Machine Learning Techniques for Effective Pathogen Detection Based on Resonant Biosensors
Abstract
:1. Introduction
2. Materials and Methods
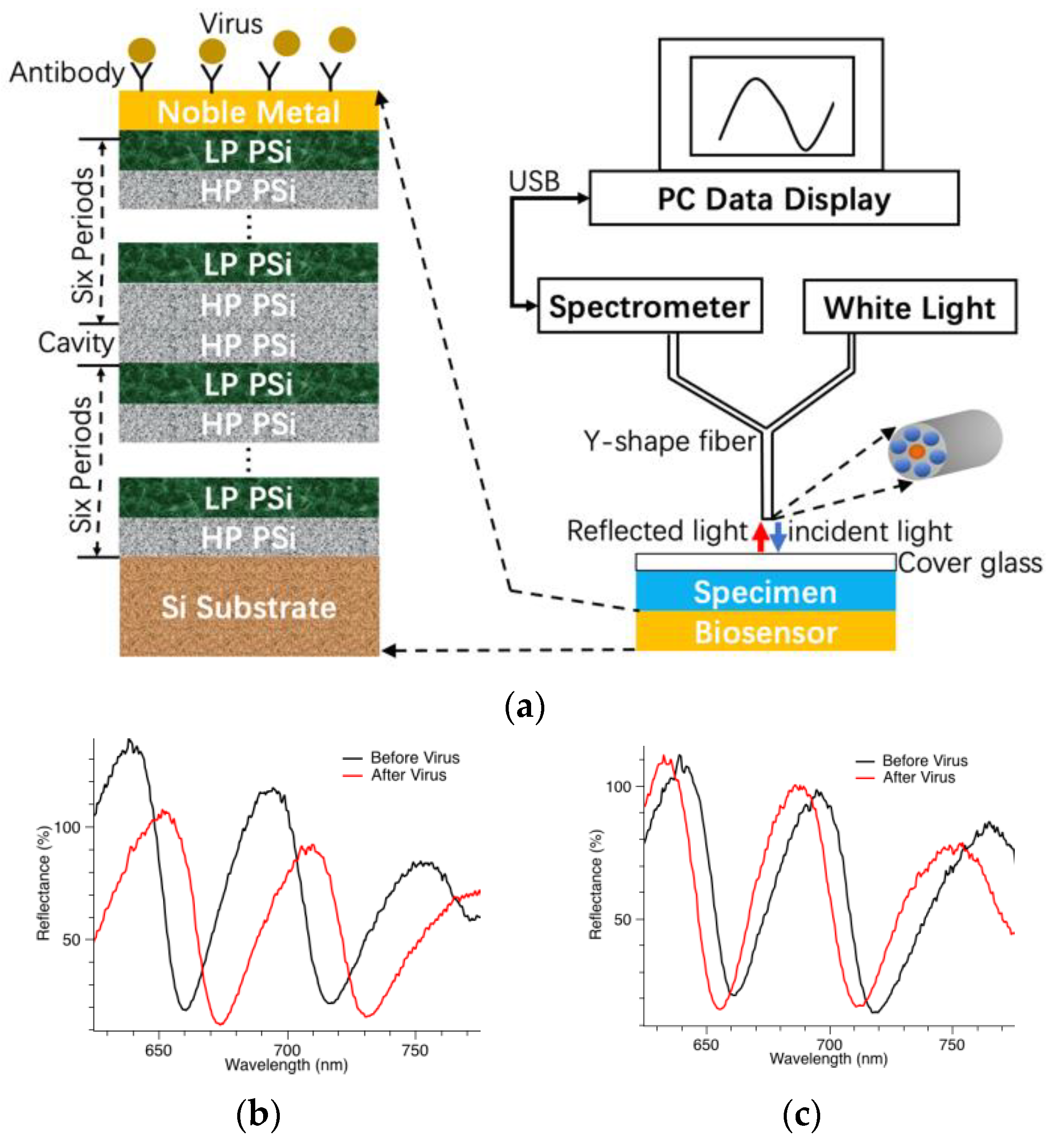
2.1. Biosensor Working Principal and Measurement Setup
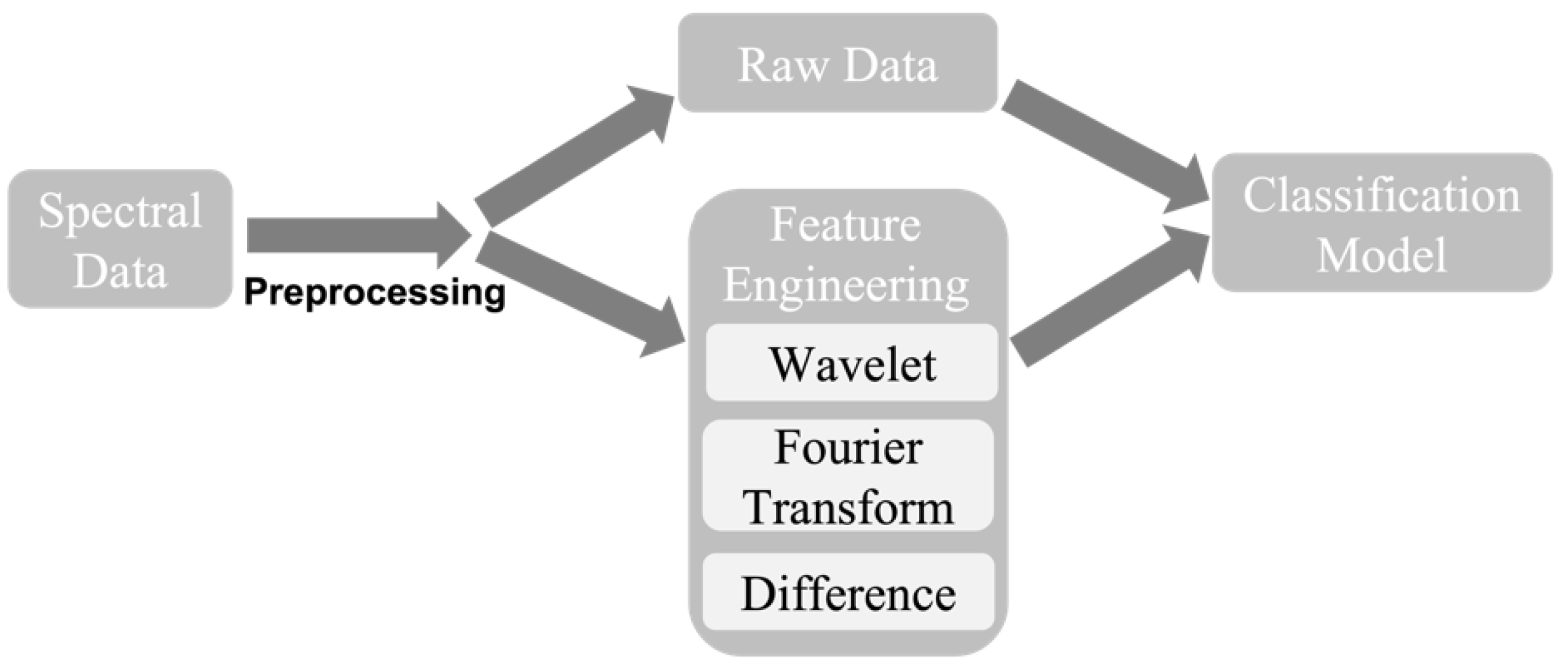
2.2. Data Preprocessing
2.3. Feature Engineering
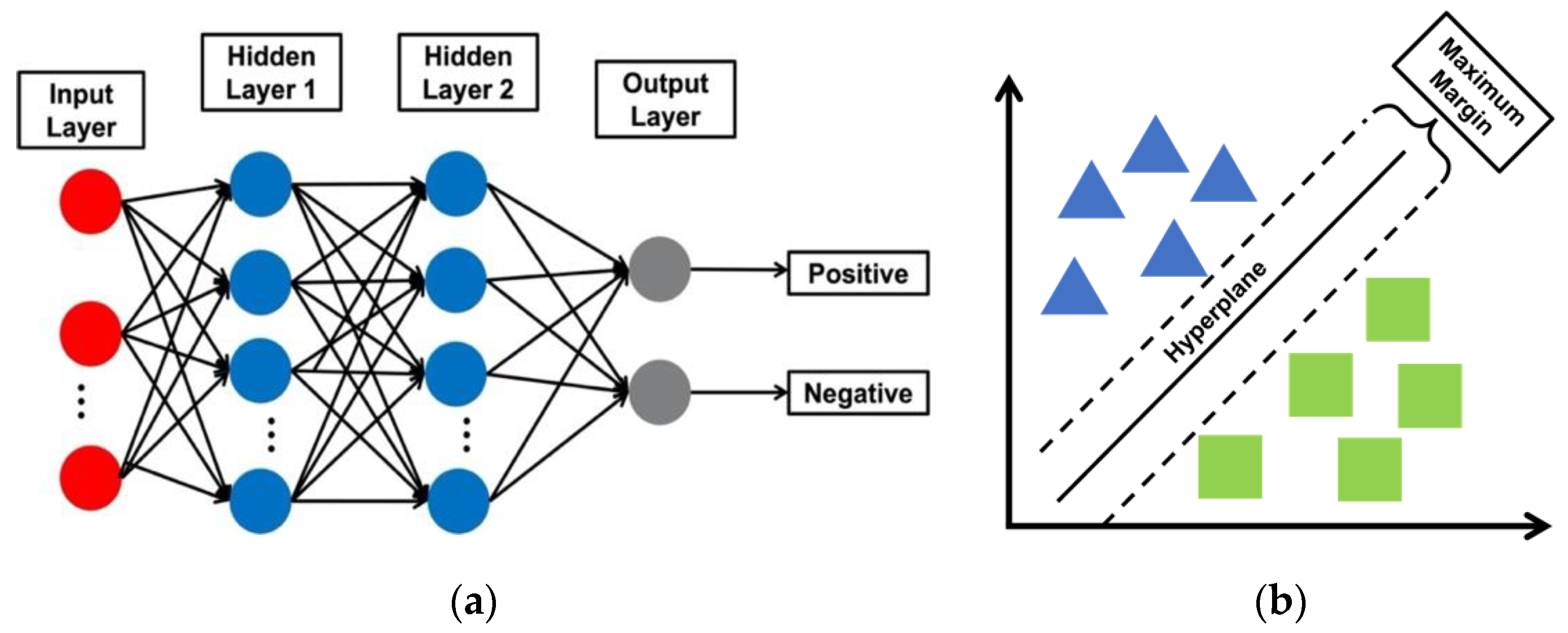
2.4. Classification Models
2.5. Control Experiments
2.6. Dataset Distinguishability Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, Y.; Yang, C.; Wang, T.; Coltey, E.; Jin, Y.; Liu, Y.; Jiang, R.; Fan, Z.; Song, X.; Shibasaki, R.; et al. A Survey on Data-driven COVID-19 and Future Pandemic Management. ACM Comput. Surv. 2022, 55, 1–36. [Google Scholar] [CrossRef]
- Tenali, N.; Babu, G.R.M. A Systematic Literature Review and Future Perspectives for Handling Big Data Analytics in COVID-19 Diagnosis. New Gener. Comput. 2023, 41, 243–280. [Google Scholar] [CrossRef] [PubMed]
- Duncan, D.B.; Mackett, K.; Ali, M.U.; Yamamura, D.; Balion, C. Performance of saliva compared with nasopharyngeal swab for diagnosis of COVID-19 by NAAT in cross-sectional studies: Systematic review and meta-analysis. Clin. Biochem. 2023, 117, 84–93. [Google Scholar] [CrossRef]
- Tng, D.J.; Yin, B.C.; Cao, J.; Ko, K.K.; Goh, K.C.; Chua, D.X.; Zhang, Y.; Chua, M.L.; Low, J.G.; Ooi, E.E.; et al. Amplified parallel antigen rapid test for point-of-care salivary detection of SARS-CoV-2 with improved sensitivity. Microchim. Acta 2022, 189, 14. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Naiyer, S.; Mansuri, S.; Soni, N.; Singh, V.; Bhat, K.H.; Singh, N.; Arora, G.; Mansuri, M.S. COVID-19 Diagnosis: A Comprehensive Review of the RT-qPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12, 1503. [Google Scholar] [CrossRef]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef]
- He, J.; Zhu, S.; Zhou, J.; Jiang, W.; Yin, L.; Su, L.; Zhang, X.; Chen, Q.; Li, X. Rapid detection of SARS-CoV-2: The gradual boom of lateral flow immunoassay. Front. Bioeng. Biotechnol. 2023, 10, 1090281. [Google Scholar] [CrossRef]
- Al-Hashimi, O.T.; AL-Ansari, W.I.; Abbas, S.A.; Jumaa, D.S.; Hammad, S.A.; Hammoudi, F.A.; Allawi, A.A. The sensitivity and specificity of COVID-19 rapid anti-gene test in comparison to RT-PCR test as a gold standard test. J. Clin. Lab. Anal. 2023, 37, e24844. [Google Scholar] [CrossRef]
- Liu, K.S.; Mao, X.D.; Ni, W.; Li, T.P. Laboratory detection of SARS-CoV-2: A review of the current literature and future perspectives. Heliyon 2022, 8, e10858. [Google Scholar] [CrossRef]
- Chong, Y.P.; Choy, K.W.; Doerig, C.; Lim, C.X. SARS-CoV-2 Testing Strategies in the Diagnosis and Management of COVID-19 Patients in Low-Income Countries: A Scoping Review. Mol. Diagn. Ther. 2023, 27, 303–320. [Google Scholar] [CrossRef]
- El-Sherif, D.M.; Abouzid, M.; Gaballah, M.S.; Ahmed, A.A.; Adeel, M.; Sheta, S.M. New approach in SARS-CoV-2 surveillance using biosensor technology: A review. Environ. Sci. Pollut. Res. 2022, 29, 1677–1695. [Google Scholar] [CrossRef] [PubMed]
- Abid, S.A.; Muneer, A.A.; Al-Kadmy, I.M.; Sattar, A.A.; Beshbishy, A.M.; Batiha, G.E.-S.; Hetta, H.F. Biosensors as a future diagnostic approach for COVID-19. Life Sci. 2021, 273, 119117. [Google Scholar] [CrossRef]
- Wei, H.; Zhang, C.; Du, X.; Zhang, Z. Research progress of biosensors for detection of SARS-CoV-2 variants based on ACE2. Talanta 2023, 251, 123813. [Google Scholar] [CrossRef]
- Rong, G.; Zheng, Y.; Li, X.; Guo, M.; Su, Y.; Bian, S.; Dang, B.; Chen, Y.; Zhang, Y.; Shen, L.; et al. A high-throughput fully automatic biosensing platform for efficient COVID-19 detection. Biosens. Bioelectron. 2023, 220, 114861. [Google Scholar] [CrossRef]
- Rong, G.; Zheng, Y.; Yang, X.; Bao, K.; Xia, F.; Ren, H.; Bian, S.; Li, L.; Zhu, B.; Sawan, M. A Closed-Loop Approach to Fight Coronavirus: Early Detection and Subsequent Treatment. Biosensors 2022, 12, 900. [Google Scholar] [CrossRef] [PubMed]
- Takemura, K. Surface Plasmon Resonance (SPR)- and Localized SPR (LSPR)-Based Virus Sensing Systems: Optical Vibration of Nano- and Micro-Metallic Materials for the Development of Next-Generation Virus Detection Technology. Biosensors 2021, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hu, W.; Fang, Y. Direct excitation of the Tamm plasmon-polaritons on a dielectric Bragg reflector coated with a metal film. Opto-Electron. Rev. 2013, 21, 338–343. [Google Scholar] [CrossRef]
- Liu, C.; Kong, M.; Li, B. Tamm plasmon-polariton with negative group velocity induced by a negative index meta-material capping layer at metal-Bragg reflector interface. Opt. Express 2014, 22, 11376–11383. [Google Scholar] [CrossRef]
- Zheng, Y.; Bian, S.; Sun, J.; Wen, L.; Rong, G.; Sawan, M. Label-Free LSPR-Vertical Microcavity Biosensor for On-Site SARS-CoV-2 Detection. Biosensors 2022, 12, 151. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, Y.; Rong, G.; Lin, Z.; Wang, G.; Gu, Z.; Sawan, M. Efficient Optical Pattern Detection for Microcavity Sensors Based Lab-on-a-Chip. IEEE Sens. J. 2012, 12, 2121–2128. [Google Scholar] [CrossRef]
- Mariani, S.; Pino, L.; Strambini, L.M.; Tedeschi, L.; Barillaro, G. 10 000-Fold Improvement in Protein Detection Using Nanostructured Porous Silicon Interferometric Aptasensors. ACS Sens. 2016, 1, 1471–1479. [Google Scholar] [CrossRef]
- Wu, C.; Rong, G.; Xu, J.; Pan, S.; Zhu, Y. Physical analysis of the response properties of porous silicon microcavity biosensor. Phys. E Low-Dimens. Syst. Nanostructures 2012, 44, 1787–1791. [Google Scholar] [CrossRef]
- Kotsiantis, S.B.; Zaharakis, I.D.; Pintelas, P.E. Machine learning: A review of classification and combining techniques. Artif. Intell. Rev. 2006, 26, 159–190. [Google Scholar] [CrossRef]
- Kliegr, T.; Bahník, Š.; Fürnkranz, J. A review of possible effects of cognitive biases on interpretation of rule-based machine learning models. Artif. Intell. 2021, 295, 103458. [Google Scholar] [CrossRef]
- Metri-Ojeda, J.; Solana-Lavalle, G.; Rosas-Romero, R.; Palou, E.; Rodrigues, M.R.; Baigts-Allende, D. Rapid screening of mayonnaise quality using computer vision and machine learning. J. Food Meas. Charact. 2023, 17, 2792–2804. [Google Scholar] [CrossRef]
- Mughaid, A.; Obeidat, I.; AlZu’bi, S.; Abu Elsoud, E.; Alnajjar, A.; Alsoud, A.R.; Abualigah, L. A novel machine learning and face recognition technique for fake accounts detection system on cyber social networks. Multimed. Tools Appl. 2023, 82, 26353–26378. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, J.; Gao, H.; Li, R.; Jiang, Z.; Yin, Y.; Wu, Y. Machine Learning-Driven APPs Recommendation for Energy Optimization in Green Communication and Networking for Connected and Autonomous Vehicles. IEEE Trans. Green Commun. Netw. 2022, 6, 1543–1552. [Google Scholar] [CrossRef]
- Balkus, S.V.; Wang, H.; Cornet, B.D.; Mahabal, C.; Ngo, H.; Fang, H. A Survey of Collaborative Machine Learning Using 5G Vehicular Communications. IEEE Commun. Surv. Tutor. 2022, 24, 1280–1303. [Google Scholar] [CrossRef]
- Dai, Z.-H.; Wang, R.-H.; Guan, J.-H. Auxiliary Decision-Making System for Steel Plate Cold Straightening Based on Multi-Machine Learning Competition Strategies. Appl. Sci. 2022, 12, 11473. [Google Scholar] [CrossRef]
- Hoque, M.N.; Mueller, K. Outcome-Explorer: A Causality Guided Interactive Visual Interface for Interpretable Algorithmic Decision Making. Ieee Trans. Vis. Comput. Graph. 2022, 28, 4728–4740. [Google Scholar] [CrossRef]
- Fidêncio, A.X.; Klaes, C.; Iossifidis, I. Error-Related Potentials in Reinforcement Learning-Based Brain-Machine Interfaces. Front. Hum. Neurosci. 2022, 16, 806517. [Google Scholar] [CrossRef] [PubMed]
- Tabari, A.; Chan, S.M.; Omar, O.M.F.; Iqbal, S.I.; Gee, M.S.; Daye, D. Role of Machine Learning in Precision Oncology: Applications in Gastrointestinal Cancers. Cancers 2022, 15, 63. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Cuzzocrea, A.; Kresta, M.; Kristjanson, K.D.; Leung, C.K.; Tebinka, T.W. A Machine Learning System for Supporting Advanced Knowledge Discovery from Chess Game Data. In Proceedings of the 2017 16th Ieee International Conference on Machine Learning and Applications (Icmla), Cancun, Mexico, 18–21 December 2017; pp. 649–654. [Google Scholar]
- Hu, J.; Sasakawa, T.; Hirasawa, K.; Zheng, H. A hierarchical learning system incorporating with supervised, unsupervised and reinforcement learning. In Advances in Neural Networks—Isnn 2007, Pt 1, Proceedings; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4491, p. 403. [Google Scholar]
- Kumar, B.; Vyas, O.P.; Vyas, R. A comprehensive review on the variants of support vector machines. Mod. Phys. Lett. B 2019, 33, 1950303. [Google Scholar] [CrossRef]
- Champati, B.B.; Padhiari, B.M.; Ray, A.; Halder, T.; Jena, S.; Sahoo, A.; Kar, B.; Kamila, P.K.; Panda, P.C.; Ghosh, B.; et al. Application of a Multilayer Perceptron Artificial Neural Network for the Prediction and Optimization of the Andrographolide Content in Andrographis paniculata. Molecules 2022, 27, 2765. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Ghosh, M. High-dimensional properties for empirical priors in linear regression with unknown error variance. Stat. Pap. 2023. [Google Scholar] [CrossRef]
- Laria, J.C.; Clemmensen, L.H.; Ersbøll, B.K.; Delgado-Gómez, D. A Generalized Linear Joint Trained Framework for Semi-Supervised Learning of Sparse Features. Mathematics 2022, 10, 3001. [Google Scholar] [CrossRef]
- Li, M.; Yuan, B. 2D-LDA: A statistical linear discriminant analysis for image matrix. Pattern Recognit. Lett. 2005, 26, 527–532. [Google Scholar] [CrossRef]
- Graf, R.; Zeldovich, M.; Friedrich, S. Comparing linear discriminant analysis and supervised learning algorithms for binary classification—A method comparison study. Biom. J. 2022. [Google Scholar] [CrossRef]
- Valero-Mas, J.J.; Gallego, A.J.; Alonso-Jiménez, P.; Serra, X. Multilabel Prototype Generation for data reduction in K-Nearest Neighbour classification. Pattern Recognit. 2023, 135, 109190. [Google Scholar] [CrossRef]
- Cannings, T.I.; Berrett, T.B.; Samworth, R.J. Local nearest neighbor classification with applications to semi-supervised learning. Ann. Stat. 2020, 48, 1789–1814. [Google Scholar] [CrossRef]
- Mohanty, S.K.; Swetapadma, A.; Nayak, P.K.; Malik, O.P. Decision tree approach for fault detection in a TCSC compensated line during power swing. Int. J. Electr. Power Energy Syst. 2023, 146, 108758. [Google Scholar] [CrossRef]
- Borchert, S.; Mathilakathu, A.; Nath, A.; Wessolly, M.; Mairinger, E.; Kreidt, D.; Steinborn, J.; Walter, R.F.H.; Christoph, D.C.; Kollmeier, J.; et al. Cancer-Associated Fibroblasts Influence Survival in Pleural Mesothelioma: Digital Gene Expression Analysis and Supervised Machine Learning Model. Int. J. Mol. Sci. 2023, 24, 12426. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Lee, J.-S. Maximizing AUC to learn weighted naive Bayes for imbalanced data classification. Expert Syst. Appl. 2023, 217, 119564. [Google Scholar] [CrossRef]
- Askari, A.; D’aspremont, A.; El Ghaoui, L. Naive Feature Selection: A Nearly Tight Convex Relaxation for Sparse Naive Bayes. Math. Oper. Res. 2023. [Google Scholar] [CrossRef]
- Meniailov, I.; Krivtsov, S.; Chumachenko, T. Dimensionality Reduction of Diabetes Mellitus Patient Data Using the T-Distributed Stochastic Neighbor Embedding; Springer: Cham, Switzerland, 2022; pp. 86–95. [Google Scholar] [CrossRef]
- Wu, B.; Rong, G.; Zhao, J.; Zhang, S.; Zhu, Y.; He, B. A Nanoscale Porous Silicon Microcavity Biosensor for Novel Label-Free Tuberculosis Antigen–Antibody Detection. Nano 2012, 7, 1250049. [Google Scholar] [CrossRef]
- Kaliteevski, M.; Iorsh, I.; Brand, S.; Abram, R.A.; Chamberlain, J.M.; Kavokin, A.V.; Shelykh, I.A. Tamm plasmon-polaritons: Possible electromagnetic states at the interface of a metal and a dielectric Bragg mirror. Phys. Rev. B 2007, 76, 165415. [Google Scholar] [CrossRef]
- van Erven, T.; Harremoes, P. Renyi Divergence and Kullback-Leibler Divergence. Ieee Trans. Inf. Theory 2014, 60, 3797–3820. [Google Scholar] [CrossRef]

| Method | Raw Data | Feature Engineering | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | SVM | MLP | SVM | MLP | ||||||||
| Parameter | SEN | SPE | AUC | SEN | SPE | AUC | SEN | SPE | AUC | SEN | SPE | AUC |
| Performance on Control Detection Data | 100% | 46% | 60.1% | 86% | 46% | 63.4% | 29% | 33% | 38.8% | 27% | 32% | 38.1% |
| p-Value for Control Detection Data | <0.05 | |||||||||||
| Performance on Valid Detection Data | 100% | |||||||||||
| p-Value for Valid Detection Data | <0.05 | |||||||||||
| Factor | Need Data Filtering and Denoising | Need to Take Care of Shift Direction | Need Stable Light Source and Low Noise Spectroscopy System | Needed Researcher Work | |
|---|---|---|---|---|---|
| Technique | |||||
| Find peaks and calculate spectral shift | Yes | Yes | No | Algorithm design and test | |
| Interferogram average over wavelength | Yes | No | Yes | Algorithm design and test | |
| Intensity interrogation | Yes | No | Yes | Algorithm design and test | |
| Machine learning | Yes | No | No | Model training from data | |
| Specimen Collection Location | qPCR Result | Biosensor with ML Result | |
|---|---|---|---|
| Vaccination Site 1 | Operation Desktop | Weak positive | Positive |
| Vaccination Site 1 | Vaccination Station | Strong positive | Positive |
| Vaccination Site 2 | Operation Desktop | Weak positive | Positive |
| Vaccination Site 2 | Vaccination Station | Weak positive | Positive |
| Vaccination Site 2 | Ventilation Plate | Strong positive | Positive |
| Vaccination Site 2 | Inoculation Table Handle | Weak positive | Positive |
| Vaccination Site 4 | Keyboard and Mouse | Negative | Negative |
| Vaccination Site 5 | Pen and White Board | Strong positive | Positive |
| Vaccination Site 55 | Inoculation Table Handle | Negative | Negative |
| No. 4 and No. 5 Inoculation Desk Room | Door Handle and Switch | Negative | Negative |
| Other | Hemostatic Swab | Weak positive | Positive |
| Other | Cleaner’s Hand | Negative | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rong, G.; Xu, Y.; Sawan, M. Machine Learning Techniques for Effective Pathogen Detection Based on Resonant Biosensors. Biosensors 2023, 13, 860. https://doi.org/10.3390/bios13090860
Rong G, Xu Y, Sawan M. Machine Learning Techniques for Effective Pathogen Detection Based on Resonant Biosensors. Biosensors. 2023; 13(9):860. https://doi.org/10.3390/bios13090860
Chicago/Turabian StyleRong, Guoguang, Yankun Xu, and Mohamad Sawan. 2023. "Machine Learning Techniques for Effective Pathogen Detection Based on Resonant Biosensors" Biosensors 13, no. 9: 860. https://doi.org/10.3390/bios13090860
APA StyleRong, G., Xu, Y., & Sawan, M. (2023). Machine Learning Techniques for Effective Pathogen Detection Based on Resonant Biosensors. Biosensors, 13(9), 860. https://doi.org/10.3390/bios13090860







