Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives
Abstract
1. Introduction
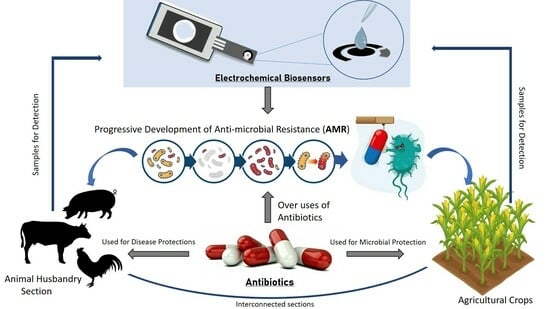
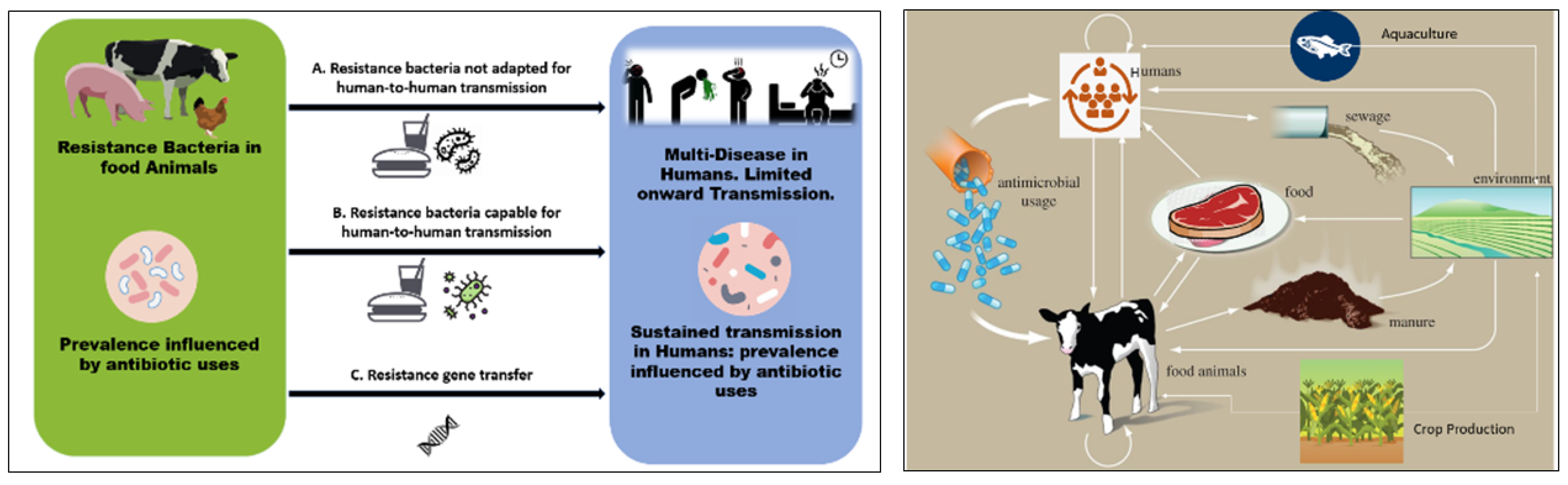
2. Antibiotic Use and AMR Development
- Develop and implement on-farm antibiotic stewardship programs: On-farm antibiotic stewardship programs could be developed and implemented to promote the appropriate use of antibiotics in food-producing animals. These programs could involve education and training for farmers and veterinarians, guidelines for antibiotic use, monitoring of and feedback regarding antibiotic use, and the use of alternative approaches to disease prevention, such as improved animal husbandry practices and vaccination.
- Implement regulations to restrict use of antibiotics for growth promotion: Regulations could be put in place to prohibit the use of antibiotics for growth promotion and other non-therapeutic purposes in food-producing animals. This could help to reduce the overall use of antibiotics in animals and agriculture and prevent the development of AMR. Pharmacists and veterinary officers should adhere strictly to the regulations and policies governing prescriptions. Policymakers should enact strict controls and regulations to ensure that antibiotics used in farming are only purchased legally (legitimately). Antibiotic usage in animals should be decreased and limited by utilising plant-derived extracts and probiotics/prebiotics for disease prevention and treatment.
- A reduction in the antibiotics required via improvements in animal health and welfare: Efforts could be made to improve animal health and welfare through better housing, nutrition, and disease prevention measures. This could help in reducing the need for the use of antibiotics in food-producing animals. By implementing such measures, it would be possible to control use of antibiotics in agricultural animals and reduce the development of antibiotic resistance. The government and financially stable companies should encourage and support the employment of regular, efficient and appropriate veterinarian services.
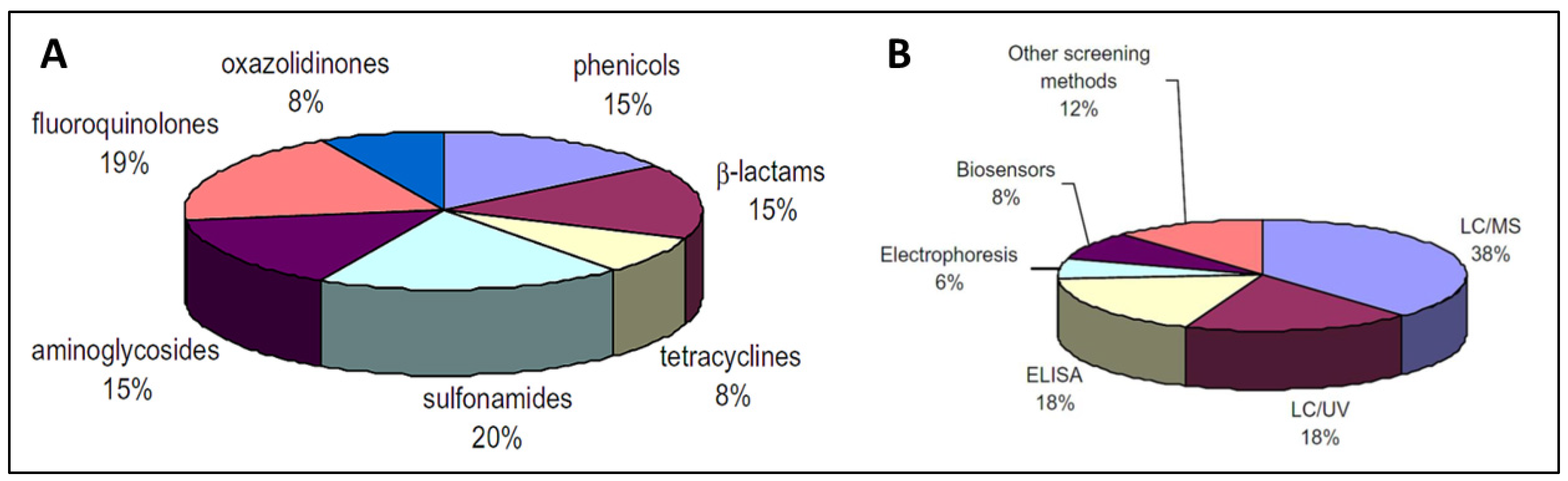
3. Detection Methods (Antibiotics)
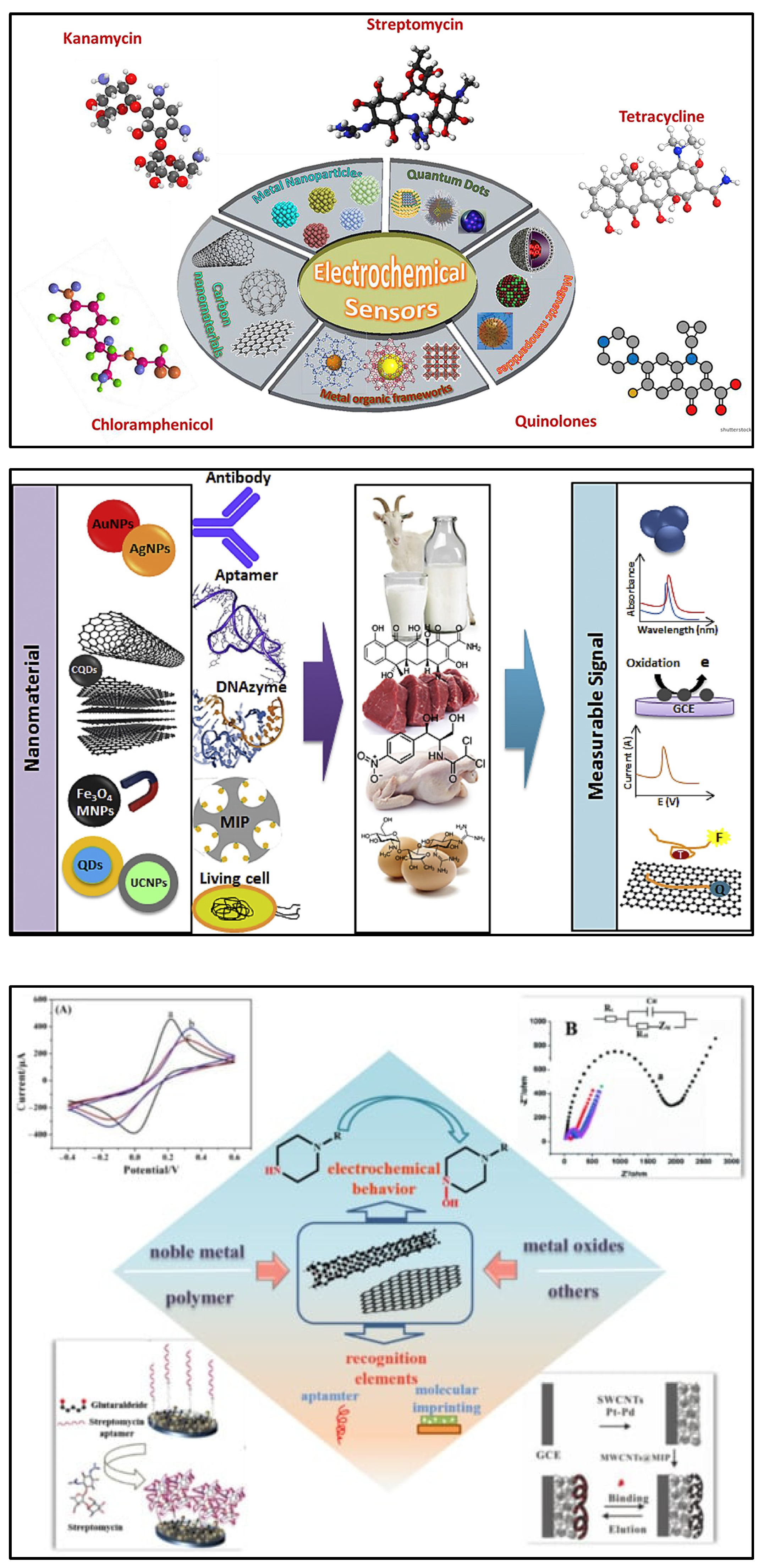
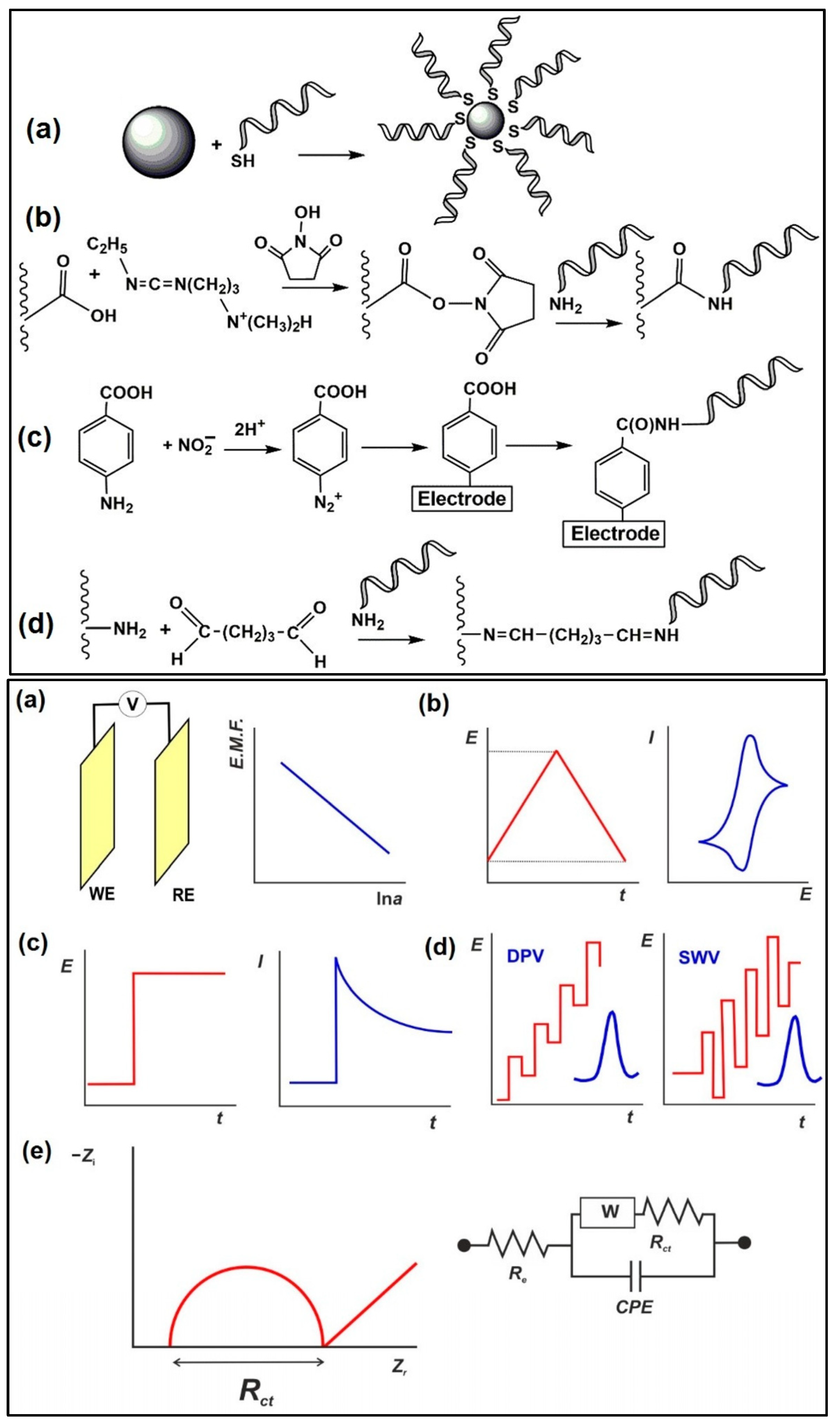
4. Electrochemical Instrumentation and Nanomaterials
5. Electrochemical Biosensors
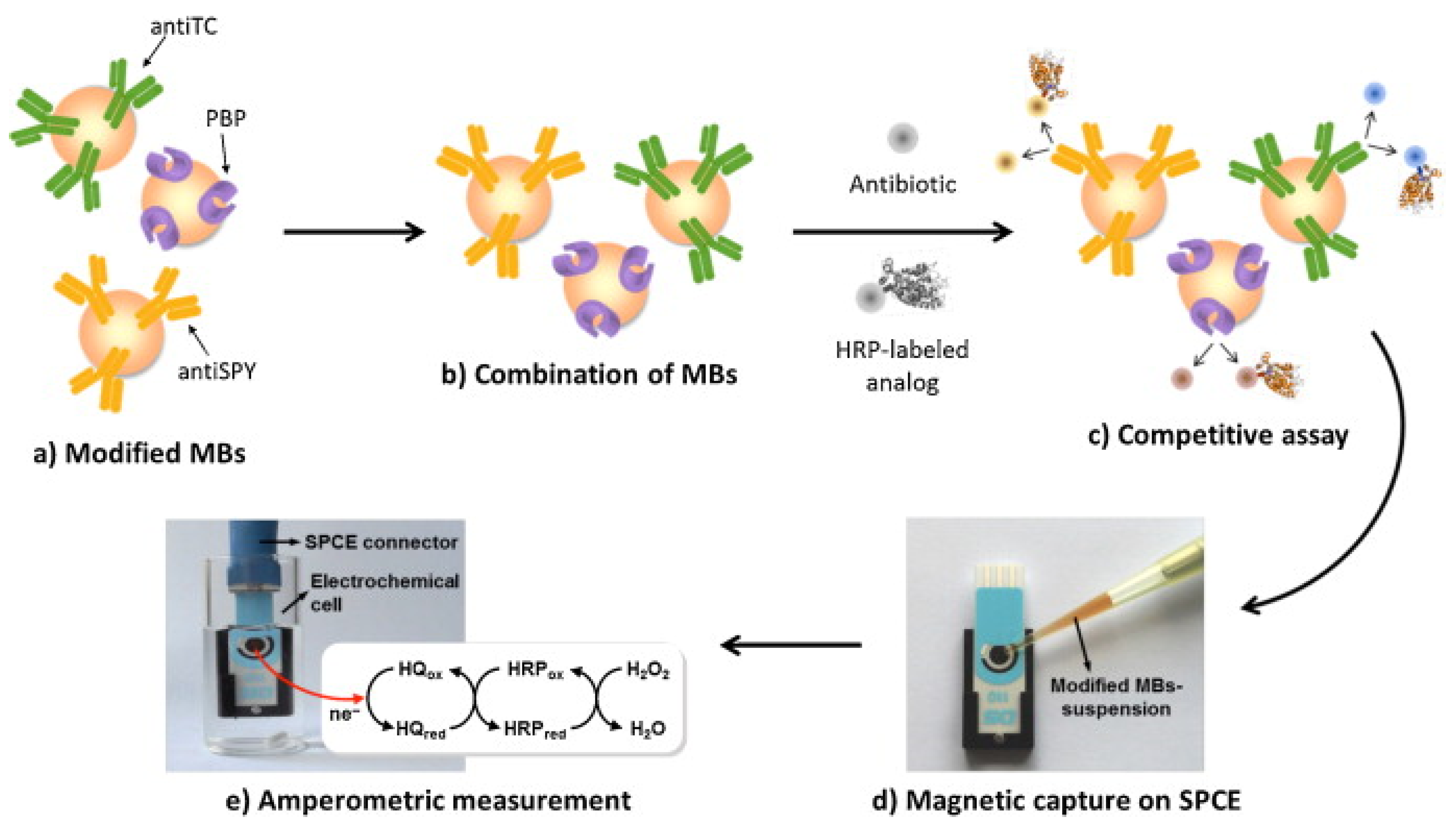
5.1. Electrochemical Immunosensors
| Antibiotics | Bio Recognition Component | Working Electrode | Detection Method/Technique | Linear/Dynamic Range (ppb) | LOD (ppb) | Label | Sample Type | Ref. |
|---|---|---|---|---|---|---|---|---|
| Sulfapyridine | Polyclonal antiserum As167 | Ab covalently immobilized on 4-ABA-modified SPCEs | Amperometry | 1.6 to 118.6 | 0.44 | HRP | Dilutedwhole milk | [97] |
| Sulfamethoxazole | anti-sulfamethoxazole polyclonal antibody | antiSMX/nanoCeO2—CS/GCE | DPV | 0.5 to 500 | 0.325 | HRP | Buffer/ food samples | [101] |
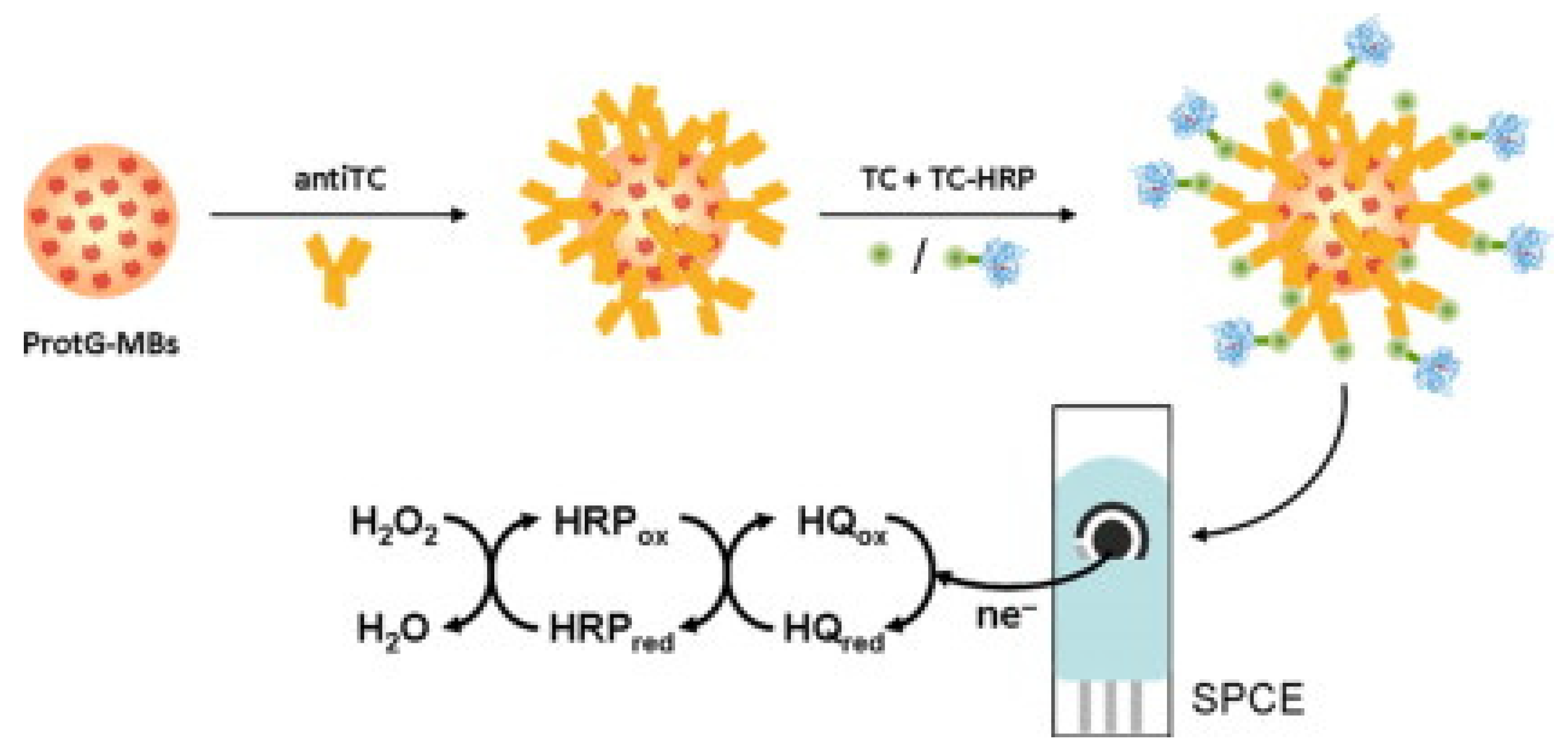
| Tetracyclines | Polyclonal sheep antiTC antibody. Competitive immunoassay using TC-HRP on antiTC-modified MBs | ProtG-MBs /SPCE/H2O2 in the presence of HQ | Amperometry | 5.0 to 202.5 | 1.9 | HRP | Undiluted milk | [95] |
| Cefquinome | BlaR-CTD | GO/TH/GCE | CV & EIS | 0.1 to 8 | 0.16 | HRP-AMP | PBS/milk | [102] |
| Ampicillin | Electrochemical immunosensor | BSA/anti-AMP/APTES/nMoS2/ITOimmunoelectrode | DPV | 32.5 to 64 × 103 | 28 | Label-free | PBS/spiked milk, orange juice & tap water | [99] |
| Ciprofloxacin | 11-BSA-MB, 11-HRP and Ab171-MB | Amperometric magneto-immunosensor (AMIS) | Amperometry | 0.043 to 7.38 | 0.009 | HRP | Whole milk | [103] |
| Enrofloxacin | Electrochemical immunosensor | rGO-TEPA/SPE | DPV | 0.1 to 1000 | 1.897 × 10−2 | Dendritic mesoporous Au@Pt nano-probe | PBS/spiked milk samples | [98] |
| Chloramphenicol | Antibody/immunosensor | PVA-co-PE NFM/Anti-CAP/SPCE | Amperometry | 0.01–10 | 0.0047 | label-free | PBS/spiked milk | [96] |
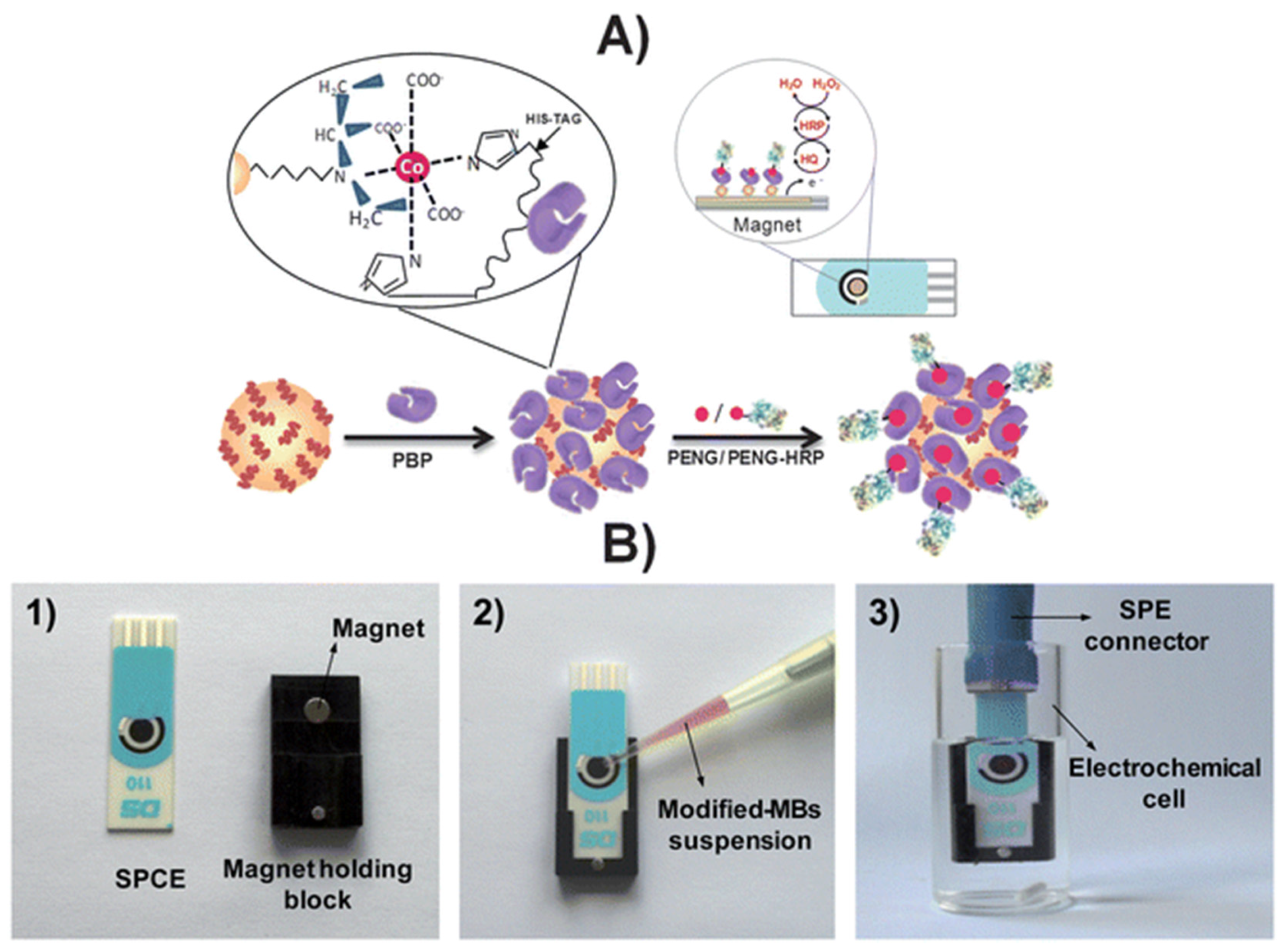
| Penicillin G | Antibody/immunosensor | AP/gold/s-BLM/GC | EIS | 3.34 × 10−6 to 3.34 | 2.7 × 10−7 | gold/s-BLM | Diluted milk | [104] |
5.2. Aptamer-Based Electrochemical Biosensors
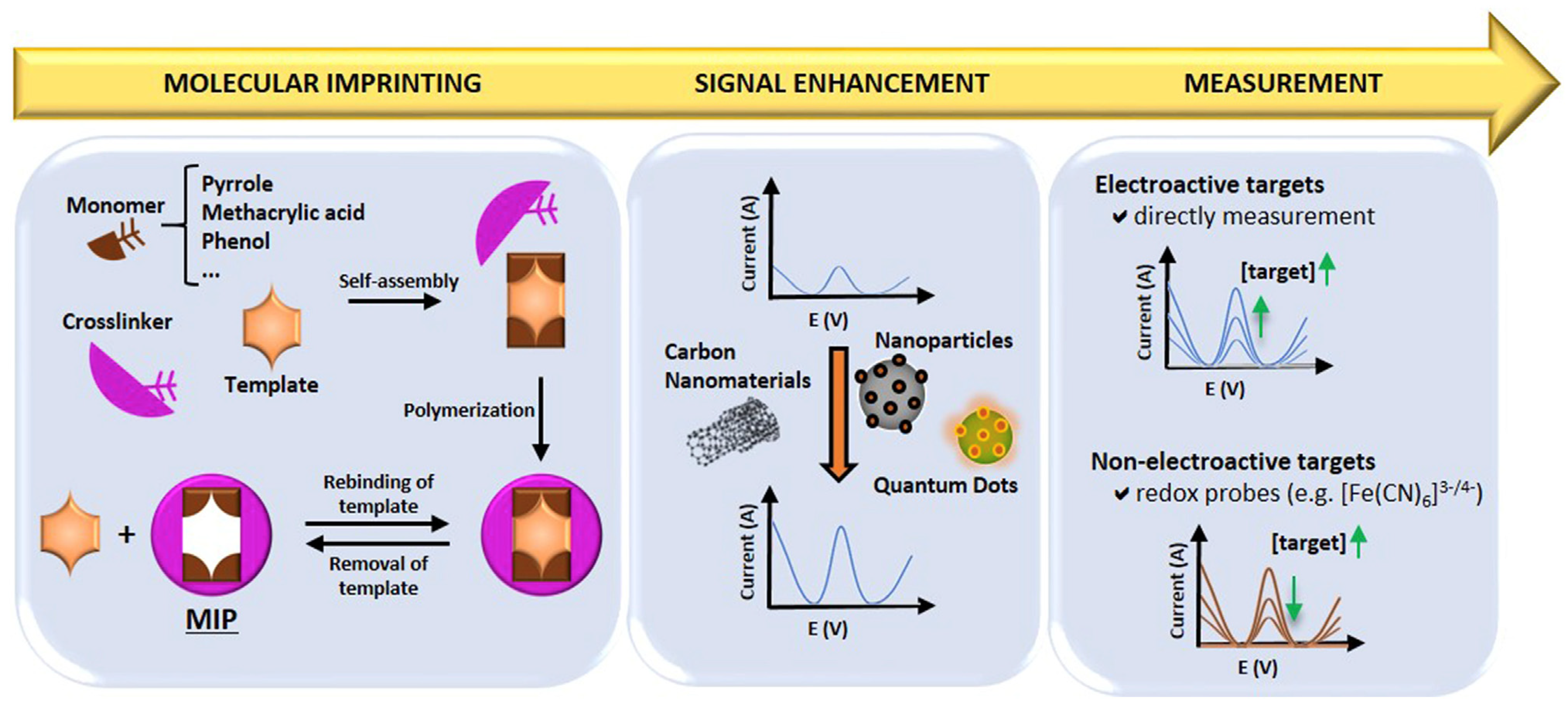
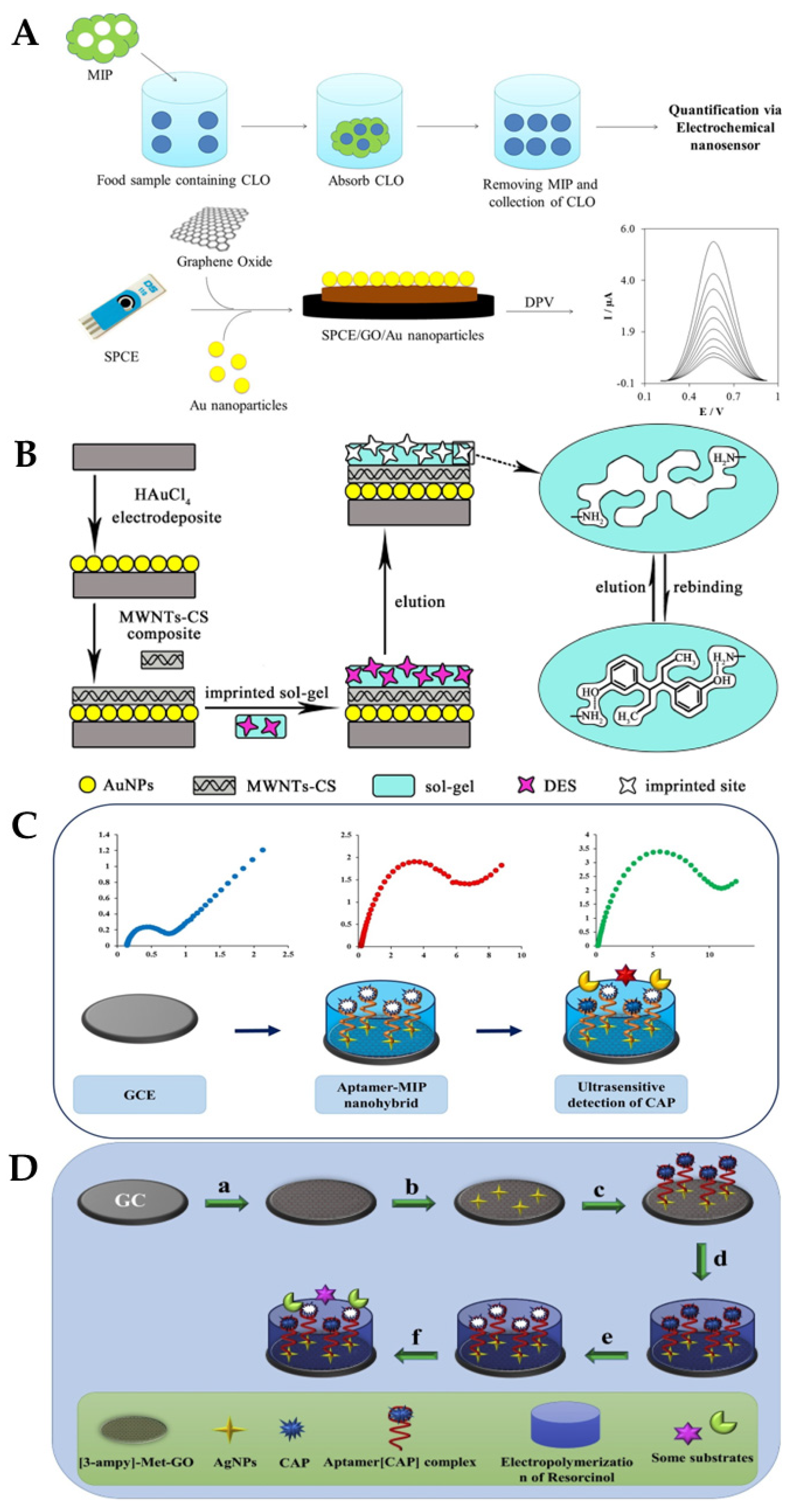
5.3. Molecularly Imprinted Polymer (MIP)-Based Electrochemical Biosensors
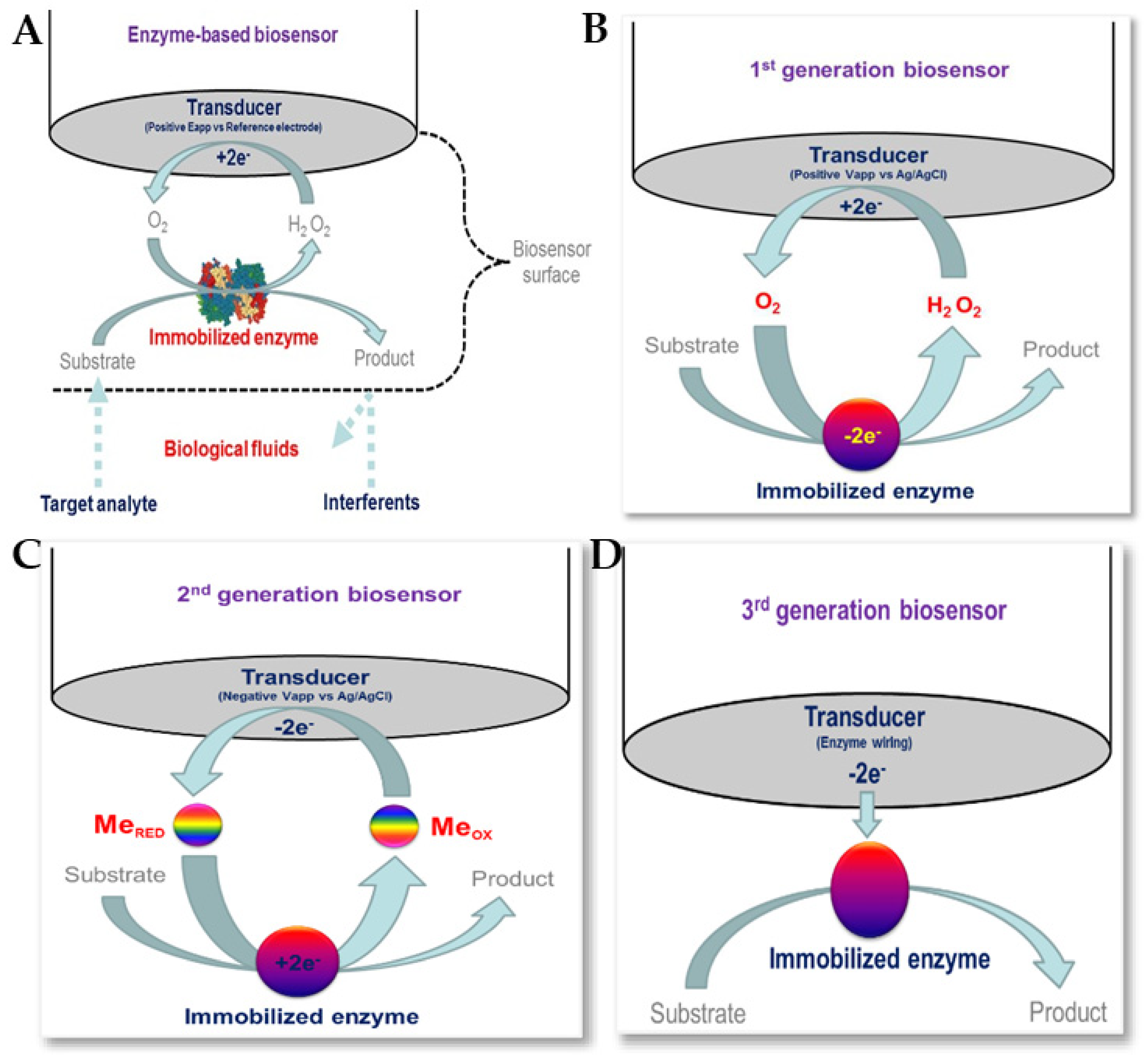
5.4. Enzyme-Based Electrochemical Biosensors
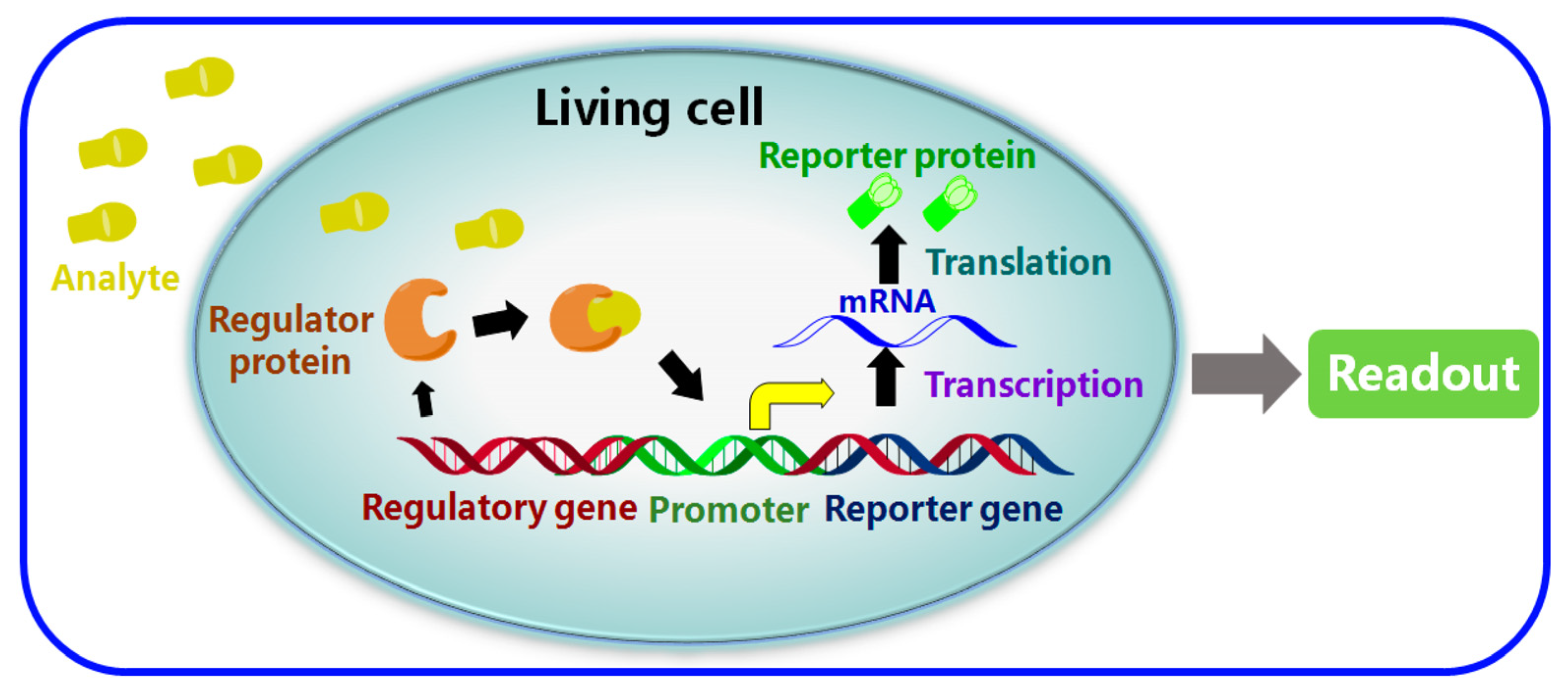
5.5. Whole-Cell-Based Electrochemical Biosensors (WCBs)
5.6. Direct Electrochemistry-Based Biosensors
6. Challenges and Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WHO: | World Health Organization |
| EMA: | European Medicines Agency |
| AMR: | Antimicrobial resistance |
| CIAs: | Critically important antibiotics |
| MRL: | Maximum residue limit |
| LOD: | Limit of detection |
| RSD: | Relative standard deviation |
| SPR: | Surface-plasmon resonance |
| PEN: | Penicillin |
| PCN: | Penicillinase |
| PENG: | Penicillin G |
| PBP: | Penicillin-binding protein |
| CAP: | Chloramphenicol |
| CLO/CLOX: | Cloxacillin |
| TOB: | Tobramycin |
| KAN: | Kanamycin |
| AMP/AMPI: | Ampicillin |
| AMOX: | Amoxicillin |
| CEF: | Cefapirin |
| OXA: | Oxacillin |
| TCs: | Tetracyclines |
| Qs: | Quinolones |
| DES: | Diethylstilbestrol |
| HRP: | Horseradish peroxidase |
| HRP-AMP: | Horseradish peroxidase-labelled ampicillin |
| GOx: | Glucose oxidase |
| GCE: | Glassy carbon electrode |
| SPE: | Screen-printed electrode |
| SPCEs: | Screen-printed carbon electrodes |
| SPGEs: | Screen-printed gold electrodes |
| m-GEC: | Magnetic graphite–epoxy composite |
| WE: | Working electrode |
| RE: | Reference electrode |
| CV: | Cyclic voltammetry |
| DPV: | Differential pulse voltammetry |
| SWV: | Square-wave voltammetry |
| EIS: | Electrochemical impedance spectroscopy |
| MIP: | Molecularly imprinted polymer |
| rGO: | Reduced graphene oxide |
| NFT: | Nitrofurantoin |
| AuNPs: | Gold nanoparticles |
| MWCNTs: | Multi-walled carbon nanotubes |
| SGCs: | Single-graphene nanosheets |
| MNP: | Magnetic nanoparticles |
| MoS2: | Molybdenum disulfide |
| MOF: | Metal–organic framework |
| SAM: | Self-assembled monolayer |
| WCBs: | Whole cell-based biosensor |
| HPLC: | High performance liquid chromatography |
| LC-MS: | Liquid chromatography-mass spectroscopy |
| GC: | Gas chromatography-mass spectroscopy |
| HQ: | Hydroquinone |
| DFR: | Dry film photoresist |
| QDs: | Quantum dots |
| MBs: | Magnetic beads |
| MB: | Methylene blue |
| ELAs: | Enzyme-linked assays |
| ELISA: | Enzyme-linked immunosorbent assay |
| EDC: | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| PPy: | Polypyrrole |
| PAMAM: | Poly(amidoamine) |
| SM2: | Sulfadimidine |
| ppm: | Parts per million |
| ppb: | Parts per billion |
| ppt: | Parts per trillion |
| Rct: | Charge transfer resistance |
References
- Sharma, A.; Istamboulie, G.; Hayat, A.; Catanante, G.; Bhand, S.; Marty, J.L. Disposable and portable aptamer functionalized impedimetric sensor for detection of kanamycin residue in milk sample. Sens. Actuators B Chem. 2017, 245, 507–515. [Google Scholar]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar]
- You, Y.; Silbergeld, E.K. Learning from agriculture: Understanding low-dose antimicrobials as drivers of resistome expansion. Front. Microbiol. 2014, 5, 284. [Google Scholar] [PubMed]
- Hong, P.-Y.; Al-Jassim, N.; Ansari, M.I.; Mackie, R.I. Environmental and public health implications of water reuse: Antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes. Antibiotics 2013, 2, 367–399. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [PubMed]
- Ngangom, B.L.; Tamunjoh, S.S.A.; Boyom, F.F. Antibiotic residues in food animals: Public health concern. Acta Ecol. Sin. 2019, 39, 411–415. [Google Scholar]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Bester, L.A.; Essack, S.Y. Observational study of the prevalence and antibiotic resistance of Campylobacter spp. from different poultry production systems in KwaZulu-Natal, South Africa. J. Food Prot. 2012, 75, 154–159. [Google Scholar] [CrossRef]
- Maron, D.F.; Smith, T.J.; Nachman, K.E. Restrictions on antimicrobial use in food animal production: An international regulatory and economic survey. Glob. Health 2013, 9, 1–11. [Google Scholar]
- Islam, K.S.; Shiraj-Um-Mahmuda, S.; Hazzaz-Bin-Kabir, M. Antibiotic usage patterns in selected broiler farms of Bangladesh and their public health implications. J. Public Health Dev. Ctries. 2016, 2, 276–284. [Google Scholar]
- Kaplan, J.; Bowman, M.; Cordova, C.; Kar, A. Pharming Chickens: It’s Time For The U.S. Poultry Industry to Demonstrate Antibiotic Stewardship. Natural Resources Defence Council: New York, NY, USA 2014. Available online: https://www.nrdc.org/sites/default/files/poultry-industry-antibiotic-stewardship-IB.pdf (accessed on 3 August 2022).
- Cogliani, C.; Goossens, H.; Greko, C. Restricting antimicrobial use in food animals: Lessons from Europe. Microbe 2011, 6, 274. [Google Scholar] [CrossRef]
- Bashahun, D.; Odoch, T. Assessment of antibiotic usage in intensive poultry farms in Wakiso District, Uganda. Livest. Res. Rural Dev. 2015, 27, 247. [Google Scholar]
- Mann, A.; Nehra, K.; Rana, J.; Dahiya, T. Antibiotic resistance in agriculture: Perspectives on upcoming strategies to overcome upsurge in resistance. Curr. Res. Microb. Sci. 2021, 2, 100030. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.P.; Saegerman, C.; Douny, C.; Dinh, T.V.; Xuan, B.H.; Vu, B.D.; Hong, N.P.; Scippo, M.-L. First survey on the use of antibiotics in pig and poultry production in the Red River Delta region of Vietnam. Food Public Health 2013, 3, 247–256. [Google Scholar]
- Braykov, N.P.; Eisenberg, J.N.; Grossman, M.; Zhang, L.; Vasco, K.; Cevallos, W.; Muñoz, D.; Acevedo, A.; Moser, K.A.; Marrs, C.F. Antibiotic resistance in animal and environmental samples associated with small-scale poultry farming in northwestern Ecuador. Msphere 2016, 1, e00021-15. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J. Antibiotic Types and Handling Practices in Disease Management among Pig Farms in Ashanti Region, Ghana. J. Vet. Med. 2014, 2014, 531952. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, C.I.; Joo, E.J.; Ha, Y.E.; Cho, S.Y.; Gwak, G.Y.; Chung, D.R.; Peck, K.R.; Song, J.H. Risk factor of community-onset spontaneous bacterial peritonitis caused by fluoroquinolone-resistant Escherichia coli in patients with cirrhosis. Liver Int. 2014, 34, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Adebowale, O.O.; Adeyemo, O.K.; Awoyomi, O.; Dada, R.; Adebowale, O. Antibiotic use and practices in commercial poultry laying hens in Ogun State Nigeria. Rev. D’élevage Méd. Vét. Pays Trop. 2016, 69, 41–45. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Sachi, S.; Ferdous, J.; Sikder, M.H.; Hussani, S.A.K. Antibiotic residues in milk: Past, present, and future. J. Adv. Vet. Anim. Res. 2019, 6, 315. [Google Scholar] [CrossRef]
- Edqvist, L.-E.; Pedersen, K.B. Antimicrobials as growth promoters: Resistance to common sense. In The Precautionary Principle in the 20th Century; Routledge: London, UK, 2013; Chapter 9; pp. 100–110. [Google Scholar]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7. [Google Scholar] [CrossRef]
- Resistance, R.O.A. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014; Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 3 August 2022).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015, ISBN: 9789241509763. Available online: https://www.who.int/publications/i/item/9789241509763 (accessed on 3 August 2022).
- World Health Organization. Critically Important Antimicrobials for Human Medicine. 2019, ISBN: 9789241515528. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 3 August 2022).
- EMA. Categorisation of Antibiotics in the European Union. 2019. Available online: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf (accessed on 16 February 2023).
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Spec. J. 2019, 4, e002104. [Google Scholar] [CrossRef] [PubMed]
- FDA-US. Antimicrobials Sold or Distributed for Use in Food-Producing Animals. 2017. Available online: https://www.fda.gov/media/119332 (accessed on 2 September 2022).
- Agency, E.M. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017. 2017. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2017_en.pdf (accessed on 3 August 2022).
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 3 August 2022).
- Carrique-Mas, J.J.; Choisy, M.; Van Cuong, N.; Thwaites, G.; Baker, S. An estimation of total antimicrobial usage in humans and animals in Vietnam. Antimicrob. Resist. Infect. Control 2020, 9, 1–6. [Google Scholar] [CrossRef]
- Uberoi, E. UK Dairy Industry Statistics. 2021. Available online: https://researchbriefings.files.parliament.uk/documents/SN02721/SN02721.pdf (accessed on 10 February 2023).
- DFAM-Ireland. Department of Agriculture, Food and the Marine Annual Review and Outlook Published for 2022. 2022. Available online: https://www.gov.ie/en/press-release/c32cb-department-of-agriculture-food-and-the-marine-annual-review-and-outlook-published-for-2022/ (accessed on 10 February 2023).
- Bacanlı, M.; Başaran, N. Importance of antibiotic residues in animal food. Food Chem. Toxicol. 2019, 125, 462–466. [Google Scholar] [CrossRef]
- Majdinasab, M.; Yaqub, M.; Rahim, A.; Catanante, G.; Hayat, A.; Marty, J.L. An overview on recent progress in electrochemical biosensors for antimicrobial drug residues in animal-derived food. Sensors 2017, 17, 1947. [Google Scholar] [CrossRef]
- Leibovici, L.; Paul, M.; Garner, P.; Sinclair, D.J.; Afshari, A.; Pace, N.L.; Cullum, N.; Williams, H.C.; Smyth, A.; Skoetz, N. Addressing resistance to antibiotics in systematic reviews of antibiotic interventions. J. Antimicrob. Chemother. 2016, 71, 2367–2369. [Google Scholar] [CrossRef]
- Kivirand, K.; Kagan, M.; Rinken, T. Biosensors for the detection of antibiotic residues in milk. Biosens. Micro Nanoscale Appl. 2015, 425–456. [Google Scholar] [CrossRef]
- Commission Regulation (EU) No. 37/2010. 2010. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2010:015:0001:0072:EN:PDF (accessed on 9 February 2023).
- EMEA. Commitee for Veterinary Medicinal Products-Note for Guidance for the Determination of Withdrawal Periods for Milk. 2000. EMEA/CVMP/473/98-FINAL. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/note-guidance-determination-withdrawal-periods-milk_en.pdf (accessed on 9 February 2023).
- Albright, J.; Tuckey, S.; Woods, G. Antibiotics in milk—A review. J. Dairy Sci. 1961, 44, 779–807. [Google Scholar] [CrossRef]
- Chen, T.; Cheng, G.; Ahmed, S.; Wang, Y.; Wang, X.; Hao, H.; Yuan, Z. New methodologies in screening of antibiotic residues in animal-derived foods: Biosensors. Talanta 2017, 175, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, V. Advances in biosensor development for the screening of antibiotic residues in food products of animal origin–A comprehensive review. Biosens. Bioelectron. 2017, 90, 363–377. [Google Scholar] [CrossRef]
- Chiesa, L.M.; Nobile, M.; Panseri, S.; Arioli, F. Antibiotic use in heavy pigs: Comparison between urine and muscle samples from food chain animals analysed by HPLC-MS/MS. Food Chem. 2017, 235, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hlabangana, L.; Memeza, S. Ion-pair isocratic simultaneous determination of broad spectrum antibiotics in environmental samples by HPLC with UV detection. Environ. Nanotechnol. Monit. Manag. 2018, 10, 104–111. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef]
- Pateraki, M.; Fysarakis, K.; Sakkalis, V.; Spanoudakis, G.; Varlamis, I.; Maniadakis, M.; Lourakis, M.; Ioannidis, S.; Cummins, N.; Schuller, B. Biosensors and Internet of Things in smart healthcare applications: Challenges and opportunities. Wearable Implant. Med. Devices 2020, 7, 25–53. [Google Scholar] [CrossRef]
- Planta, M.B. The role of poverty in antimicrobial resistance. J. Am. Board Fam. Med. 2007, 20, 533–539. [Google Scholar] [CrossRef]
- Doron, S.; Davidson, L.E. Antimicrobial stewardship. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1113–1123. [Google Scholar]
- Gyssens, I.C. Antibiotic policy. Int. J. Antimicrob. Agents 2011, 38, 11–20. [Google Scholar] [CrossRef]
- Jamison, D.T.; Breman, J.G.; Measham, A.R.; Alleyne, G.; Claeson, M.; Evans, D.B.; Jha, P.; Mills, A.; Musgrove, P. Disease Control Priorities in Developing Countries. 2006. Available online: https://www.google.com/books?hl=zh-CN&lr=&id=Ds93H98Z6D0C&oi=fnd&pg=PR7&dq=Disease+control+priorities+in+developing+countries&ots=rjF0-P4Gg5&sig=bJdDUJBKzs1HT68_PL6VtNuV6SE (accessed on 24 August 2022).
- Kakkar, M.; Sharma, A.; Vong, S. Developing a situation analysis tool to assess containment of antimicrobial resistance in South East Asia. BMJ 2017, 358, j3760. [Google Scholar] [CrossRef]
- Adhikari, B.; Pokharel, S.; Mishra, S.R. Shrinking urban Greenspace and the rise in non-communicable diseases in South Asia: An urgent need for an advocacy. Front. Sustain. Cities 2019, 1, 5. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Dey, S.; Gupta, T.; Dhaliwal, R.; Brauer, M.; Cohen, A.J.; Stanaway, J.D.; Beig, G.; Joshi, T.K.; Aggarwal, A.N. The impact of air pollution on deaths, disease burden, and life expectancy across the states of India: The Global Burden of Disease Study 2017. Lancet Planet. Health 2019, 3, e26–e39. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; Van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef] [PubMed]
- Acar, J.; Moulin, G. Antimicrobial resistance at farm level. Rev. Sci. Tech. 2006, 25, 775–792. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Moutafis, G.; Tran, L.T.; Coloe, P.J. Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl. Environ. Microbiol. 2007, 73, 7906–7911. [Google Scholar] [CrossRef]
- Wegener, H.C. A15 Antibiotic resistance—Linking human and animal health. In Improving Food Safety through a One Health Approach: Workshop Summary; Choffnes, E.R., Relman, D.A., Olsen, L., Hutton, R., Eds.; National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Franklin, A.M.; Aga, D.S.; Cytryn, E.; Durso, L.M.; McLain, J.E.; Pruden, A.; Roberts, M.C.; Rothrock, M.J., Jr.; Snow, D.D.; Watson, J.E. Antibiotics in agroecosystems: Introduction to the special section. J. Environ. Qual. 2016, 45, 377–393. [Google Scholar] [CrossRef]
- Amin, F.R.; Khalid, H.; Zhang, H.; Zhang, R.; Liu, G.; Chen, C. Pretreatment methods of lignocellulosic biomass for anaerobic digestion. Amb Express 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Moyane, J.N.; Jideani, A.I.O.; Aiyegoro, O.A. Antibiotics usage in food-producing animals in South Africa and impact on human: Antibiotic resistance. Afr. J. Microbiol. Res. 2013, 7, 2990–2997. [Google Scholar]
- Cháfer-Pericás, C.; Maquieira, A.; Puchades, R. Fast screening methods to detect antibiotic residues in food samples. TrAC Trends Anal. Chem. 2010, 29, 1038–1049. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, Q.; Chen, T.; Li, J.; Liu, Y.; Shan, X.; Liu, F.; Jia, J. Recent advances in electrochemical sensors for antibiotics and their applications. Chin. Chem. Lett. 2021, 32, 609–619. [Google Scholar] [CrossRef]
- Lopes, L.C.; Santos, A.; Bueno, P.R. An outlook on electrochemical approaches for molecular diagnostics assays and discussions on the limitations of miniaturized technologies for point-of-care devices. Sens. Actuators Rep. 2022, 4, 100087. [Google Scholar] [CrossRef]
- Raykova, M.R.; Corrigan, D.K.; Holdsworth, M.; Henriquez, F.L.; Ward, A.C. Emerging electrochemical sensors for real-time detection of tetracyclines in milk. Biosensors 2021, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Fredj, Z.; Singh, B.; Bahri, M.; Qin, P.; Sawan, M. Enzymatic Electrochemical Biosensors for Neurotransmitters Detection: Recent Achievements and Trends. Chemosensors 2023, 11, 388. [Google Scholar] [CrossRef]
- Joshi, A.; Kim, K.-H. Recent advances in nanomaterial-based electrochemical detection of antibiotics: Challenges and future perspectives. Biosens. Bioelectron. 2020, 153, 112046. [Google Scholar] [CrossRef]
- Majdinasab, M.; Mishra, R.K.; Tang, X.; Marty, J.L. Detection of antibiotics in food: New achievements in the development of biosensors. TrAC Trends Anal. Chem. 2020, 127, 115883. [Google Scholar] [CrossRef]
- Aihaiti, A.; Li, Z.; Qin, Y.; Meng, F.; Li, X.; Huangfu, Z.; Chen, K.; Zhang, M. Construction of Electrochemical Sensors for Antibiotic Detection Based on Carbon Nanocomposites. Nanomaterials 2022, 12, 2789. [Google Scholar] [CrossRef]
- Zhou, C.; Zou, H.; Sun, C.; Li, Y. Recent advances in biosensors for antibiotic detection: Selectivity and signal amplification with nanomaterials. Food Chem. 2021, 361, 130109. [Google Scholar] [CrossRef]
- Wilson, J.S. Sensor Technology Handbook; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Ping, J.; Wang, Y.; Fan, K.; Tang, W.; Wu, J.; Ying, Y. High-performance flexible potentiometric sensing devices using free-standing graphene paper. J. Mater. Chem. B 2013, 1, 4781–4791. [Google Scholar] [CrossRef]
- Bhat, A.; Hara, T.O.; Tian, F.; Singh, B. Review of analytical techniques for arsenic detection and determination in drinking water. Environ. Sci. Adv. 2023, 2, 171–195. [Google Scholar] [CrossRef]
- Hara, T.O.; Singh, B. Electrochemical biosensors for detection of pesticides and heavy metal toxicants in water: Recent trends and progress. ACS EST Water 2021, 1, 462–478. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, X.; Guo, J. Functionalized Carbon-Based Electrochemical Sensors for Food and Alcoholic Beverage Safety. Appl. Sci. 2022, 12, 9082. [Google Scholar] [CrossRef]
- Ribeiro, B.V.; Ferreira, L.F.; Franco, D.L. Advances in biosensor development for the determination of antibiotics in cow’s milk-a review. Talanta Open 2022, 6, 100145. [Google Scholar] [CrossRef]
- Madhu, S.; Ramasamy, S.; Choi, J. Recent Developments in Electrochemical Sensors for the Detection of Antibiotic-Resistant Bacteria. Pharmaceuticals 2022, 15, 1488. [Google Scholar] [CrossRef]
- Kling, A.; Chatelle, C.; Armbrecht, L.; Qelibari, E.; Kieninger, J.; Dincer, C.; Weber, W.; Urban, G. Multianalyte antibiotic detection on an electrochemical microfluidic platform. Anal. Chem. 2016, 88, 10036–10043. [Google Scholar] [CrossRef]
- Cho, I.-H.; Lee, J.; Kim, J.; Kang, M.-S.; Paik, J.K.; Ku, S.; Cho, H.-M.; Irudayaraj, J.; Kim, D.-H. Current technologies of electrochemical immunosensors: Perspective on signal amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef]
- Wilson, G.S.; Hu, Y. Enzyme-based biosensors for in vivo measurements. Chem. Rev. 2000, 100, 2693–2704. [Google Scholar] [CrossRef]
- Singh, B.; Flampouri, E.; Dempsey, E. Electrochemical enzyme-linked immunosorbent assay (e-ELISA) for parasitic nematode Ostertagia ostertagi (brown stomach worm) infections in dairy cattle. Analyst 2019, 144, 5748–5754. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, Q.; Xu, C.; Li, H.; Wu, D.; Cai, Y.; Mao, K.; Cui, Z.; Du, B. Label-free electrochemical immunosensor for sensitive detection of kanamycin. Sens. Actuators B Chem. 2011, 155, 618–625. [Google Scholar] [CrossRef]
- Wu, X.; Kuang, H.; Hao, C.; Xing, C.; Wang, L.; Xu, C. Paper supported immunosensor for detection of antibiotics. Biosens. Bioelectron. 2012, 33, 309–312. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, F.; Wang, H.; Gao, L.; Wang, Z. A sensor based on Au nanoparticles/carbon nitride/graphene composites for the detection of chloramphenicol and ciprofloxacin. ECS J. Solid State Sci. Technol. 2018, 7, M201. [Google Scholar] [CrossRef]
- Giroud, F.; Gorgy, K.; Gondran, C.; Cosnier, S.; Pinacho, D.G.; Marco, M.-P.; Sánchez-Baeza, F.J. Impedimetric immunosensor based on a polypyrrole—Antibiotic model film for the label-free picomolar detection of ciprofloxacin. Anal. Chem. 2009, 81, 8405–8409. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, S.; Hu, Y.; Li, Z.; Luo, F.; He, Z. Electrochemical immunosensor based on the chitosan-magnetic nanoparticles for detection of tetracycline. Food Anal. Methods 2016, 9, 2972–2978. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, B.; Chen, G.; Tang, D. Biotin-avidin-conjugated metal sulfide nanoclusters for simultaneous electrochemical immunoassay of tetracycline and chloramphenicol. Microchim. Acta 2014, 181, 257–262. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, L.; Huang, L.; Han, Z.; Wang, J.; Pan, H. A low detection limit penicillin biosensor based on single graphene nanosheets preadsorbed with hematein/ionic liquids/penicillinase. Mater. Sci. Eng. C 2014, 39, 92–99. [Google Scholar] [CrossRef]
- Wu, H.; Fan, S.; Zhang, W.; Chen, H.; Peng, L.; Jin, X.; Ma, J.; Zhang, H. Amperometric immunosensor based on covalent immobilization of new methylene blue and penicillin polyclonal antibody for determination of penicillin G in milk. Anal. Methods 2014, 6, 497–502. [Google Scholar] [CrossRef]
- Conzuelo, F.; Gamella, M.; Campuzano, S.; Reviejo, A.J.; Pingarrón, J.M. Disposable amperometric magneto-immunosensor for direct detection of tetracyclines antibiotics residues in milk. Anal. Chim. Acta 2012, 737, 29–36. [Google Scholar] [CrossRef]
- El-Moghazy, A.Y.; Zhao, C.; Istamboulie, G.; Amaly, N.; Si, Y.; Noguer, T.; Sun, G. Ultrasensitive label-free electrochemical immunosensor based on PVA-co-PE nanofibrous membrane for the detection of chloramphenicol residues in milk. Biosens. Bioelectron. 2018, 117, 838–844. [Google Scholar] [CrossRef]
- Conzuelo, F.; Gamella, M.; Campuzano, S.; Pinacho, D.G.; Reviejo, A.J.; Marco, M.P.; Pingarrón, J.M. Disposable and integrated amperometric immunosensor for direct determination of sulfonamide antibiotics in milk. Biosens. Bioelectron. 2012, 36, 81–88. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, J.; Pan, M.; Yun, Y.; Wen, W.; Fang, G.; Wang, S. On-chip multiplex electrochemical immunosensor based on disposable 24-site fluidic micro-array screen printing analytical device for multi-component quantitative analysis. Sens. Actuators B Chem. 2018, 260, 499–507. [Google Scholar] [CrossRef]
- Yadav, A.K.; Verma, D.; Lakshmi, G.; Eremin, S.; Solanki, P.R. Fabrication of label-free and ultrasensitive electrochemical immunosensor based on molybdenum disulfide nanoparticles modified disposable ITO: An analytical platform for antibiotic detection in food samples. Food Chem. 2021, 363, 130245. [Google Scholar] [CrossRef] [PubMed]
- Gamella, M.; Campuzano, S.; Conzuelo, F.; Esteban-Torres, M.; De Las Rivas, B.; Reviejo, A.; Muñoz, R.; Pingarrón, J.M. An amperometric affinity penicillin-binding protein magnetosensor for the detection of β-lactam antibiotics in milk. Analyst 2013, 138, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Zhu, L.; Ding, Y.; Wang, J.; Li, J.; Du, X. Determination of sulfamethoxazole in foods based on CeO2/chitosan nanocomposite-modified electrodes. Mater. Sci. Eng. C 2012, 32, 2623–2627. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L.; Wang, Y.; Ou, Y.; Wang, X.; Pan, Y.; Wang, Y.; Huang, L.; Cheng, G.; Xie, S. Construction of an Electrochemical Receptor Sensor Based on Graphene/Thionine for the Sensitive Determination of β-Lactam Antibiotics Content in Milk. Int. J. Mol. Sci. 2020, 21, 3306. [Google Scholar] [CrossRef]
- Pinacho, D.G.; Sánchez-Baeza, F.; Pividori, M.-I.; Marco, M.-P. Electrochemical detection of fluoroquinolone antibiotics in milk using a magneto immunosensor. Sensors 2014, 14, 15965–15980. [Google Scholar] [CrossRef]
- Li, H.; Xu, B.; Wang, D.; Zhou, Y.; Zhang, H.; Xia, W.; Xu, S.; Li, Y. Immunosensor for trace penicillin G detection in milk based on supported bilayer lipid membrane modified with gold nanoparticles. J. Biotechnol. 2015, 203, 97–103. [Google Scholar] [CrossRef]
- Huang, Y.; Yan, X.; Zhao, L.; Qi, X.; Wang, S.; Liang, X. An aptamer cocktail-based electrochemical aptasensor for direct capture and rapid detection of tetracycline in honey. Microchem. J. 2019, 150, 104179. [Google Scholar] [CrossRef]
- Su, Z.; Xu, H.; Xu, X.; Zhang, Y.; Ma, Y.; Li, C.; Xie, Q. Effective covalent immobilization of quinone and aptamer onto a gold electrode via thiol addition for sensitive and selective protein biosensing. Talanta 2017, 164, 244–248. [Google Scholar] [CrossRef]
- An, K.; Lu, X.; Wang, C.; Qian, J.; Chen, Q.; Hao, N.; Wang, K. Porous gold nanocages: High atom utilization for thiolated aptamer immobilization to well balance the simplicity, sensitivity, and cost of disposable aptasensors. Anal. Chem. 2019, 91, 8660–8666. [Google Scholar] [CrossRef]
- Istamboulié, G.; Paniel, N.; Zara, L.; Granados, L.R.; Barthelmebs, L.; Noguer, T. Development of an impedimetric aptasensor for the determination of aflatoxin M1 in milk. Talanta 2016, 146, 464–469. [Google Scholar] [CrossRef]
- Rabai, S.; Benounis, M.; Catanante, G.; Baraket, A.; Errachid, A.; Renault, N.J.; Marty, J.-L.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 113956. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, F.; Noorbakhsh, A. Sensitive electrochemical prostate specific antigen aptasensor: Effect of carboxylic acid functionalized carbon nanotube and glutaraldehyde linker. Electroanalysis 2016, 28, 1134–1145. [Google Scholar] [CrossRef]
- Evtugyn, G.; Porfireva, A.; Tsekenis, G.; Oravczova, V.; Hianik, T. Electrochemical Aptasensors for Antibiotics Detection: Recent Achievements and Applications for Monitoring Food Safety. Sensors 2022, 22, 3684. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef]
- Saberian-Borujeni, M.; Johari-Ahar, M.; Hamzeiy, H.; Barar, J.; Omidi, Y. Nanoscaled aptasensors for multi-analyte sensing. BioImpacts BI 2014, 4, 205. [Google Scholar] [CrossRef]
- Chen, M.; Gan, N.; Zhou, Y.; Li, T.; Xu, Q.; Cao, Y.; Chen, Y. An electrochemical aptasensor for multiplex antibiotics detection based on metal ions doped nanoscale MOFs as signal tracers and RecJf exonuclease-assisted targets recycling amplification. Talanta 2016, 161, 867–874. [Google Scholar] [CrossRef]
- Chen, M.; Gan, N.; Zhang, H.; Yan, Z.; Li, T.; Chen, Y.; Xu, Q.; Jiang, Q. Electrochemical simultaneous assay of chloramphenicol and PCB72 using magnetic and aptamer-modified quantum dot-encoded dendritic nanotracers for signal amplification. Microchim. Acta 2016, 183, 1099–1106. [Google Scholar] [CrossRef]
- Huang, S.; Gan, N.; Li, T.; Zhou, Y.; Cao, Y.; Dong, Y. Electrochemical aptasensor for multi-antibiotics detection based on endonuclease and exonuclease assisted dual recycling amplification strategy. Talanta 2018, 179, 28–36. [Google Scholar] [CrossRef]
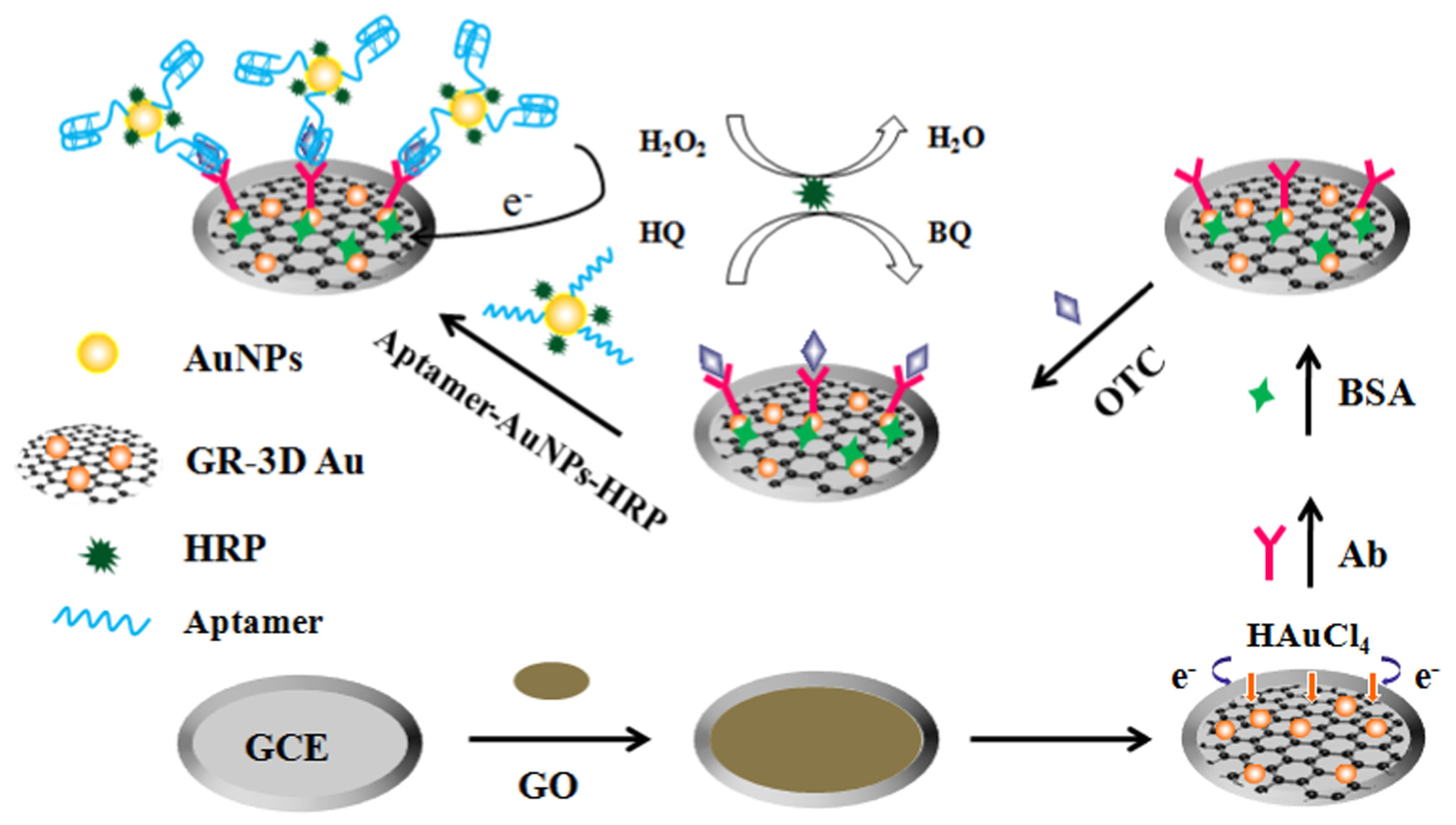
- Liu, S.; Wang, Y.; Xu, W.; Leng, X.; Wang, H.; Guo, Y.; Huang, J. A novel sandwich-type electrochemical aptasensor based on GR-3D Au and aptamer-AuNPs-HRP for sensitive detection of oxytetracycline. Biosens. Bioelectron. 2017, 88, 181–187. [Google Scholar] [CrossRef]
- Mohammad-Razdari, A.; Ghasemi-Varnamkhasti, M.; Rostami, S.; Izadi, Z.; Ensafi, A.A. Magnetic and gold nanocomposite as a novel aptasensor for early detection of tetracycline residues. J. Food Meas. Charact. 2021, 15, 3387–3396. [Google Scholar] [CrossRef]
- Li, F.; Wu, Y.; Chen, D.; Guo, Y.; Wang, X.; Sun, X. Sensitive dual-labeled electrochemical aptasensor for simultaneous detection of multi-antibiotics in milk. Int. J. Hydrogen Energy 2021, 46, 23301–23309. [Google Scholar] [CrossRef]
- Yue, F.; Liu, M.; Bai, M.; Hu, M.; Li, F.; Guo, Y.; Vrublevsky, I.; Sun, X. Novel Electrochemical Aptasensor Based on Ordered Mesoporous Carbon/2D Ti3C2 MXene as Nanocarrier for Simultaneous Detection of Aminoglycoside Antibiotics in Milk. Biosensors 2022, 12, 626. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.; Duan, F.; Hu, B.; He, L.; Wang, M.; Zhou, N.; Jia, Q.; Zhang, Z. Bimetallic cerium/copper organic framework-derived cerium and copper oxides embedded by mesoporous carbon: Label-free aptasensor for ultrasensitive tobramycin detection. Anal. Chim. Acta 2019, 1047, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Guo, W.; Qin, X.; Pei, M.; Wang, L.; Ding, F. A regular “signal attenuation” electrochemical aptasensor for highly sensitive detection of streptomycin. New J. Chem. 2016, 40, 9711–9718. [Google Scholar] [CrossRef]
- Ebrahimi Vafaye, S.; Rahman, A.; Safaeian, S.; Adabi, M. An electrochemical aptasensor based on electrospun carbon nanofiber mat and gold nanoparticles for the sensitive detection of Penicillin in milk. J. Food Meas. Charact. 2021, 15, 876–882. [Google Scholar] [CrossRef]
- Mahmoudpour, M.; Kholafazad-Kordasht, H.; Dolatabadi, J.E.N.; Hasanzadeh, M.; Rad, A.H.; Torbati, M. Sensitive aptasensing of ciprofloxacin residues in raw milk samples using reduced graphene oxide and nanogold-functionalized poly (amidoamine) dendrimer: An innovative apta-platform towards electroanalysis of antibiotics. Anal. Chim. Acta 2021, 1174, 338736. [Google Scholar] [CrossRef]
- Yu, Z.; Cui, P.; Xiang, Y.; Li, B.; Han, X.; Shi, W.; Yan, H.; Zhang, G. Developing a fast electrochemical aptasensor method for the quantitative detection of penicillin G residue in milk with high sensitivity and good anti-fouling ability. Microchem. J. 2020, 157, 105077. [Google Scholar] [CrossRef]
- Kulikova, T.; Gorbatchuk, V.; Stoikov, I.; Rogov, A.; Evtugyn, G.; Hianik, T. Impedimetric determination of kanamycin in milk with aptasensor based on carbon black-oligolactide composite. Sensors 2020, 20, 4738. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, W.; Guo, Y.; Wang, X.; Zhang, F.; Yu, L.; Guo, C.; Fang, G. Sensitive and selective electrochemical aptasensor via diazonium-coupling reaction for label-free determination of oxytetracycline in milk samples. Sens. Actuators Rep. 2020, 2, 100009. [Google Scholar] [CrossRef]
- Rosati, G.; Ravarotto, M.; Scaramuzza, M.; De Toni, A.; Paccagnella, A. Silver nanoparticles inkjet-printed flexible biosensor for rapid label-free antibiotic detection in milk. Sens. Actuators B Chem. 2019, 280, 280–289. [Google Scholar] [CrossRef]
- Li, F.; Guo, Y.; Wang, X.; Sun, X. Multiplexed aptasensor based on metal ions labels for simultaneous detection of multiple antibiotic residues in milk. Biosens. Bioelectron. 2018, 115, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Nameghi, M.A.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. An ultrasensitive electrochemical sensor for 17β-estradiol using split aptamers. Anal. Chim. Acta 2019, 1065, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef]
- Madikizela, L.M.; Tavengwa, N.T.; Tutu, H.; Chimuka, L. Green aspects in molecular imprinting technology: From design to environmental applications. Trends Environ. Anal. Chem. 2018, 17, 14–22. [Google Scholar] [CrossRef]
- Rebelo, P.; Costa-Rama, E.; Seguro, I.; Pacheco, J.G.; Nouws, H.P.; Cordeiro, M.N.D.; Delerue-Matos, C. Molecularly imprinted polymer-based electrochemical sensors for environmental analysis. Biosens. Bioelectron. 2021, 172, 112719. [Google Scholar] [CrossRef]
- Lian, W.; Liu, S.; Yu, J.; Xing, X.; Li, J.; Cui, M.; Huang, J. Electrochemical sensor based on gold nanoparticles fabricated molecularly imprinted polymer film at chitosan–platinum nanoparticles/graphene–gold nanoparticles double nanocomposites modified electrode for detection of erythromycin. Biosens. Bioelectron. 2012, 38, 163–169. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Zhang, Y.; Wei, G. Highly sensitive molecularly imprinted electrochemical sensor based on the double amplification by an inorganic prussian blue catalytic polymer and the enzymatic effect of glucose oxidase. Anal. Chem. 2012, 84, 1888–1893. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, L.; Luo, Z.; Tang, H. Fabrication of molecular imprinted polymer sensor for chlortetracycline based on controlled electrochemical reduction of graphene oxide. Sens. Actuators B Chem. 2013, 185, 438–444. [Google Scholar] [CrossRef]
- Liu, B.; Tang, D.; Zhang, B.; Que, X.; Yang, H.; Chen, G. Au (III)-promoted magnetic molecularly imprinted polymer nanospheres for electrochemical determination of streptomycin residues in food. Biosens. Bioelectron. 2013, 41, 551–556. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, L.; Zhang, J. Electrochemical sensor for levofloxacin based on molecularly imprinted polypyrrole–graphene–gold nanoparticles modified electrode. Sens. Actuators B Chem. 2014, 192, 642–647. [Google Scholar] [CrossRef]
- Yola, M.L.; Eren, T.; Atar, N. Molecularly imprinted electrochemical biosensor based on Fe@ Au nanoparticles involved in 2-aminoethanethiol functionalized multi-walled carbon nanotubes for sensitive determination of cefexime in human plasma. Biosens. Bioelectron. 2014, 60, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Jafari, S.; Dehghani, M.; Nasirizadeh, N.; Baghersad, M.H.; Azimzadeh, M. Label-free electrochemical detection of Cloxacillin antibiotic in milk samples based on molecularly imprinted polymer and graphene oxide-gold nanocomposite. Measurement 2019, 145, 22–29. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, X.; Peng, Y.; Hong, X.; Liu, Y.; Jiang, S.; Ning, B.; Gao, Z. Ultrasensitive sensing of diethylstilbestrol based on AuNPs/MWCNTs-CS composites coupling with sol-gel molecularly imprinted polymer as a recognition element of an electrochemical sensor. Sens. Actuators B Chem. 2017, 238, 420–426. [Google Scholar] [CrossRef]
- Roushani, M.; Rahmati, Z.; Hoseini, S.J.; Fath, R.H. Impedimetric ultrasensitive detection of chloramphenicol based on aptamer MIP using a glassy carbon electrode modified by 3-ampy-RGO and silver nanoparticle. Colloids Surf. B Biointerfaces 2019, 183, 110451. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Z.; Zhang, L.; Xu, X. Synthesis of molecularly imprinted polymers/NiCo2O4 nanoneedle arrays on 3D graphene electrode for determination of sulfadimidine residue in food. J. Mater. Sci. 2019, 54, 2066–2078. [Google Scholar] [CrossRef]
- Surya, S.G.; Khatoon, S.; Lahcen, A.A.; Nguyen, A.T.; Dzantiev, B.B.; Tarannum, N.; Salama, K.N. A chitosan gold nanoparticles molecularly imprinted polymer based ciprofloxacin sensor. RSC Adv. 2020, 10, 12823–12832. [Google Scholar] [CrossRef]
- Long, F.; Zhang, Z.; Yang, Z.; Zeng, J.; Jiang, Y. Imprinted electrochemical sensor based on magnetic multi-walled carbon nanotube for sensitive determination of kanamycin. J. Electroanal. Chem. 2015, 755, 7–14. [Google Scholar] [CrossRef]
- Wei, X.; Xu, X.; Qi, W.; Wu, Y.; Wang, L. Molecularly imprinted polymer/graphene oxide modified glassy carbon electrode for selective detection of sulfanilamide. Prog. Nat. Sci. Mater. Int. 2017, 27, 374–379. [Google Scholar] [CrossRef]
- Que, X.; Liu, B.; Fu, L.; Zhuang, J.; Chen, G.; Tang, D. Molecular imprint for electrochemical detection of streptomycin residues using enzyme signal amplification. Electroanalysis 2013, 25, 531–537. [Google Scholar] [CrossRef]
- Lian, W.; Liu, S.; Yu, J.; Li, J.; Cui, M.; Xu, W.; Huang, J. Electrochemical sensor using neomycin-imprinted film as recognition element based on chitosan-silver nanoparticles/graphene-multiwalled carbon nanotubes composites modified electrode. Biosens. Bioelectron. 2013, 44, 70–76. [Google Scholar] [CrossRef]
- Wen, T.; Wang, M.; Luo, M.; Yu, N.; Xiong, H.; Peng, H. A nanowell-based molecularly imprinted electrochemical sensor for highly sensitive and selective detection of 17β-estradiol in food samples. Food Chem. 2019, 297, 124968. [Google Scholar] [CrossRef]
- Turco, A.; Corvaglia, S.; Mazzotta, E.; Pompa, P.P.; Malitesta, C. Preparation and characterization of molecularly imprinted mussel inspired film as antifouling and selective layer for electrochemical detection of sulfamethoxazole. Sens. Actuators B Chem. 2018, 255, 3374–3383. [Google Scholar] [CrossRef]
- Wang, S.-X.; Ma, R.-R.; Mazzu, Y.Z.; Zhang, J.-W.; Li, W.; Tan, L.; Zhou, L.-D.; Xia, Z.-N.; Zhang, Q.-H.; Yuan, C.-S. Specific adsorption of tetracycline from milk by using biocompatible magnetic molecular imprinting material and evaluation by ECD. Food Chem. 2020, 326, 126969. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, S.B.; Ramasamy, R.; Gopal, N.; Kuzhandaivelu, V. Biosensors in clinical chemistry: An overview. Adv. Biomed. Res. 2014, 3, 67. [Google Scholar] [CrossRef]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V. Enzyme biosensors for biomedical applications: Strategies for safeguarding analytical performances in biological fluids. Sensors 2016, 16, 780. [Google Scholar] [CrossRef]
- Faridah, S.; Hazana, R.; Gayah, A.; Norzaili, Z.; Azima, A.; Nur Azura, M.; Zamri, I. Electrochemical sensors for detection of tetracycline antibiotics. Malays. Soc. Anim. Prod 2012, 15, 67–80. [Google Scholar]
- Gonçalves, L.M.; Callera, W.F.; Sotomayor, M.D.; Bueno, P.R. Penicillinase-based amperometric biosensor for penicillin G. Electrochem. Commun. 2014, 38, 131–133. [Google Scholar] [CrossRef]
- Drawz, S.M.; Bonomo, R.A. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef]
- Chen, B.; Ma, M.; Su, X. An amperometric penicillin biosensor with enhanced sensitivity based on co-immobilization of carbon nanotubes, hematein, and β-lactamase on glassy carbon electrode. Anal. Chim. Acta 2010, 674, 89–95. [Google Scholar] [CrossRef]
- Zhou, S.; Zhao, Y.; Mecklenburg, M.; Yang, D.; Xie, B. A novel thermometric biosensor for fast surveillance of β-lactamase activity in milk. Biosens. Bioelectron. 2013, 49, 99–104. [Google Scholar] [CrossRef]
- Ismail, F.; Adeloju, S.B. Galvanostatic entrapment of penicillinase into polytyramine films and its utilization for the potentiometric determination of penicillin. Sensors 2010, 10, 2851–2868. [Google Scholar] [CrossRef] [PubMed]
- Rinken, T.; Jaanisoo, R. On-Line System, Method of Its Calibration and Simultaneous Detection of Antibiotic Residues and Their Concentration in Milk. U.S. Patent 575,669, 10 January 2013. [Google Scholar]
- Conzuelo, F.; Montiel, V.R.-V.; Campuzano, S.; Gamella, M.; Torrente-Rodríguez, R.; Reviejo, A.; Pingarrón, J. Rapid screening of multiple antibiotic residues in milk using disposable amperometric magnetosensors. Anal. Chim. Acta 2014, 820, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef]
- Bahl, M.I.; Hansen, L.H.; Sørensen, S.J. Construction of an extended range whole-cell tetracycline biosensor by use of the tet (M) resistance gene. FEMS Microbiol. Lett. 2005, 253, 201–205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valtonen, S.J.; Kurittu, J.S.; Karp, M.T. A luminescent Escherichia coli biosensor for the high throughput detection of β-lactams. J. Biomol. Screen. 2002, 7, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Änkö, M.-L.; Kurittu, J.; Karp, M. An Escherichia coli biosensor strain for amplified and high throughput detection of antimicrobial agents. SLAS Discov. 2002, 7, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.E.; Carpico, G.; Coni, E. Electrochemical sensor for the detection and presumptive identification of quinolone and tetracycline residues in milk. Anal. Chim. Acta 2004, 520, 13–18. [Google Scholar] [CrossRef]
- Ferrini, A.M.; Mannoni, V.; Carpico, G.; Pellegrini, G.E. Detection and identification of β-lactam residues in milk using a hybrid biosensor. J. Agric. Food Chem. 2008, 56, 784–788. [Google Scholar] [CrossRef]
- Dizaji, A.N.; Ali, Z.; Ghorbanpoor, H.; Ozturk, Y.; Akcakoca, I.; Avci, H.; Guzel, F.D. Electrochemical-based ‘‘antibiotsensor’’for the whole-cell detection of the vancomycin-susceptible bacteria. Talanta 2021, 234, 122695. [Google Scholar] [CrossRef]
- Rezaei, B.; Damiri, S. Electrochemistry and adsorptive stripping voltammetric determination of amoxicillin on a multiwalled carbon nanotubes modified glassy carbon electrode. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2009, 21, 1577–1586. [Google Scholar] [CrossRef]
- Kumar, N.; Goyal, R.N. Gold-palladium nanoparticles aided electrochemically reduced graphene oxide sensor for the simultaneous estimation of lomefloxacin and amoxicillin. Sens. Actuators B Chem. 2017, 243, 658–668. [Google Scholar] [CrossRef]
- Hwa, K.-Y.; Sharma, T.S.K. Nano assembly of NiFe spheres anchored on f-MWCNT for electrocatalytic reduction and sensing of nitrofurantoin in biological samples. Sci. Rep. 2020, 10, 12256. [Google Scholar] [CrossRef] [PubMed]
- Ensafi, A.A.; Zandi-Atashbar, N.; Gorgabi-Khorzoughi, M.; Rezaei, B. Nickel-ferrite oxide decorated on reduced graphene oxide, an efficient and selective electrochemical sensor for detection of furazolidone. IEEE Sens. J. 2019, 19, 5396–5403. [Google Scholar] [CrossRef]






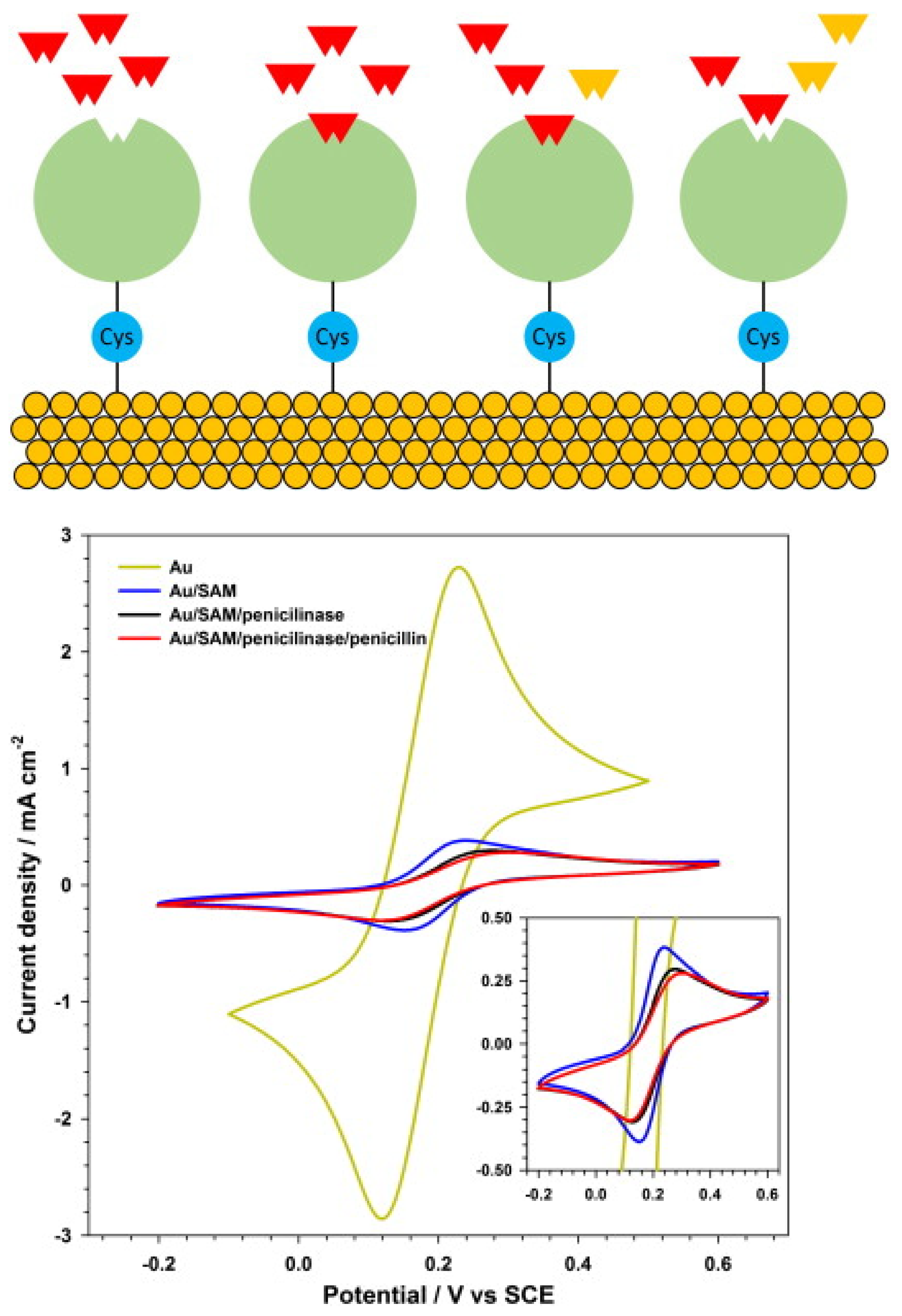

| Antibiotics | Biorecognition Component | Working Electrode | Detection Method/Mode | Linear/Dynamic Range | LOD | Sample Type | Ref. |
|---|---|---|---|---|---|---|---|
| Tetracycline | Aptamer | Aptamer/GNP/MNP/PGE | EIS | 1 pM to 1 µM | 0.03 pM | Milk of cow, sheep, goat, and buffalo | [118] |
| 5′-SH-CCC CCG GCA GGC CAC GGC TTG GGT TGG TCC CAC TGC GCG-3′ | |||||||
| Oxytetracyclines | 5′-TCA CGT TGA CGC TGG TGC CCG GTT GTG GTG GGA GTG TTG TGT- (CH2)6- NH2-3′ | GCE, capture beads (anti-ssDNA Ab/Dynabeads) & Apts-MNM | SWV | 0.5 to 5 × 104 pM | 0.18 pM | Milk | [114] |
| Kanamycin | 5′-TCT GGG GGT TGA GGC TAA GCC GAC- (CH2)6- NH2-3′ | GCE, capture beads (anti-ssDNA Ab/Dynabeads) & Apts-MNM | SWV | 0.5 to 5 × 104 pM | 0.15 pM | Milk | [114] |
| Tobramycin | 5′-Bio-GGCACGAGG UUUAGCUACACUCGUGCC-NH2-3′ | AuNSs/Gold electrode | DPV | 1 to 104 nM | 0.49 nM | Spiked milk | [119] |
| Streptomycin | 5′-TAG GGA ATT CGT CGA CGG ATC CGG GGT CTG GTG TTC TGC TTT GTT CTG TCG GGT CGT CTG CAG GTC GAC GCA TGC GCC G-thiol-3′ | MWCNTs–CuO–AuNPs/PCNRs/GCE | DPV | 0.05 to 300 ppb | 0.036 ppb | Milk & Honey | [122] |
| Penicillin | 5′-thiol-(CH2)6-CTG AAT TGG ATC TCT CTT CTT GAG CGA TCT CCA CA-3′ | pDNA/AuNPs/ECNF mat electrode | CV | 1 to 400 ppb | 0.6 ppb | Spiked fat-free milk | [123] |
| Aminoglycosides | 5′-CGGATCCCCAGCT-CGGGGTGCTATGGAGG-CTGTATCGGAGACCTGCAGG-3′ | Ti3C2 MXene/OMC-CS/SPCE | DPV | 10 to 2 × 103 nM | 3.51 nM | Spiked milk | [120] |
| Ciprofloxacin | 5′ –ATACCAGCTTATTCAA-TTGCAGGGTATCTG-AGGCTTGATCTACT-AAATGTCGTGGGGCA-TTGCTATTGGCGTTGA-TACGTACAATCGTAA TCAGTTAG-3′ | Apt/3D Au-PAMAM/rGO/GCE | DPV/SWV | 1 nM to 1 µM | 1 nM (LLOQ) | spiked milk | [124] |
| Penicillin-G | 5′GGGTCTGAGGAGTG-CGCGGTGCCAGTGAGT-3′ | Gold functionalised electrode | SWV | 5 nM to 5 µM | 1.7 nM | Spiked milk | [125] |
| Kanamycin | 3′-NH2-TGG GGG TTG AGG CTA AGC CGA-C-5’ | GCE covered with carbon black and Calix arene-bearing lactic fragments, aminated aptamer covalently attached via carbodiimide binding | EIS | 0.7–50 nM | 0.3 nM | Spiked milk | [126] |
| Oxytetracycline | 5′-NH2-GGA ATT CGC TAG CAC GTT GAC GCT GGT GCC CGG TTG TGG TGC GAG TGT TGT GTG GAT CCG AGC TCC ACG TG-3′ | GCE grafted with diazonium salt, followed by aptamer attachment by carbodiimide binding | DPV | 10−3 to 100 ppm | 0.229 ppb | Spiked milk | [127] |
| Tobramycin | 5′-ACUUGGUUUAGGUAAUGAGU-3′ | CeO2/CuOx@mC nanocomposite | EIS | 0.01 to 104 ppt | 2.0× 10−3 ppt | Spiked milk/human serum | [121] |
| Ampicillin | 3′-thiol-modified 40 nucleotides (ATW0001-GO3-GN3-100) with a 10 bases 3′ spacer and without any 5′ modification | Inkjet-printed AgNPs | EIS | 102 to 104 ppm | 10 ppm | Milk | [128] |
| Kanamycin & Streptomycin | 5′-NH2-ACGACCCGACAGAACAAAGCAGAACA CCAGACCCCAAAAAAAAAATCGGCTTAGCCTCAACCCCCATCT-3′ | Multiplexed graphitised multi-walled carbon nanotubes/carbon nanofibers-gold nanoparticles aptasensor (MWCNTGr/CNFs-AuNPs/SPCE) | DPV | 102 to 105 pM | 74.50 pM (KAN) & 36.45 pM (STR) | Diluted milk | [129] |
| 17β-estradiol (E2) | 5′-Thiol-TTTTTTTTTTTTTTTGCTTCCAGCTTATTGAATTACACGCAGAGGGTA-3′ (split1) and 5′- GCGGCTCTGCGCATTCAATTGCTGCGCGCTGAAGCGCGGAAGCTTTTTTTTTTTT-Thiol-3′ (split2) | Screen-printed gold electrode | DPV | 3 to 300 pM 300-9000 pM | 0.7 pM | Diluted spiked milk samples | [130] |
| Analyte | Sensing Scheme | Detection Method | Linear Range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|
| Ciprofloxacin | Ch-AuMIP/GCE | DPV | 1 to 100 µM | 0.21 µM | Mineral & tap water, milk and pharmaceuticals | [144] |
| Cloxacillin | MIP-GO-AuNPs/SPE | DPV | 0.11 to 0.75 µM | 0.036 µM | PBS/milk | [140] |
| Kanamycin | MMIP/CE (MIP-MWCNTs-Fe3O4/CE) | DPV | 10−4 to 1 µM | 2.3 × 10−5 µM | PBS/milk/liver | [145] |
| Sulfanilamide | MIP/GO/GCE | SWV | 10 to 1000 ppb | - | Buffer | [146] |
| Streptomycin | MIP/Gold electrode | DPV | 0.01 to 10 ppb | 0.007 ppb | PBS/milk/honey | [147] |
| Neomycin | MIPs/GR-MWCNTs/CS-SNP/gold electrode. | Amperometry | 0.009 to 7 µM | 7.63 × 10−3 µM | Standard solution/milk/honey | [148] |
| Chloramphenicol | aptamer-MIP/AgNP/3-ampy-RGO/GCE | EIS | 1 × 10−6 to 1 × 10−3 µM | 0.3 × 10−6 µM | PBS/milk | [142] |
| 17β-estradiol (Steroid) | MIP/NPGL/Au | CV | 1 × 10−6 to 10 µM | 1 × 10−7 µM | Food samples | [149] |
| Sulfamethoxasole | PDA-MIP/gold electrode | Amperometry | 0.8 to 170 µM | 0.8 µM | PBS/milk | [150] |
| Sulfadimidine | MIP-NiCo2O4/3D graphene sensor | DPV | 0.2 to 1000 ppb | 0.169 ppb | Spiked milk samples | [143] |
| Tetracycline | BMMIP/GCE | DPV | 0.025 to 500 ppm | 0.025 ppm | Buffer/milk | [151] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, B.; Bhat, A.; Dutta, L.; Pati, K.R.; Korpan, Y.; Dahiya, I. Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives. Biosensors 2023, 13, 867. https://doi.org/10.3390/bios13090867
Singh B, Bhat A, Dutta L, Pati KR, Korpan Y, Dahiya I. Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives. Biosensors. 2023; 13(9):867. https://doi.org/10.3390/bios13090867
Chicago/Turabian StyleSingh, Baljit, Abhijnan Bhat, Lesa Dutta, Kumari Riya Pati, Yaroslav Korpan, and Isha Dahiya. 2023. "Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives" Biosensors 13, no. 9: 867. https://doi.org/10.3390/bios13090867
APA StyleSingh, B., Bhat, A., Dutta, L., Pati, K. R., Korpan, Y., & Dahiya, I. (2023). Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives. Biosensors, 13(9), 867. https://doi.org/10.3390/bios13090867