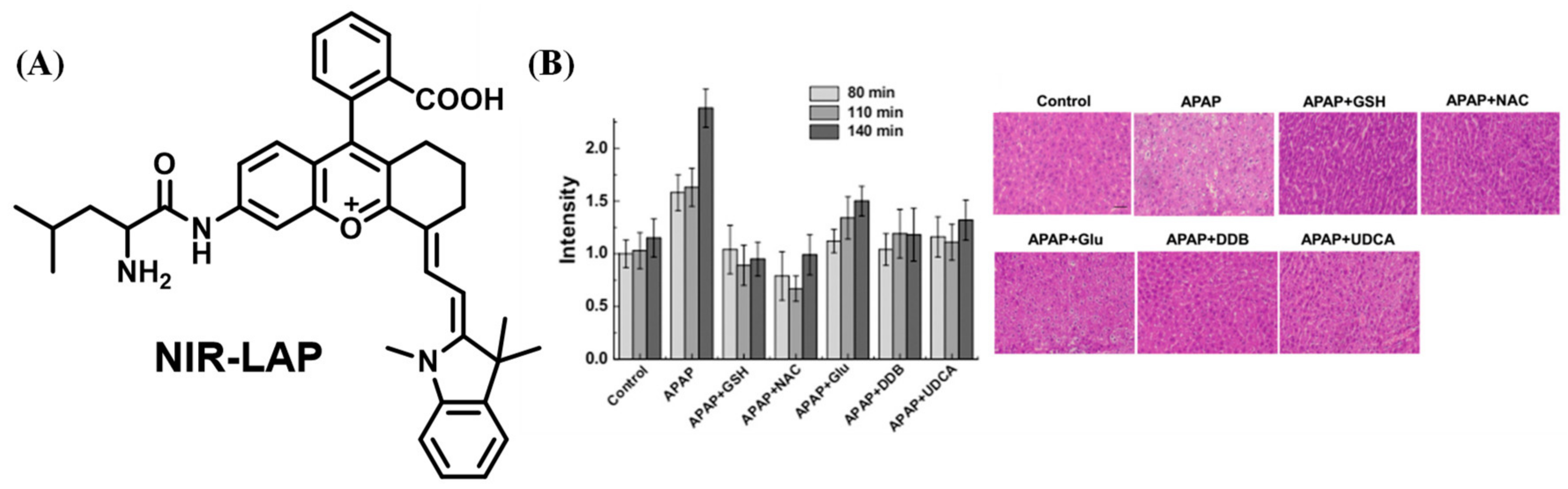
2.1.8. NIR Probes for Detecting LAP
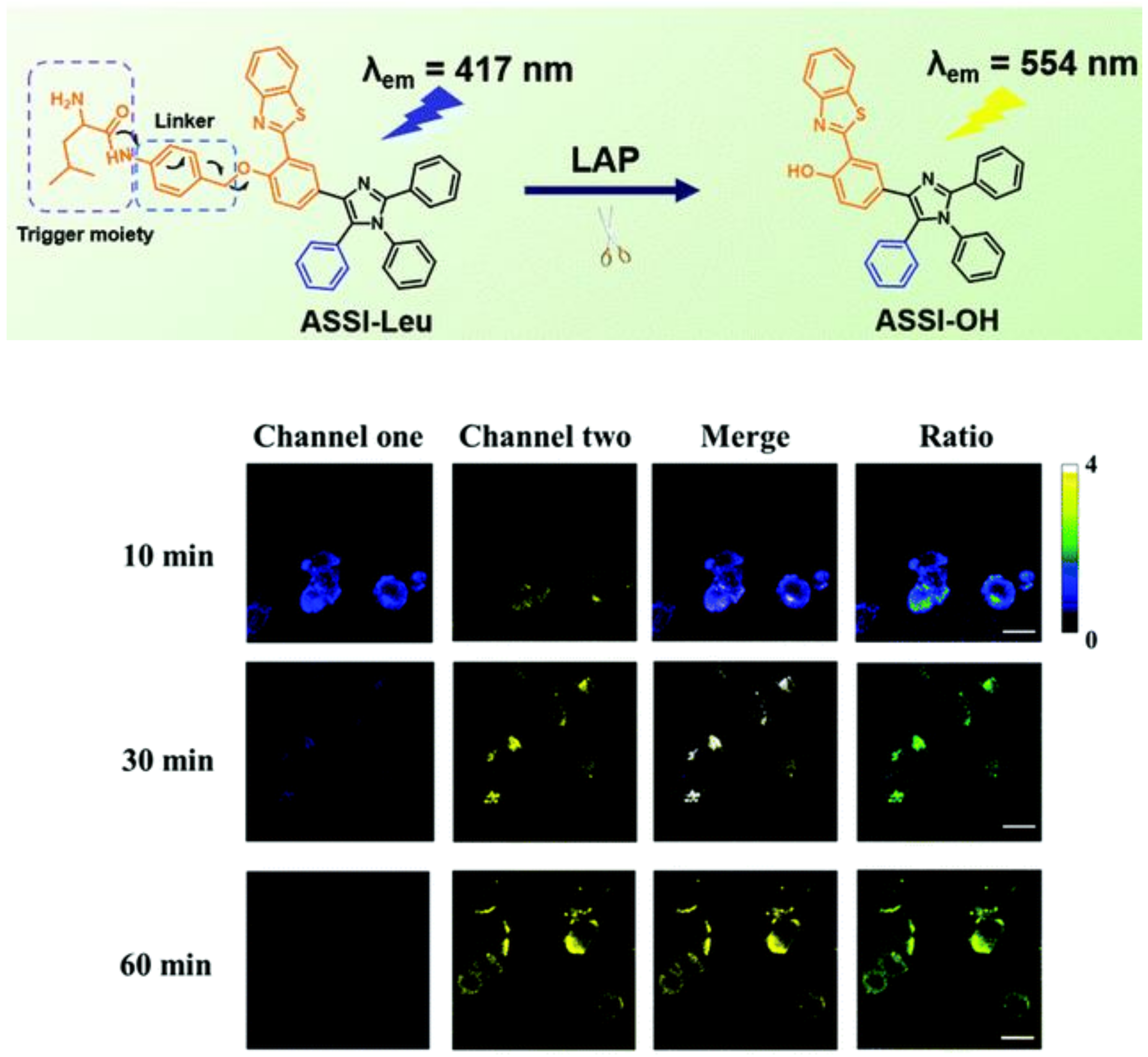
2.1.9. Other Types of Leu Residue Hydrolysis Probes
2.2. Intramolecular Ring Formation
3. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizutani, S.; Shibata, K.; Kikkawa, F.; Hattori, A.; Tsujimoto, M.; Ishii, M.; Kobayashi, H. Essential role of placental leucine aminopeptidase in gynecologic malignancy. Expert Opin. Ther. Targets 2007, 11, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A. Aminopeptidases: Structure and function. FASEB J. 1993, 7, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Fowler, J.H.; Walling, L. Leucine aminopeptidases: Diversity in structure and function. Biol. Chem. 2006, 387, 1535–1544. [Google Scholar] [CrossRef]
- Yamazaki, T.; Akada, T.; Niizeki, O.; Suzuki, T.; Miyashita, H.; Sato, Y. Puromycin-insensitive leucyl-specific aminopeptidase (PILSAP) binds and catalyzes PDK1, allowing VEGF-stimulated activation of S6K for endothelial cell proliferation and angiogenesis. Blood 2004, 104, 2345–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burley, S.; David, P.; Lipscomb, W. Leucine aminopeptidase: Bestatin inhibition and a model for enzyme-catalyzed peptide hydrolysis. Proc. Natl. Acad. Sci. USA 1991, 88, 6916–6920. [Google Scholar] [CrossRef]
- Harbut, M.; Velmourougane, G.; Dalal, S.; Reiss, G.; Whisstock, J.; Onder, O.; Brisson, D.; McGowan, S.; Klemba, M.; Greenbaum, D. Bestatin-based chemical biology strategy reveals distinct roles for malaria M1-and M17-family aminopeptidases. Proc. Natl. Acad. Sci. USA 2011, 108, E526–E534. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Goto, Y.; Maruyama, M.; Hattori, A. Biochemical and enzymatic properties of the M1 family of aminopeptidases involved in the regulation of blood pressure. Heart Fail. Rev. 2008, 13, 285–291. [Google Scholar] [CrossRef]
- Carroll, R.; Robison, T.; Rivera, F.; Davenport, J.; Jonsson, M.; Florczyk, D.; Tarkowski, A.; Potempa, J.; Koziel, J.; Shaw, L. Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus. Microbes Infect. 2012, 14, 989–999. [Google Scholar] [CrossRef] [Green Version]
- Carroll, R.; Veillard, F.; Gagne, D.; Lindenmuth, J.; Poreba, M.; Drag, M.; Potempa, J.; Shaw, L. The Staphylococcus aureus leucine aminopeptidase is localized to the bacterial cytosol and demonstrates a broad substrate range that extends beyond leucine. Biol. Chem. 2013, 394, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Shibata, K.; Kajiyama, H.; Mizokami, Y.; Ino, K.; Nomura, S.; Mizutani, S.; Terauchi, M.; Kikkawa, F. Placental leucine aminopeptidase (P-LAP) and glucose transporter 4 (GLUT4) expression in benign, borderline, and malignant ovarian epithelia. Gynecol. Oncol. 2005, 98, 11–18. [Google Scholar] [CrossRef]
- Kondo, C.; Shibata, K.; Terauchi, M.; Kajiyama, H.; Ino, K.; Nomura, S.; Nawa, A.; Mizutani, S.; Kikkawa, F. A novel role for placental leucine aminopeptidase (P-LAP) as a determinant of chemoresistance in endometrial carcinoma cells. Int. J. Cancer 2006, 118, 1390–1394. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yu, W.; Kovalski, K.; Ossowski, L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell 1998, 94, 353–362. [Google Scholar] [CrossRef] [Green Version]
- Shibata, K.; Kikkawa, F.; Mizokami, Y.; Kajiyama, H.; Ino, K.; Nomura, S.; Mizutani, S. Possible involvement of adipocyte-derived leucine aminopeptidase via angiotensin II in endometrial carcinoma. Tumour Biol. 2005, 26, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Pearse, A.; Tremblay, G. Leucine aminopeptidase in rat parathyroid and its relation to parathyroid hormone production. Nature 1958, 181, 1532–1533. [Google Scholar] [CrossRef] [PubMed]
- Bardawill, C.; Chang, C. Serum lactic dehydrogenase, leucine aminopeptidase and 5-nucleotidase activity: Observations in patients with carcinoma of the pancreas and hepatobiliary disease. Can. Med. Assoc. J. 1963, 89, 755. [Google Scholar]
- Reichling, J.; Kaplan, M. Clinical use of serum enzymes in liver disease. Dig. Dis. Sci. 1988, 33, 1601–1614. [Google Scholar] [CrossRef]
- Su, M.; Wei, M.; Zhou, Z.; Liu, S. Application of capillary electrophoresis coupling with electrochemiluminescence detection to estimate activity of leucine aminopeptidas. Biomed. Chromatogr. 2013, 27, 946–952. [Google Scholar] [CrossRef]
- Xiong, X.; Barathi, A.; Beuerman, R.; Tan, D. Assay of leucine aminopeptidase activity in vitro using large-pore reversed-phase chromatography with fluorescence detection. J. Chromatogr. B 2003, 796, 63–70. [Google Scholar] [CrossRef]
- Huang, H.; Tanaka, H.; Hammock, B.; Morisseau, C. Novel and highly sensitive fluorescent assay for leucine aminopeptidases. Anal. Biochem. 2009, 391, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fan, J.; Wang, K.; Li, J.; Wang, C.; Nie, Y.; Jiang, T.; Mu, H.; Peng, X.; Jiang, K. Highly sensitive naphthalene-based two-photon fluorescent probe for in situ real-time bioimaging of ultratrace cyclooxygenase-2 in living biosystems. Anal. Chem. 2014, 86, 9131–9138. [Google Scholar] [CrossRef]
- HeeáLee, M.; SeungáKim, J. Small molecule-based ratiometric fluorescence probes for cations, anions, and biomolecules. Chem. Soc. Rev. 2015, 44, 4185–4191. [Google Scholar]
- Geng, Y.; Wang, Z.; Zhou, J.; Zhu, M.; Liu, J.; James, T. Recent progress in the development of fluorescent probes for imaging pathological oxidative stress. Chem. Soc. Rev. 2023, 52, 3873–3926. [Google Scholar] [CrossRef]
- Van de Bittner, G.; Bertozzi, C.; Chang, C. Strategy for dual-analyte luciferin imaging: In vivo bioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute inflammation. J. Am. Chem. Soc. 2013, 135, 1783–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, D.; Teoh, C.; Wang, L.; Liu, X.; Chang, Y. Motion-induced change in emission (MICE) for developing fluorescent probes. Chem. Soc. Rev. 2017, 46, 4833–4844. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Dong, B.; Zhang, Z.; Yin, J.; Lin, W. Permeability-Controlled Probe for Directly Visualizing the Opening of Mitochondrial Permeability Transition Pore in Native Status. Anal. Chem. 2022, 94, 5255–5264. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, S.; Dong, B.; Kong, X.; Tian, M. Chameleon-Like Fluorescent Probe for Monitoring Interplays between Three Organelles and Reporting Cell Damage Processes through Dramatic Color Change. Small 2022, 18, 2205026. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Shim, S.; Baek, L.; Hong, J. Small-molecule probe using dual signals to monitor leucine aminopeptidase activity. Bioorg. Med. Chem. Lett. 2011, 21, 2403–2405. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Li, J.; Wang, J.; Cao, T.; Zheng, L.; Cao, Y.; Qin, W.; Liu, Y. In vivo imaging via a red-emitting fluorescent probe to diagnosing liver cancer or drug-induced liver disease. Anal. Chim. Acta 2021, 1168, 338621. [Google Scholar] [CrossRef]
- Sakabe, M.; Asanuma, D.; Kamiya, M.; Iwatate, R.; Hanaoka, K.; Terai, T.; Nagano, T.; Urano, Y. Rational design of highly sensitive fluorescence probes for protease and glycosidase based on precisely controlled spirocyclization. J. Am. Chem. Soc. 2013, 135, 409–414. [Google Scholar] [CrossRef]
- Kushida, Y.; Hanaoka, K.; Komatsu, T.; Terai, T.; Ueno, T.; Yoshida, K.; Uchiyama, M.; Nagano, T. Red fluorescent scaffold for highly sensitive protease activity probes. Bioorg. Med. Chem. Lett. 2012, 22, 3908–3911. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Wang, T.; Fan, M.; Wang, N.; Ma, D.; Hu, T.; Cui, X. Asymmetric Si-rhodamine scaffolds: Rational design of pH-durable protease-activated NIR probes in vivo. Chem. Commun. 2020, 56, 2455–2458. [Google Scholar] [CrossRef]
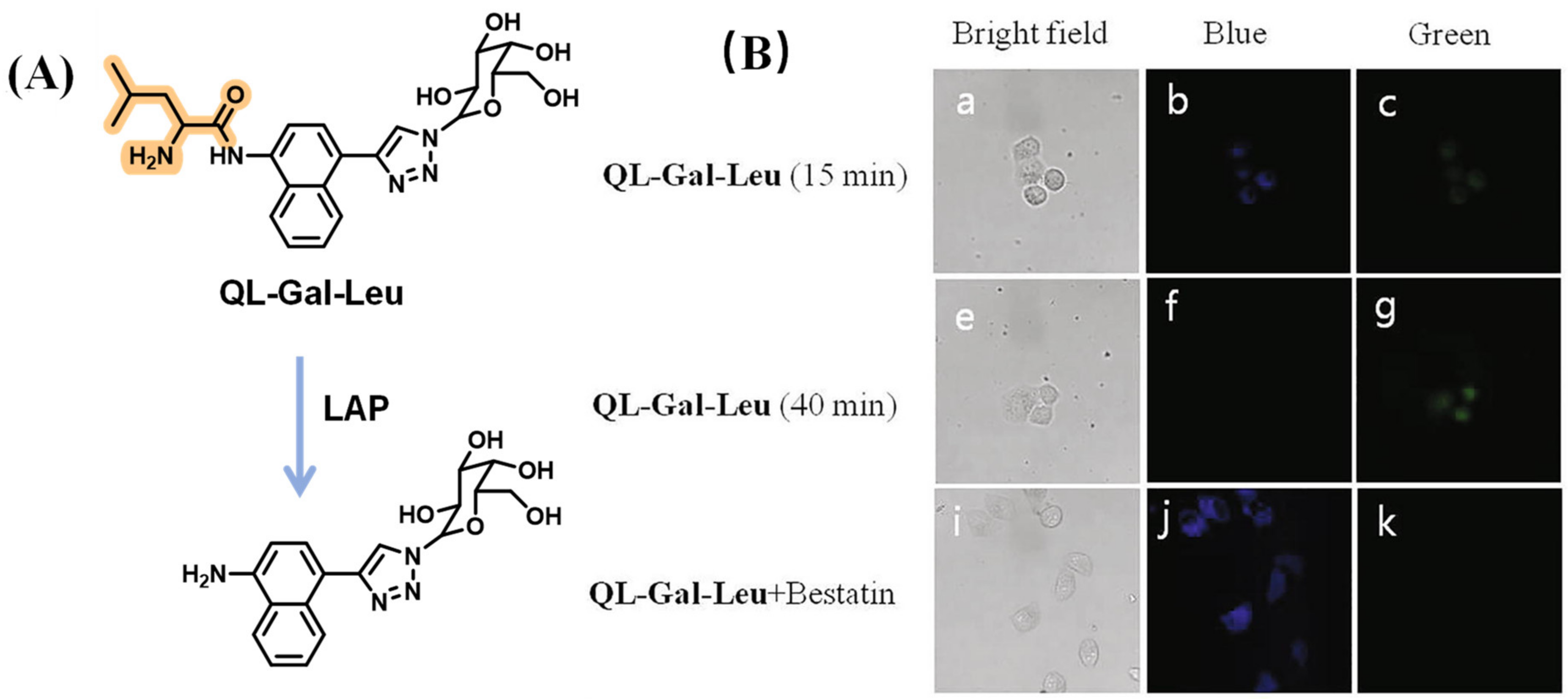
- Kang, J.; Mun, S.; Choi, E.; Kim, J.; Yee, S.; Chang, D. A preliminary study for the development of cleavable linkers using activatable fluorescent probes targeting leucine aminopeptidase. Analyst 2022, 147, 5386–5394. [Google Scholar] [CrossRef] [PubMed]
- Haris, U.; Kagalwala, H.; Kim, Y.; Lippert, A. Seeking Illumination: The Path to Chemiluminescent 1,2-Dioxetanes for Quantitative Measurements and In Vivo Imaging. Acc. Chem. Res. 2021, 54, 2844–2857. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zeng, Z.; Lam, J.; Fan, J.; Pu, K.; Tang, B. State-of-the-art self-luminescence: A win-win situation. Chem. Soc. Rev. 2022, 51, 8815–8831. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Huang, J.; Fan, J.; Du, J.; Pu, K.; Peng, X. Chemiluminescence for bioimaging and therapeutics: Recent advances and challenges. Chem. Soc. Rev. 2020, 49, 6800–6815. [Google Scholar] [CrossRef]
- Wang, B.; Chen, Z.; Cen, X.; Liang, Y.; Tan, L.; Liang, E.; Zheng, L.; Zheng, Y.; Zhan, Z.; Cheng, K. A highly selective and sensitive chemiluminescent probe for leucine aminopeptidase detection in vitro, in vivo and in human liver cancer tissue. Chem. Sci. 2022, 13, 2324–2330. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xie, Z.; Lam, J.; Cheng, L.; Chen, H.; Qiu, C.; Kwok, H.; Zhan, X.; Liu, Y.; Zhu, D. Aggregation-induced emission of 1-methyl-1, 2, 3, 4, 5-pentaphenylsilole. Chem. Commun. 2001, 18, 1740–1741. [Google Scholar] [CrossRef]
- Mei, J.; Leung, N.; Kwok, R.; Lam, J.; Tang, B. Aggregation-induced emission: Together we shine, united we soar! Chem. Rev. 2015, 115, 11718–11940. [Google Scholar] [CrossRef] [PubMed]
- Alam, P.; Leung, N.; Zhang, J.; Kwok, R.; Lam, J.; Tang, B. AIE-based luminescence probes for metal ion detection. Coord. Chem. Rev. 2021, 429, 213693. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Wang, P.; Yu, L.; An, J.; Deng, G.; Sun, Y.; Kim, J. Reactive oxygen species, thiols and enzymes activable AIEgens from single fluorescence imaging to multifunctional theranostics. Coord. Chem. Rev. 2021, 427, 213559. [Google Scholar] [CrossRef]
- Li, X.; Li, M.; Yang, M.; Xiao, H.; Wang, L.; Chen, Z.; Liu, S.; Li, J.; Li, S.; James, T. “Irregular” aggregation-induced emission luminogens. Coord. Chem. Rev. 2020, 418, 213358. [Google Scholar] [CrossRef]
- Ouyang, J.; Sun, L.; Zeng, F.; Wu, S. Biomarker-activatable probes based on smart AIEgens for fluorescence and optoacoustic imaging. Coord. Chem. Rev. 2022, 458, 214438. [Google Scholar] [CrossRef]
- Kwok, R.; Leung, C.; Lam, J.; Tang, B. Biosensing by luminogens with aggregation-induced emission characteristics. Chem. Soc. Rev. 2015, 44, 4228–4238. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wu, Y.; Zeng, F.; Chen, J.; Wu, S. A turn-on fluorescence probe based on aggregation-induced emission for leucine aminopeptidase in living cells and tumor tissue. Anal. Chim. Acta 2018, 1031, 169–177. [Google Scholar] [CrossRef]
- Huang, X.; Lei, Q.; Huang, S.; Zeng, H.; Feng, B.; Zeng, Q.; Hu, Y.; Zeng, W. Construction of a novel asymmetric imidazole-cored AIE probe for ratiometric imaging of endogenous leucine aminopeptidase. Chem. Commun. 2021, 57, 6608–6611. [Google Scholar] [CrossRef]
- Lin, V.; Chen, W.; Xian, M.; Chang, C. Chemical probes for molecular imaging and detection of hydrogen sulfide and reactive sulfur species in biological systems. Chem. Soc. Rev. 2015, 44, 4596–4618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Ma, X.; Chen, H.; Liu, S.; Yin, J. Fluorescent probes for pH and alkali metal ions. Coord. Chem. Rev. 2021, 427, 213584. [Google Scholar] [CrossRef]
- Kaur, A.; New, E. Bioinspired small-molecule tools for the imaging of redox biology. Acc. Chem. Res. 2019, 52, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Li, P.; Tang, B. Recent progresses in fluorescent probes for detection of polarity. Coord. Chem. Rev. 2021, 427, 213582. [Google Scholar] [CrossRef]
- Liu, Y.; Teng, L.; Xu, C.; Liu, H.; Xu, S.; Guo, H.; Yuan, L.; Zhang, X. A “Double-Locked” and enzyme-activated molecular probe for accurate bioimaging and hepatopathy differentiation. Chem. Sci. 2019, 10, 10931–10936. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Wang, C.; Ding, Z.; Wang, H.; Wang, Y.; Liu, Z. A Molecular Logic Gate for Developing "AND" Logic Probes and the Application in Hepatopathy Differentiation. ACS Cent. Sci. 2022, 8, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Prost, M.; Hasserodt, J. “Double gating”—A concept for enzyme-responsive imaging probes aiming at high tissue specificity. Chem. Commun. 2014, 50, 14896–14899. [Google Scholar] [CrossRef]
- Li, R.; Guo, J.; Duan, Y.; Liu, X.; Gui, L.; Xu, Y.; Kong, X.; Li, Y.; Chen, H.; Yuan, Z. Monitoring inflammation-cancer progression by cell viscosity, polarity and leucine aminopeptidase using multicolor fluorescent probe. Chem. Eng. J. 2022, 435, 135043. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, H.; Zhang, Y.; Su, M.; Liu, M.; Zhang, X.; Wang, X.; Rong, X.; Wang, K.; Li, X. Recent advances in Golgi-targeted small-molecule fluorescent probes. Coord. Chem. Rev. 2022, 462, 214504. [Google Scholar] [CrossRef]
- Zhu, H.; Fan, J.; Du, J.; Peng, X. Fluorescent probes for sensing and imaging within specific cellular organelles. Acc. Chem. Res. 2016, 49, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Klymchenko, A. Fluorescent Probes for Lipid Membranes: From the Cell Surface to Organelles. Acc. Chem. Res. 2023, 56, 1–12. [Google Scholar] [CrossRef]
- Yue, Y.; Huo, F.; Cheng, F.; Zhu, X.; Mafireyi, T.; Strongin, R.; Yin, C. Functional synthetic probes for selective targeting and multi-analyte detection and imaging. Chem. Soc. Rev. 2019, 48, 4155–4177. [Google Scholar] [CrossRef]
- Han, H.; Tian, H.; Zang, Y.; Sedgwick, A.; Li, J.; Sessler, J.; He, X.; James, T. Small-molecule fluorescence-based probes for interrogating major organ diseases. Chem. Soc. Rev. 2021, 50, 9391–9429. [Google Scholar] [CrossRef]
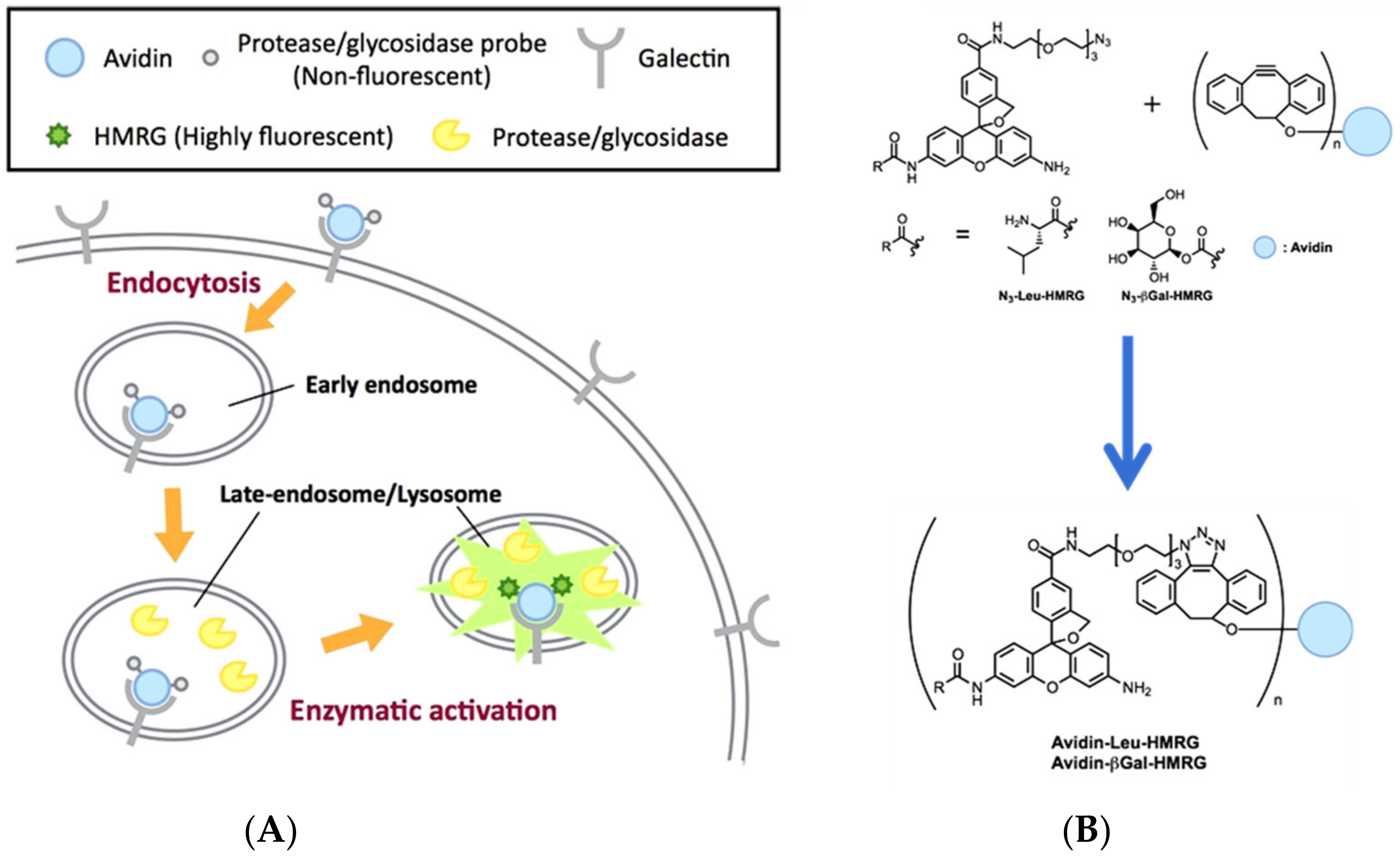
- Yamamoto, K.; Kamiya, M.; Urano, Y. Highly sensitive fluorescence imaging of cancer with avidin-protease probe conjugate. Bioorg. Med. Chem. Lett. 2019, 29, 126663. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Yuan, Q.; Bian, Y.; Li, M.; Wang, Y.; Gao, X.; Su, D. Enzyme-activated near-infrared fluorogenic probe with high-efficiency intrahepatic targeting ability for visualization of drug-induced liver injury. Chem. Sci. 2021, 12, 14855–14862. [Google Scholar] [CrossRef]
- Du, K.; Sheng, L.; Luo, X.; Fan, G.; Shen, D.; Wu, C.; Shen, R. A ratiometric fluorescent probe based on quinoline for monitoring and imaging of Leucine aminopeptidase in liver tumor cells. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2021, 249, 119328. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, J.; Li, P.; Tang, B.; James, T. Two-photon small-molecule fluorescence-based agents for sensing, imaging, and therapy within biological systems. Chem. Soc. Rev. 2021, 50, 702–734. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Song, J.; Qu, J.; Cheng, Z. Crucial breakthrough of second near-infrared biological window fluorophores: Design and synthesis toward multimodal imaging and theranostics. Chem. Soc. Rev. 2018, 47, 4258–4278. [Google Scholar] [CrossRef]
- Kenry; Chong, K.; Liu, B. Reactivity-based organic theranostic bioprobes. Acc. Chem. Res. 2019, 52, 3051–3063. [Google Scholar] [CrossRef]
- Ji, C.; Cheng, W.; Yuan, Q.; Mullen, K.; Yin, M. From dyestuff chemistry to cancer theranostics: The rise of rylenecarboximides. Acc. Chem. Res. 2019, 52, 2266–2277. [Google Scholar] [CrossRef]
- Li, H.; Kim, Y.; Jung, H.; Hyun, J.; Shin, I. Near-infrared (NIR) fluorescence-emitting small organic molecules for cancer imaging and therapy. Chem. Soc. Rev. 2022, 51, 8957–9008. [Google Scholar] [CrossRef]
- Li, K.; Xu, S.; Xiong, M.; Huan, S.; Yuan, L.; Zhang, X. Molecular engineering of organic-based agents for in situ bioimaging and phototherapeutics. Chem. Soc. Rev. 2021, 50, 11766–11784. [Google Scholar] [CrossRef]
- Jung, H.; Verwilst, P.; Sharma, A.; Shin, J.; Sessler, J.; Kim, J. Organic molecule-based photothermal agents: An expanding photothermal therapy universe. Chem. Soc. Rev. 2018, 47, 2280–2297. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, H.; Jangili, P.; Li, M.; Wu, L.; Zang, Y.; Sedgwick, A.; Li, J.; He, X.; James, T. The design of small-molecule prodrugs and activatable phototherapeutics for cancer therapy. Chem. Soc. Rev. 2023, 52, 879–920. [Google Scholar] [CrossRef]
- Jin, W.; Fan, B.; Qin, X.; Liu, Y.; Qian, C.; Tang, B.; James, T.; Chen, G. Structure-activity of chlormethine fluorescent prodrugs: Witnessing the development of trackable drug delivery. Coord. Chem. Rev. 2023, 480, 214999. [Google Scholar] [CrossRef]
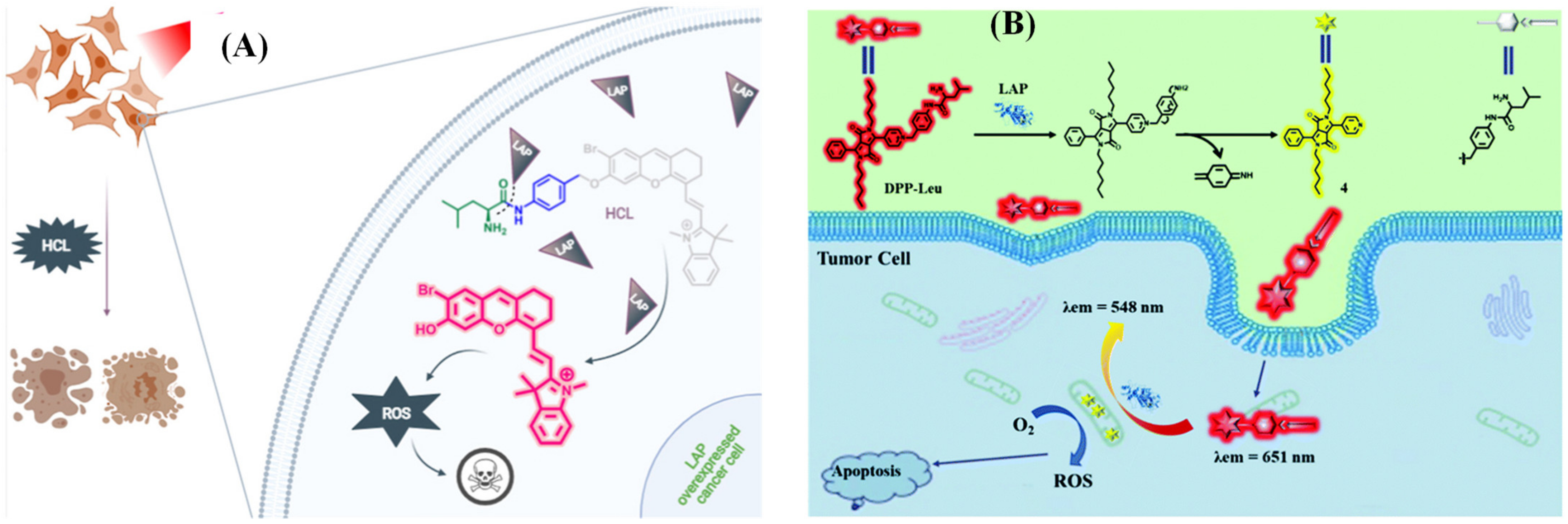
- Arslan, B.; Bilici, K.; Demirci, G.; Almammadov, T.; Khan, M.; Sennaroglu, A.; Acar, H.; Kolemen, S. A leucine aminopeptidase activatable photosensitizer for cancer cell selective photodynamic therapy action. Dye. Pigment. 2021, 195, 109735. [Google Scholar] [CrossRef]
- Xu, W.; Wang, J.; Xu, C.; Hua, J.; Wang, Y. A diketopyrrolopyrrole-based ratiometric fluorescent probe for endogenous leucine aminopeptidase detecting and imaging with specific phototoxicity in tumor cells. J Mater Chem B 2021, 9, 8842–8850. [Google Scholar] [CrossRef]
- Wang, F.; Hu, S.; Sun, Q.; Fei, Q.; Ma, C.; Lu, C.; Nie, J.; Chen, Z.; Ren, J.; Chen, G.; et al. A Leucine Aminopeptidase-Activated Theranostic Prodrug for Cancer Diagnosis and Chemotherapy. ACS Appl Bio Mater 2019, 2, 4904–4910. [Google Scholar] [CrossRef]
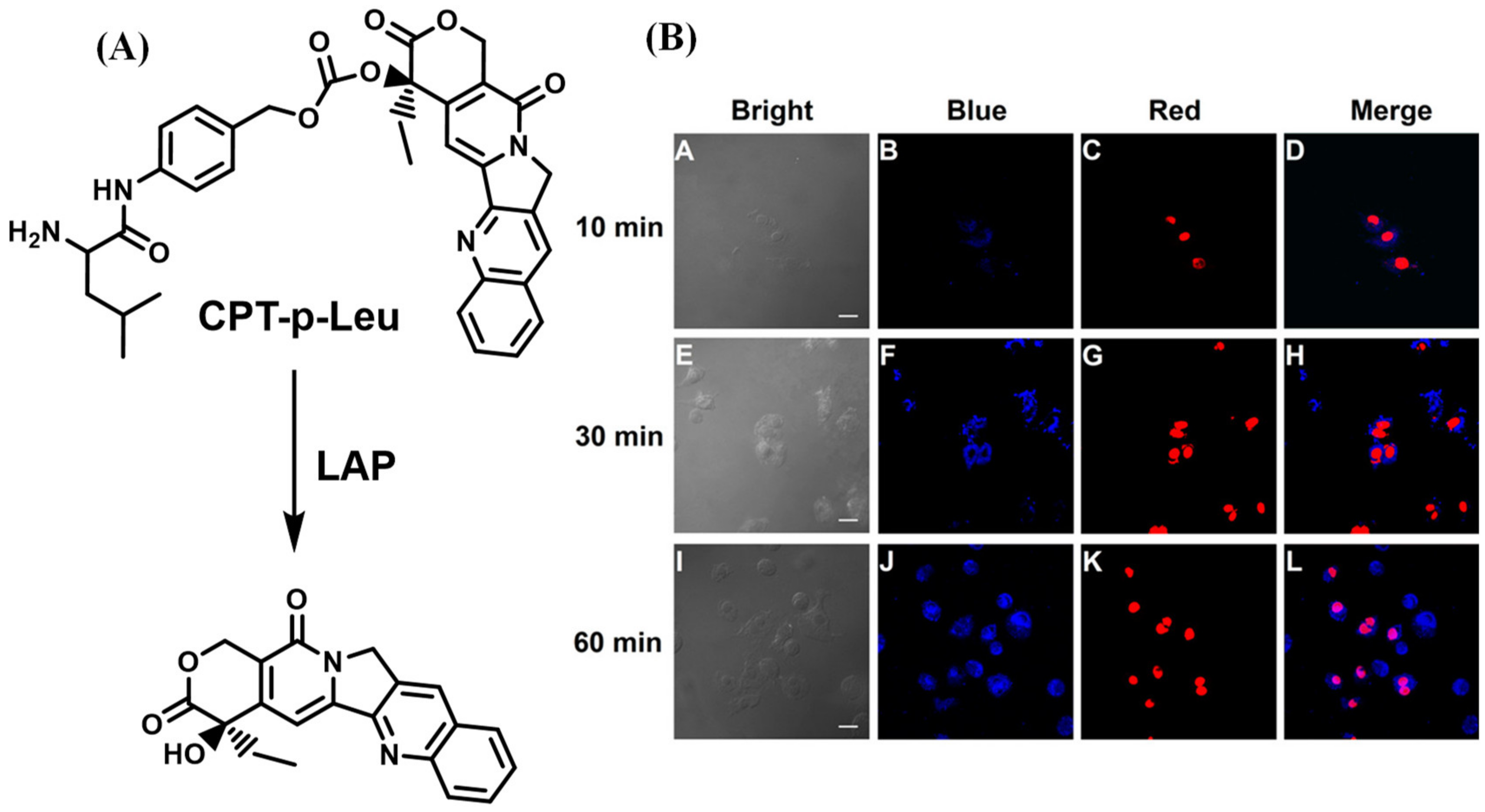
- He, X.; Li, L.; Fang, Y.; Shi, W.; Li, X.; Ma, H. In vivo imaging of leucine aminopeptidase activity in drug-induced liver injury and liver cancer via a near-infrared fluorescent probe. Chem. Sci. 2017, 8, 3479–3483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Qi, Y.; Zhan, C.; Zeng, F.; Wu, S. Diagnosing Drug-Induced Liver Injury by Multispectral Optoacoustic Tomography and Fluorescence Imaging Using a Leucine-Aminopeptidase-Activated Probe. Anal. Chem. 2019, 91, 8085–8092. [Google Scholar] [CrossRef]
- Cheng, D.; Peng, J.; Lv, Y.; Su, D.; Liu, D.; Chen, M.; Yuan, L.; Zhang, X. De Novo Design of Chemical Stability Near-Infrared Molecular Probes for High-Fidelity Hepatotoxicity Evaluation In Vivo. J. Am. Chem. Soc. 2019, 141, 6352–6361. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.; Liu, Y.; Guo, Z.; Lian, C.; Yan, C.; Shi, P.; Tian, H.; Zhu, W. In Situ Ratiometric Quantitative Tracing of Intracellular Leucine Aminopeptidase Activity via an Activatable Near-Infrared Fluorescent Probe. ACS Appl. Mater. Interfaces 2016, 8, 26622–26629. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, F.; Zhang, C.; Luo, J.; Luo, J.; Yu, W.; Kong, L. Near-Infrared Fluorescent Probe with Remarkable Large Stokes Shift and Favorable Water Solubility for Real-Time Tracking Leucine Aminopeptidase in Living Cells and In Vivo. Anal. Chem. 2017, 89, 12319–12326. [Google Scholar] [CrossRef]
- Wu, B.; Lin, Y.; Li, B.; Zhan, C.; Zeng, F.; Wu, S. Oligo(ethylene glycol)-Functionalized Squaraine Fluorophore as a Near-Infrared-Fluorescent Probe for the In Vivo Detection of Diagnostic Enzymes. Anal. Chem. 2018, 90, 9359–9365. [Google Scholar] [CrossRef]
- Chai, Y.; Gao, Y.; Xiong, H.; Lv, W.; Yang, G.; Lu, C.; Nie, J.; Ma, C.; Chen, Z.; Ren, J.; et al. A near-infrared fluorescent probe for monitoring leucine aminopeptidase in living cells. Analyst 2019, 144, 463–467. [Google Scholar] [CrossRef]
- Esoda, C.; Kuehn, M. Pseudomonas aeruginosa leucine aminopeptidase influences early biofilm composition and structure via vesicle-associated antibiofilm activity. MBio 2019, 10, e02548-19. [Google Scholar] [CrossRef] [Green Version]
- Jarocki, V.; Santos, J.; Tacchi, J.; Raymond, B.; Deutscher, A.; Jenkins, C.; Padula, M.; Djordjevic, S. MHJ_0461 is a multifunctional leucine aminopeptidase on the surface of Mycoplasma hyopneumoniae. Open Biol. 2015, 5, 140175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, C.; Liu, L.; Li, C.; He, L.; Li, T.; Shen, Y.; Gao, C.; Wang, N.; Xia, Y.; Zhu, Y. Structure–Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 2385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa, A.; Bastos, I.; Neves, D.; Kipnis, A.; Junqueira-Kipnis, A.; De Santana, J. The activity of a hexameric M17 metallo-aminopeptidase is associated with survival of Mycobacterium tuberculosis. Front. Microbiol. 2017, 8, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Tian, Z.; Wang, J.; Tian, X.; Wang, C.; Cui, J.; Huo, X.; Feng, L.; Yu, Z.; Ma, X. Visual Analysis and Inhibitor Screening of Leucine Aminopeptidase, a Key Virulence Factor for Pathogenic Bacteria-Associated Infection. ACS Sens 2021, 6, 3604–3610. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, M.; Alonso, C.; Pérez, J. Biochemical modulation of cisplatin mechanisms of action: Enhancement of antitumor activity and circumvention of drug resistance. Chem. Rev. 2003, 103, 645–662. [Google Scholar] [CrossRef]
- Siddik, Z. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Q.; Shi, W.; Li, L.; Ma, H. Leucine aminopeptidase may contribute to the intrinsic resistance of cancer cells toward cisplatin as revealed by an ultrasensitive fluorescent probe. Chem. Sci. 2016, 7, 788–792. [Google Scholar] [CrossRef] [Green Version]
- Lei, Z.; Yang, Y. A concise colorimetric and fluorimetric probe for sarin related threats designed via the "covalent-assembly" approach. J. Am. Chem. Soc. 2014, 136, 6594–6597. [Google Scholar] [CrossRef]
- Kévin, R.; Sylvain, D.; Jean-Alexandre, R.; Anthony, R. Deeper insight into protease-sensitive “covalent-assembly” fluorescent probes for practical biosensing applications. Org. Biomol. Chem. 2019, 17, 8918–8932. [Google Scholar]
- Yuan, D.; Xu, Z.; Zhang, B.; Yin, X.; Ye, J.; Zhou, X.; Wang, L. A ratiometric fluorescence probe for selective and sensitive detection of leucine aminopeptidase in lysosome. Chem. Commun. 2022, 58, 8364–8367. [Google Scholar] [CrossRef]
- Liu, T.; Tian, M.; Wang, J.; Tian, X.; Liu, J.; Feng, L.; Ma, X.; Cui, J. Rational design of a fluorescent probe for the detection of LAP and its application in drug-induced liver injury. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2021, 251, 119362. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, F.; Yang, G.; Lu, C.; Nie, J.; Chen, Z.; Ren, J.; Sun, Q.; Zhao, C.; Zhu, W. A Ratiometric Fluorescent Probe for Monitoring Leucine Aminopeptidase in Living Cells and Zebrafish Model. Anal. Chem. 2017, 89, 11576–11582. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sun, Q.; Xiong, H.; Ma, C.; Lu, C.; Nie, J.; Yang, G.; Chen, Z.; Zhang, Y.; Ren, J.; et al. Rational design of fluorescent probes: Improving hydrophilicity, ratiometric and NIR trapping of endogenous leucine aminopeptidase. Sens. Actuators B. Chem 2020, 321, 128631. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, Z.; Wang, F.; Yang, G.; Zhang, Y.; Zhang, X. Multi-dimensional imaging of endogenous leucine aminopeptidase via fast response fluorescent read-out probe. Dye. Pigment. 2021, 187, 109145. [Google Scholar] [CrossRef]
- Tao, L.; Liu, S.; Xia, X.; Chai, Y.; Cai, S.; Liu, H.; Lu, C.; Ma, C.; Nie, J.; Zeng, F.; et al. Near-infrared fluorescent read-out probe for ultra-sensitive imaging of leucine aminopeptidase in vitro and in vivo. Tetrahedron 2021, 99, 132449. [Google Scholar] [CrossRef]










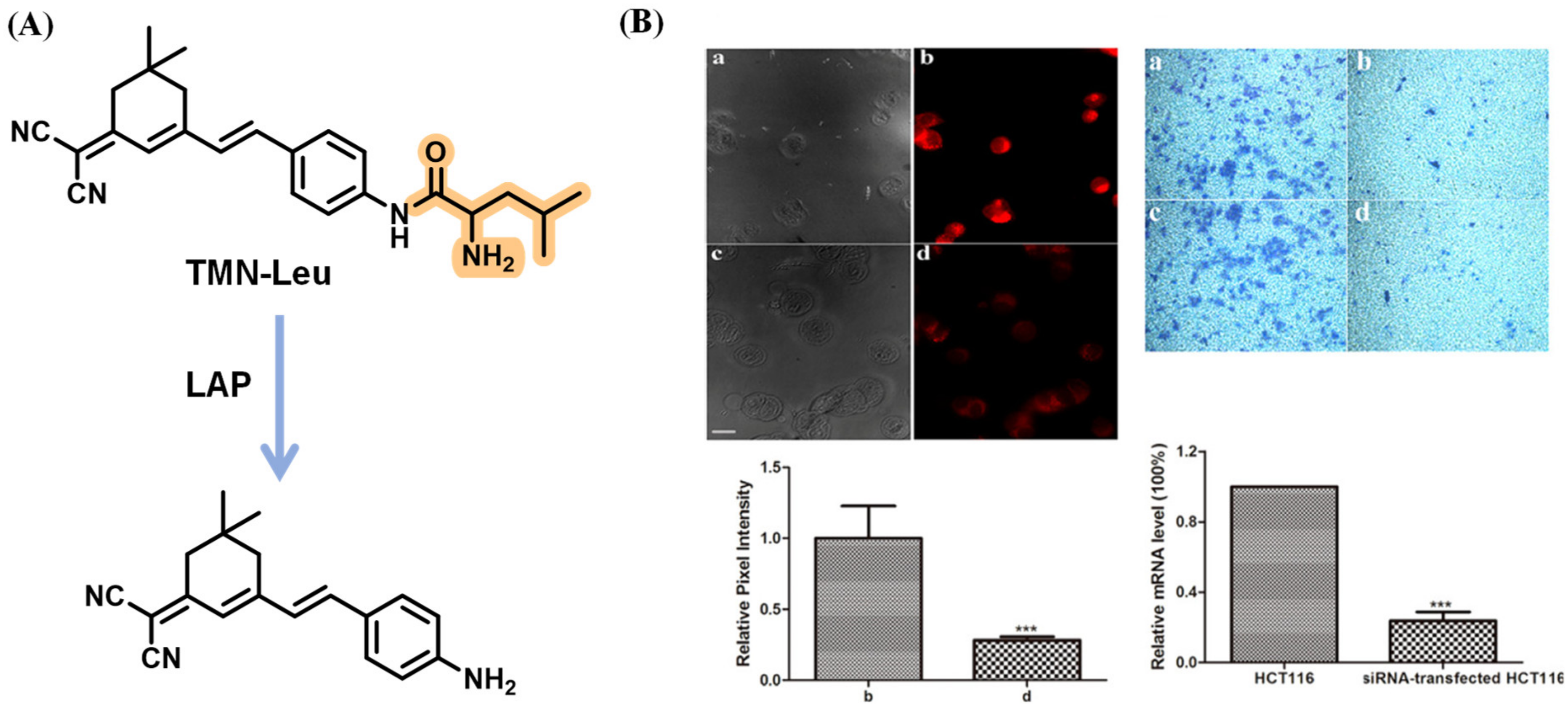


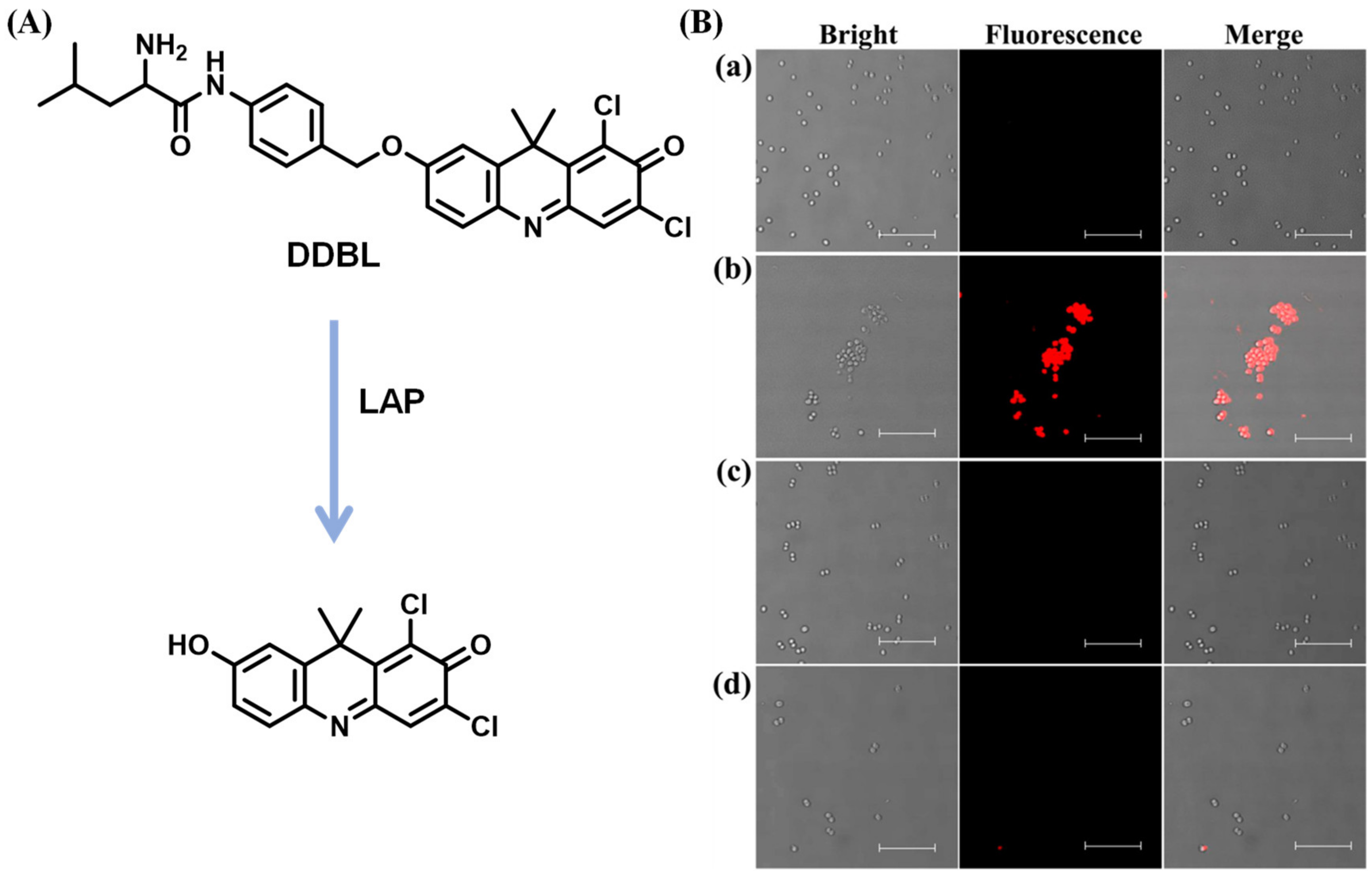
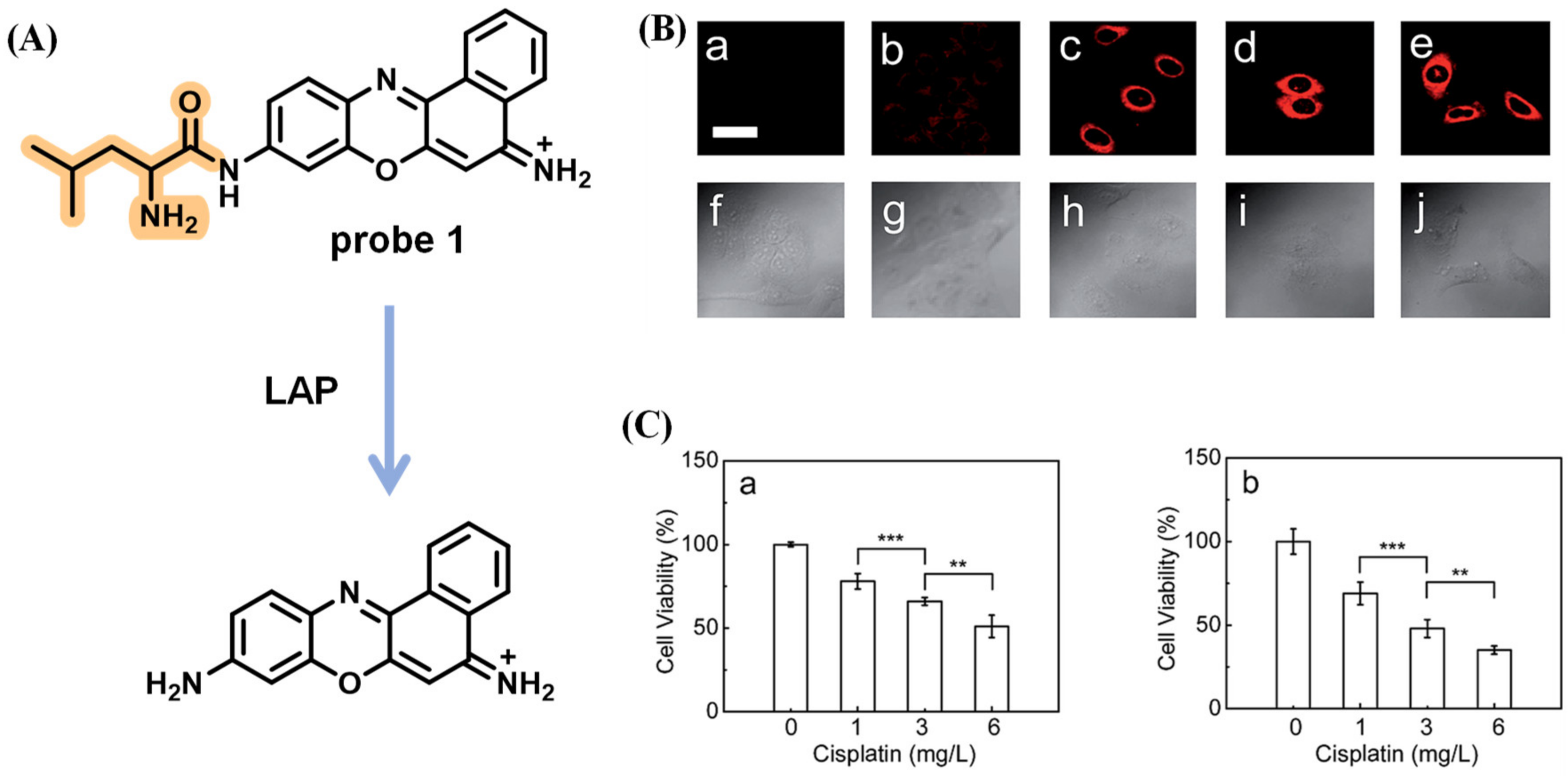

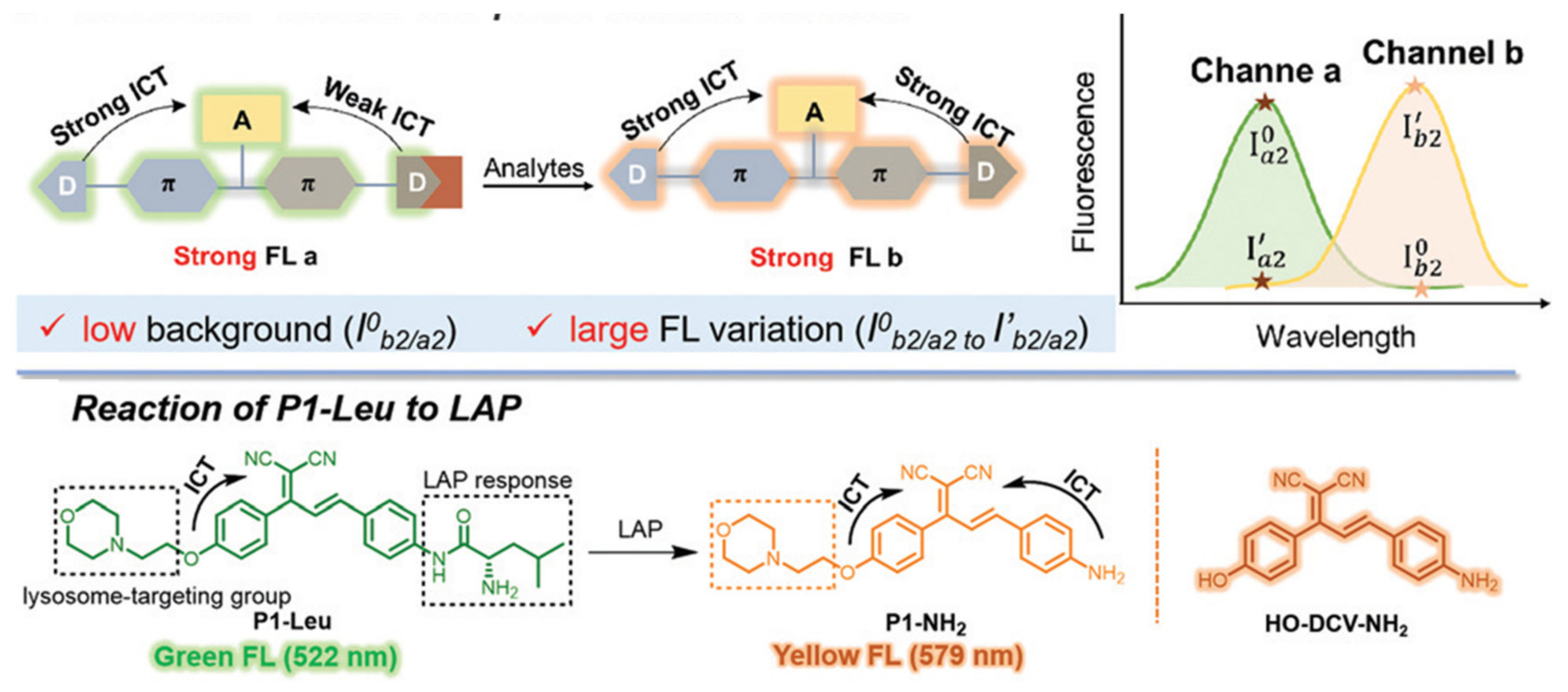


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.-J.; Wang, C.-Y.; Xu, L.; Zhang, Z.-Y.; Tang, Y.-H.; Qin, T.-Y.; Wang, Y.-L. Recent Progress of Activity-Based Fluorescent Probes for Imaging Leucine Aminopeptidase. Biosensors 2023, 13, 752. https://doi.org/10.3390/bios13070752
Li Z-J, Wang C-Y, Xu L, Zhang Z-Y, Tang Y-H, Qin T-Y, Wang Y-L. Recent Progress of Activity-Based Fluorescent Probes for Imaging Leucine Aminopeptidase. Biosensors. 2023; 13(7):752. https://doi.org/10.3390/bios13070752
Chicago/Turabian StyleLi, Ze-Jun, Cai-Yun Wang, Liang Xu, Zhen-Yu Zhang, Ying-Hao Tang, Tian-Yi Qin, and Ya-Long Wang. 2023. "Recent Progress of Activity-Based Fluorescent Probes for Imaging Leucine Aminopeptidase" Biosensors 13, no. 7: 752. https://doi.org/10.3390/bios13070752
APA StyleLi, Z. -J., Wang, C. -Y., Xu, L., Zhang, Z. -Y., Tang, Y. -H., Qin, T. -Y., & Wang, Y. -L. (2023). Recent Progress of Activity-Based Fluorescent Probes for Imaging Leucine Aminopeptidase. Biosensors, 13(7), 752. https://doi.org/10.3390/bios13070752




