Simultaneous Detection of SARS-CoV-2 Nucleoprotein and Receptor Binding Domain by a Multi-Area Reflectance Spectroscopy Sensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation
2.3. Chemical Activation and Biofunctionalization of the Chip
2.4. Simultaneous Immunodetection of RBD and NP Biomarkers with MARS Biosensor
3. Results
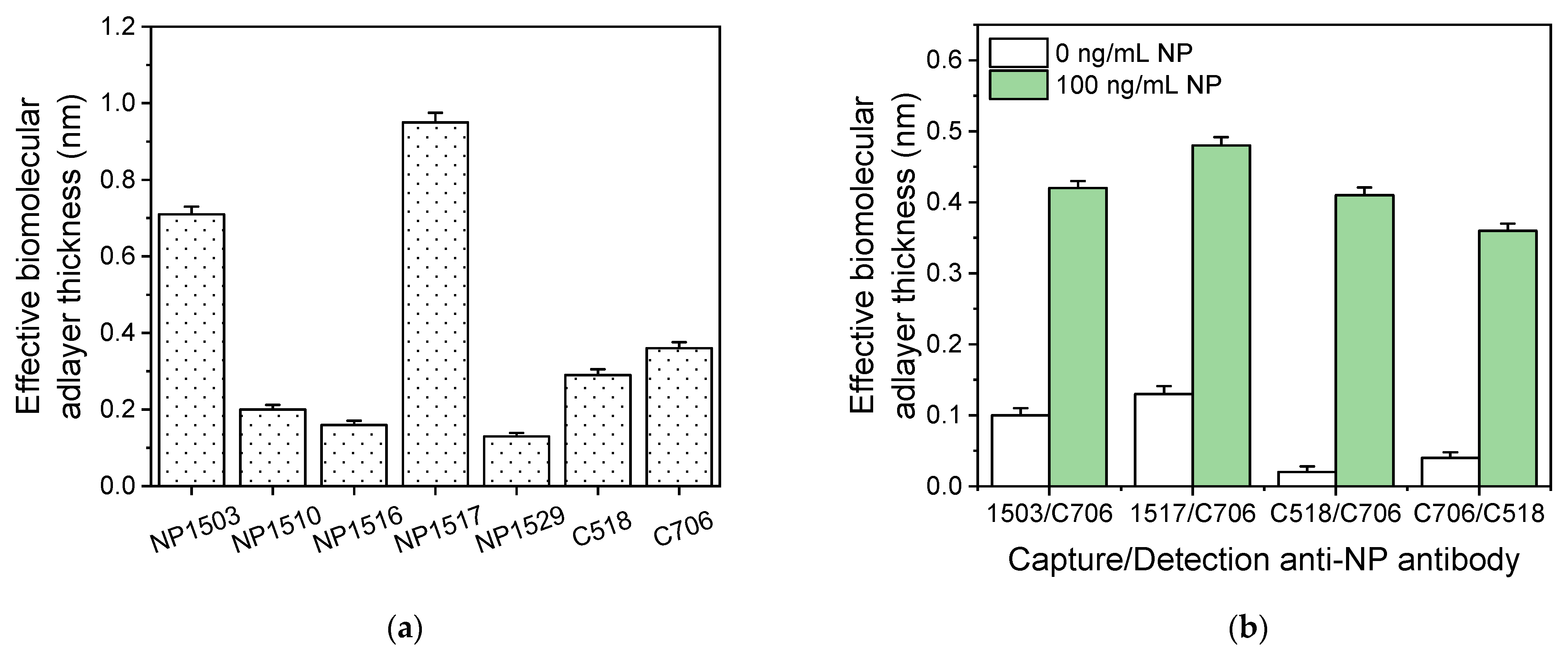
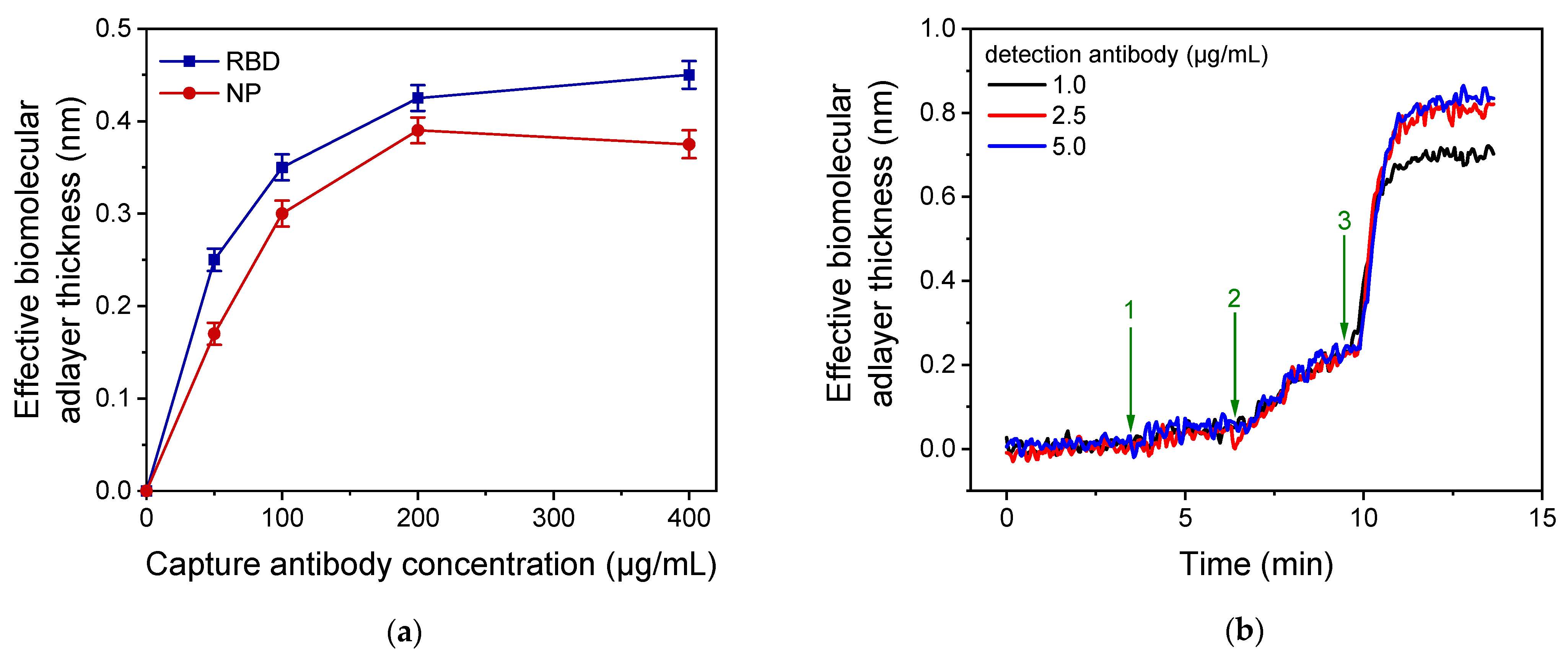
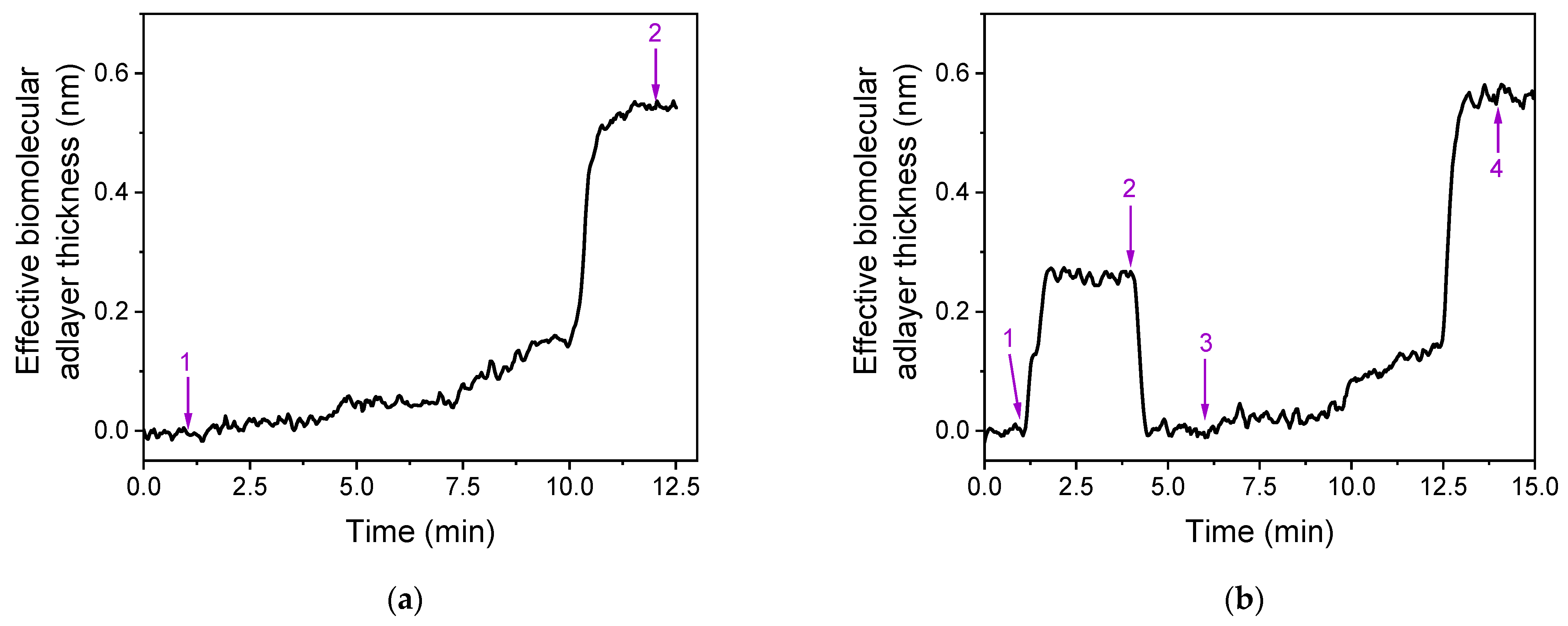
3.1. Assay Development and Optimization
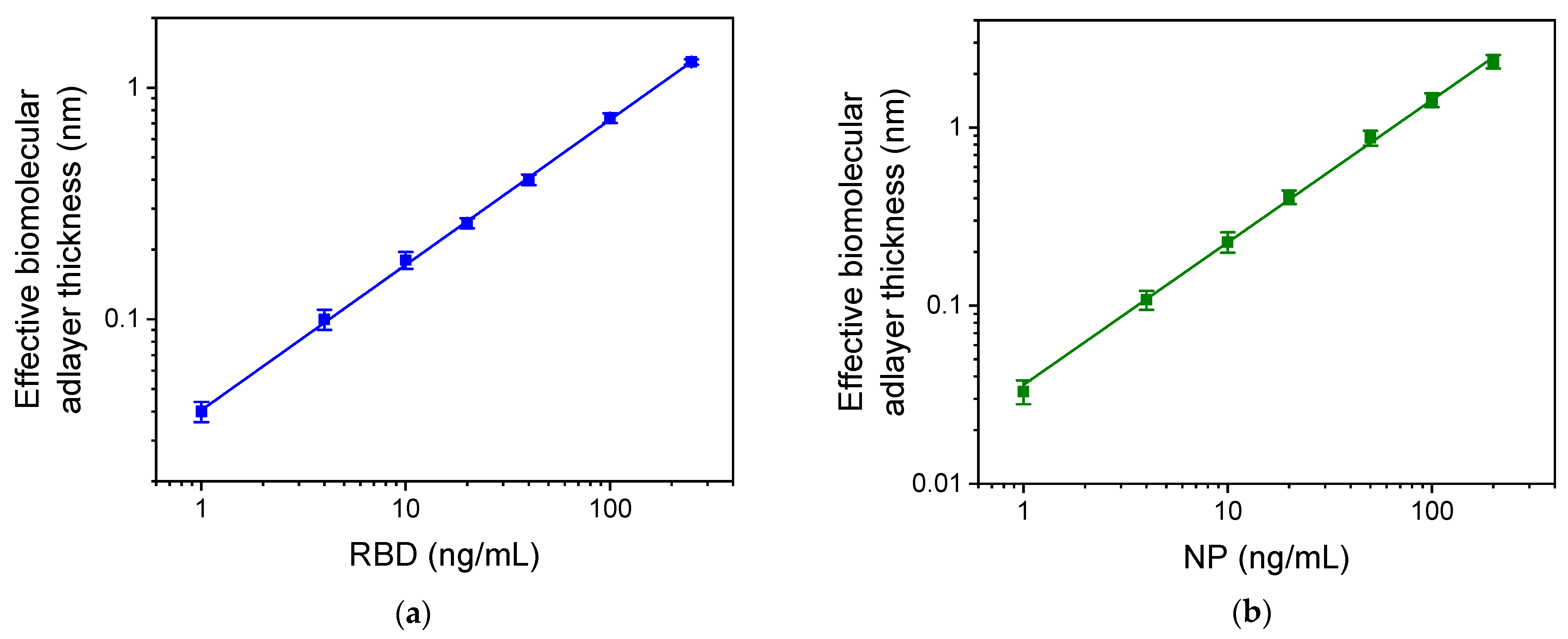
3.2. Analytical Characteristics of RBD-NP Immunosensor
3.3. Comparison with other Optical Detection Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 10 April 2023).
- Chakraborty, I.; Maity, P. COVID-19 outbreak: Migration, effects on society, global environment and prevention. Sci. Total Environ. 2020, 728, 138882. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Mattioli, I.A.; Hassan, A.; Oliveira, O.N., Jr.; Crespilho, F.N. On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sens. 2020, 5, 3655–3677. [Google Scholar] [CrossRef]
- Xu, L.; Li, D.; Ramadan, S.; Li, Y.; Klein, N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020, 170, 112673. [Google Scholar] [CrossRef]
- Suleman, S.; Shukla, S.K.; Malhotra, N.; Bukkitgar, S.D.; Shetti, N.P.; Pilloton, R.; Narang, J.; Nee Tan, Y.; Aminabhavi, T.M. Point of care detection of COVID-19: Advancement in biosensing and diagnostic methods. Chem. Eng. J. 2021, 414, 128759. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Giri, B.; Pandey, S.; Shrestha, R.; Pokharel, K.; Ligler, F.S.; Neupane, B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2021, 413, 35–48. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, 0234682. [Google Scholar] [CrossRef]
- Ge, A.; Liu, F.; Teng, X.; Cui, C.; Wu, F.; Liu, W.; Liu, Y.; Chen, X.; Xu, J.; Ma, B. A Palm Germ-Radar (PaGeR) for rapid and simple COVID-19 detection by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Biosens. Bioelectron. 2022, 200, 113925. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Eissa, S.; Ismail, A.; Zourob, M. Diagnostic techniques for COVID-19 and new developments. Talanta 2020, 220, 121392. [Google Scholar] [CrossRef]
- Ghaffari, A.; Meurant, R.; Ardakani, A. COVID-19 Point-of-Care diagnostics that satisfy global target product profiles. Diagnostics 2021, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Asghari, A.; Wang, C.; Yoo, K.M.; Rostamian, A.; Xu, X.; Shin, J.-D.; Dalir, H.; Chen, R.T. Fast, accurate, point-of-care COVID-19 pandemic diagnosis enabled through advanced lab-on-chip optical biosensors: Opportunities and challenges. Appl. Phys. Rev. 2021, 8, 031313. [Google Scholar] [CrossRef]
- He, J.; Zhu, S.; Zhou, J.; Jiang, W.; Liliang Yin, L.; Su, L.; Zhang, X.; Chen, Q.; Li, X. Rapid detection of SARS-CoV-2: The gradual boom of lateral flow immunoassay. Bioeng. Biotechnol. 2023, 10, 1090281. [Google Scholar] [CrossRef]
- Christopoulou, N.-M.; Kalogianni, D.P.; Christopoulos, T.K. Macromolecular crowding agents enhance the sensitivity of lateral flow immunoassays. Biosens. Bioelectron. 2022, 218, 114737. [Google Scholar] [CrossRef]
- Pohanka, M. Progress in Biosensors for the point-of-care diagnosis of COVID-19. Sensors 2022, 22, 7423. [Google Scholar] [CrossRef] [PubMed]
- Laghrib, F.; Saqrane, S.; El Bouabi, Y.; Farahi, A.; Bakasse, M.; Lahrich, S.; El Mhammedi, M.A. Current progress on COVID-19 related to biosensing technologies: New opportunity for detection and monitoring of viruses. Microchem. J. 2021, 160, 105606. [Google Scholar] [CrossRef]
- Drobysh, M.; Ramanaviciene, A.; Viter, R.; Chen, C.-F.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int. J. Mol. Sci. 2022, 23, 666. [Google Scholar] [CrossRef] [PubMed]
- Sadighbayan, D.; Ghafar-Zadeh, E. Portable sensing devices for detection of COVID-19: A review. IEEE Sens. J. 2021, 21, 10219–10230. [Google Scholar] [CrossRef]
- Alsalameh, S.; Alnajjar, K.; Makhzoum, T.; Al Eman, N.; Shakir, I.; Mir, T.A.; Alkattan, K.; Chinnappan, R.; Yaqinuddin, A. Advances in biosensing technologies for diagnosis of COVID-19. Biosensors 2022, 12, 898. [Google Scholar] [CrossRef]
- Flores-Contreras, E.A.; González-González, R.B.; Rodríguez-Sánchez, I.P.; Yee-de León, J.F.; Iqbal, H.M.N.; González-González, E. Microfluidics-based biosensing platforms: Emerging frontiers in point-of-care testing SARS-CoV-2 and seroprevalence. Biosensors 2022, 12, 179. [Google Scholar] [CrossRef] [PubMed]
- Zambry, N.S.; Obande, G.A.; Khalid, M.F.; Bustami, Y.; Hamzah, H.H.; Awang, M.S.; Aziah, I.; Manaf, A.A. Utilizing electrochemical-based sensing approaches for the detection of SARS-CoV-2 in clinical samples: A review. Biosensors 2022, 12, 473. [Google Scholar] [CrossRef]
- Xu, M.; Li, Y.; Lin, C.; Peng, Y.; Zhao, S.; Yang, X.; Yang, Y. Recent advances of representative optical biosensors for rapid and sensitive diagnostics of SARS-CoV-2. Biosensors 2022, 12, 862. [Google Scholar] [CrossRef]
- Zheng, Y.; Bian, S.; Sun, J.; Wen, L.; Rong, G.; Sawan, M. Label-free LSPR-vertical microcavity biosensor for on-site SARS-CoV-2 detection. Biosensors 2022, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar Khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Diao, B.; Wen, K.; Zhang, J.; Chen, J.; Han, C.; Chen, Y.; Wang, S.; Deng, G.; Zhou, H.; Wu, Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021, 27, 289.e1–289.e4. [Google Scholar] [CrossRef]
- Fabiani, L.; Saroglia, M.; Galatà, G.; De Santis, R.; Fillo, S.; Luca, V.; Faggioni, G.; D’Amore, N.; Regalbuto, E.; Salvatori, P.; et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021, 171, 112686. [Google Scholar] [CrossRef]
- Malla, P.; Liao, H.-P.; Liu, C.-H.; Wu, W.-C.; Sreearunothai, P. Voltammetric biosensor for coronavirus spike protein using magnetic bead and screen-printed electrode for point-of-care diagnostics. Microchim. Acta 2022, 189, 168. [Google Scholar] [CrossRef]
- Tao, Y.; Bian, S.; Wang, P.; Zhang, H.; Bi, W.; Zhu, P.; Sawan, M. Rapid optical biosensing of SARS-CoV-2 spike proteins in artificial samples. Sensors 2022, 22, 3768. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, W.; Chen, F.; Ren, P. Highly sensitive and selective surface plasmon resonance biosensor for the detection of SARS-CoV-2 spike S1 protein. Analyst 2022, 147, 2809–2818. [Google Scholar] [CrossRef]
- Kawasaki, D.; Yamada, H.; Sueyoshi, K.; Hisamoto, H.; Endo, T. Imprinted photonic crystal-film-based smartphone-compatible label-free optical sensor for SARS-CoV-2 testing. Biosensors 2022, 12, 200. [Google Scholar] [CrossRef]
- Agarwal, D.K.; Nandwana, V.; Henrich, S.E.; Josyula, V.P.V.N.; Thaxton, C.S.; Qi, C.; Simons, L.M.; Hultquist, J.F.; Ozer, E.A.; Shekhawat, G.S.; et al. Highly sensitive and ultra-rapid antigen-based detection of SARS-CoV-2 using nanomechanical sensor platform. Biosens. Bioelectron. 2022, 195, 113647. [Google Scholar] [CrossRef] [PubMed]
- Proll, G.; Markovic, G.; Fechner, P.; Proell, F.; Gauglitz, G. Reflectometric Interference Spectroscopy. In Biosensors and Biodetection; Methods in Molecular, Biology; Rasooly, A., Prickril, B., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1571, pp. 207–220. [Google Scholar]
- Fechner, P.; Gauglitz, G.; Proll, G. Through the looking-glass—Recent developments in reflectometry open new possibilities for biosensor applications. TrAC Trend Anal. Chem. 2022, 156, 116708. [Google Scholar] [CrossRef]
- Anastasiadis, V.; Koukouvinos, G.; Petrou, P.S.; Economou, A.; Dekker, J.; Harjanne, M.; Heimala, P.; Goustouridis, D.; Raptis, I.; Kakabakos, S.E. Multiplexed mycotoxins determination employing white light reflectance spectroscopy and silicon chips with silicon oxide areas of different thickness. Biosens. Bioelectron. 2020, 153, 112035. [Google Scholar] [CrossRef] [PubMed]
- Tsounidi, D.; Tsaousis, V.; Xenos, N.; Kroupis, C.; Moutsatsou, P.; Christianidis, V.; Goustouridis, D.; Raptis, I.; Kakabakos, S.; Petrou, P. Simultaneous determination of procalcitonin and interleukin-6 in human serum samples with a point-of-care biosensing device. Talanta 2023, 258, 124403. [Google Scholar] [CrossRef]
- Tsounidi, D.; Koukouvinos, G.; Petrou, P.; Misiakos, K.; Zisis, G.; Goustouridis, D.; Raptis, I.; Kakabakos, S.E. Rapid and sensitive label-free determination of aflatoxin M1 levels in milk through a White Light Reflectance Spectroscopy immunosensor. Sens. Actuators B Chem. 2019, 282, 104–111. [Google Scholar] [CrossRef]
- Cennamo, N.; Pasquardini, L.; Arcadio, F.; Lunelli, L.; Vanzetti, L.; Carafa, V.; Altucci, L.; Zeni, L. SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor. Talanta 2021, 233, 122532. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-L.; Kim, J.; Choi, S.; Han, J.; Seo, G.; Lee, Y.W. Fiber-optic label-free biosensor for SARS-CoV-2 spike protein detection using biofunctionalized long-period fiber grating. Talanta 2021, 235, 122801. [Google Scholar] [CrossRef]
- Xu, W.; Zhuo, Y.; Song, D.; Han, X.; Xu, J.; Long, F. Development of a novel label-free all-fiber optofluidic biosensor based on Fresnel reflection and its applications. Anal. Chim. Acta 2021, 1181, 338910. [Google Scholar] [CrossRef] [PubMed]
- Janik, M.; Gabler, T.; Koba, M.; Panasiuk, M.; Dashkevich, Y.; Łęga, T.; Dąbrowska, A.; Naskalska, A.; Żołędowska, S.; Nidzworski, D.; et al. Low-volume label-free SARS-CoV-2 detection with the microcavity-based optical fiber sensor. Sci. Rep. 2023, 13, 1512. [Google Scholar] [CrossRef] [PubMed]





| Sensing Principle | Analyte | Recognition Molecule | Label | Dynamic Range | Analysis Time (min) | Ref. No |
|---|---|---|---|---|---|---|
| Fiber-optic biolayer interferometry (FO-BLI) | RBD | antibody | 3-amino-9-ethylcarbazole | 0.5–16 fg/mL | 13 | [34] |
| 2D MXene-based SPR | RBD | antibody | PDA-AgNPs nanohybrids | 0.1 pg/mL–1 μg/mL | >60 | [35] |
| SPR D-shaped plastic optical fiber (POF) | RBD | aptamer | no | 1–40 pg/mL | >10 | [43] |
| Imprinted photonic crystal | RBD | antibody | no | 1 pg/mL–100 ng/mL | <60 | [36] |
| Phase-shifted long-period fiber grating (PS-LPFG) | RBD | antibody | no | 1 pg/mL–100 μg/mL | >20 | [44] |
| All-fiber optofluidic biosensor (LF-AOB) | RBD | antibody | no | 0.01–100 ng/mL | <10 | [45] |
| Microcantilever | NP | antibody | no | 1 ng/mL–1 μg/mL | <5 | [37] |
| Μicrocavity in-line Mach–Zehnder interferometer (μIMZI) | NP | antibody | no | 3–300 ng/mL | 30 | [46] |
| MARS | RBD NP | antibody | no | 20 ng/mL–1 μg/mL | 12 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsounidi, D.; Angelopoulou, M.; Petrou, P.; Raptis, I.; Kakabakos, S. Simultaneous Detection of SARS-CoV-2 Nucleoprotein and Receptor Binding Domain by a Multi-Area Reflectance Spectroscopy Sensor. Biosensors 2023, 13, 865. https://doi.org/10.3390/bios13090865
Tsounidi D, Angelopoulou M, Petrou P, Raptis I, Kakabakos S. Simultaneous Detection of SARS-CoV-2 Nucleoprotein and Receptor Binding Domain by a Multi-Area Reflectance Spectroscopy Sensor. Biosensors. 2023; 13(9):865. https://doi.org/10.3390/bios13090865
Chicago/Turabian StyleTsounidi, Dimitra, Michailia Angelopoulou, Panagiota Petrou, Ioannis Raptis, and Sotirios Kakabakos. 2023. "Simultaneous Detection of SARS-CoV-2 Nucleoprotein and Receptor Binding Domain by a Multi-Area Reflectance Spectroscopy Sensor" Biosensors 13, no. 9: 865. https://doi.org/10.3390/bios13090865
APA StyleTsounidi, D., Angelopoulou, M., Petrou, P., Raptis, I., & Kakabakos, S. (2023). Simultaneous Detection of SARS-CoV-2 Nucleoprotein and Receptor Binding Domain by a Multi-Area Reflectance Spectroscopy Sensor. Biosensors, 13(9), 865. https://doi.org/10.3390/bios13090865








