Unraveling the Possibilities: Recent Progress in DNA Biosensing
Abstract
:1. Introduction
2. DNA Sensing Methods and Their Applications
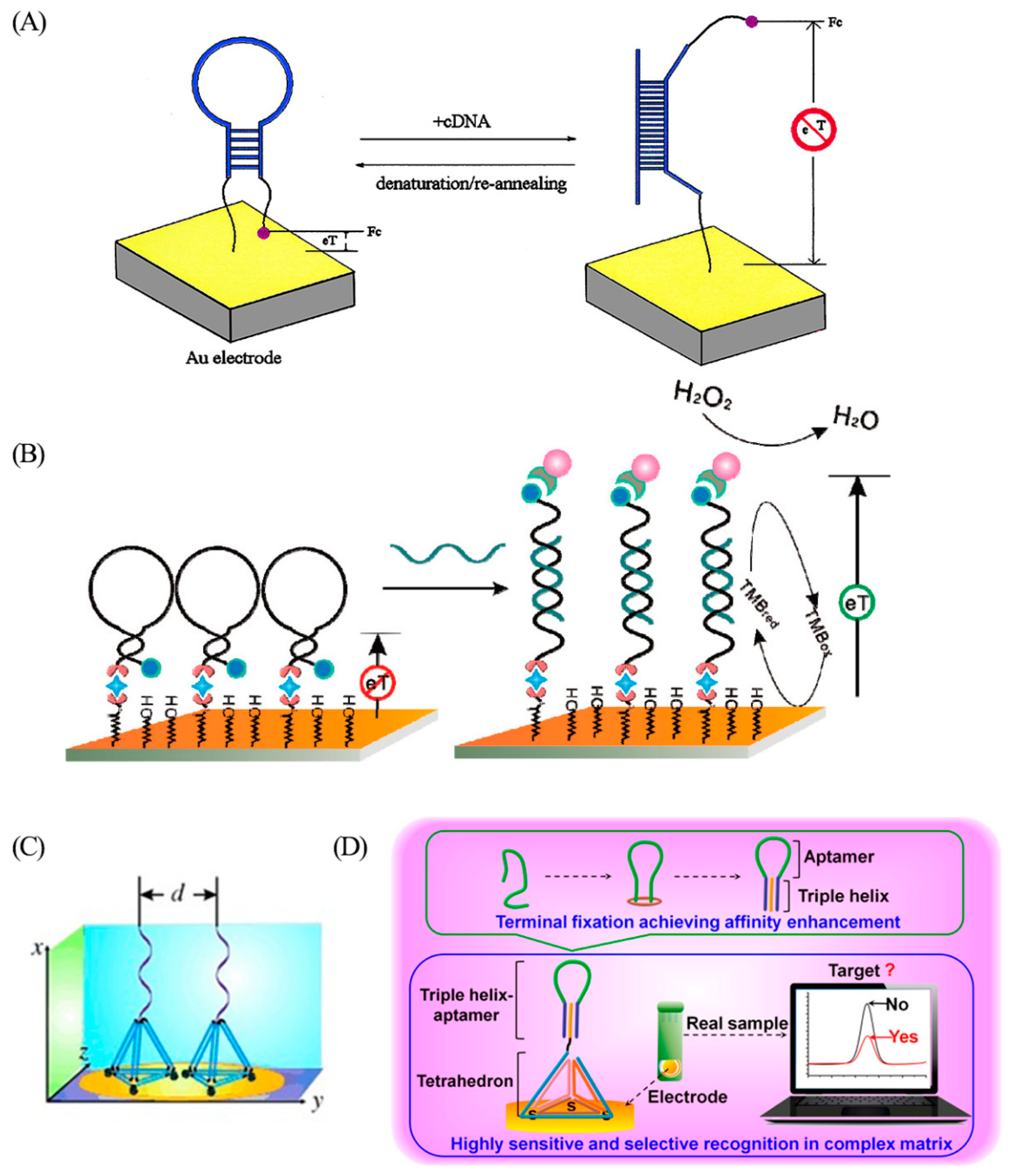
2.1. Sensing with Aptasensors

2.2. Sensing with DNAzymes

2.3. Sensing with i-Motif
2.4. Sensing with G-Quadruplexes


3. Summary and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vargas, E.; Zhang, F.; Ben Hassine, A.; Ruiz-Valdepeñas Montiel, V.; Mundaca-Uribe, R.; Nandhakumar, P.; He, P.; Guo, Z.; Zhou, Z.; Fang, R.H. Using Cell Membranes as Recognition Layers to Construct Ultrasensitive and Selective Bioelectronic Affinity Sensors. J. Am. Chem. Soc. 2022, 144, 17700–17708. [Google Scholar] [CrossRef]
- Walgama, C.; Nerimetla, R.; Materer, N.F.; Schildkraut, D.; Elman, J.F.; Krishnan, S. A simple construction of electrochemical liver microsomal bioreactor for rapid drug metabolism and inhibition assays. Anal. Chem. 2015, 87, 4712–4718. [Google Scholar] [CrossRef]
- Huang, Z.; Li, J.; Zhong, H.; Tian, B. Nucleic acid amplification strategies for volume-amplified magnetic nanoparticle detection assay. Front. Bioeng. Biotechnol. 2022, 10, 939807. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Jiang, N.; Hu, Y. Detection of proteins based on amino acid sequences by multiple aptamers against tripeptides. Anal. Biochem. 2007, 362, 126–135. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef]
- Zhao, W.; Chiuman, W.; Lam, J.C.; McManus, S.A.; Chen, W.; Cui, Y.; Pelton, R.; Brook, M.A.; Li, Y. DNA aptamer folding on gold nanoparticles: From colloid chemistry to biosensors. J. Am. Chem. Soc. 2008, 130, 3610–3618. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Csordas, A.T.; Wang, J.; Oh, S.S.; Eisenstein, M.S.; Soh, H.T. Rapid and label-free strategy to isolate aptamers for metal ions. ACS Nano 2016, 10, 7558–7565. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wan, Y.; Gau, V.; Zhang, J.; Wang, L.; Song, S.; Fan, C. An enzyme-based E-DNA sensor for sequence-specific detection of femtomolar DNA targets. J. Am. Chem. Soc. 2008, 130, 6820–6825. [Google Scholar] [CrossRef]
- Lin, M.; Wang, J.; Zhou, G.; Wang, J.; Wu, N.; Lu, J.; Gao, J.; Chen, X.; Shi, J.; Zuo, X. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. 2015, 54, 2151–2155. [Google Scholar] [CrossRef]
- Zhao, L.; Qi, X.; Yan, X.; Huang, Y.; Liang, X.; Zhang, L.; Wang, S.; Tan, W. Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J. Am. Chem. Soc. 2019, 141, 17493–17497. [Google Scholar] [CrossRef]
- Feagin, T.A.; Maganzini, N.; Soh, H.T. Strategies for creating structure-switching aptamers. ACS Sens. 2018, 3, 1611–1615. [Google Scholar] [CrossRef]
- Victorious, A.; Zhang, Z.; Chang, D.; Maclachlan, R.; Pandey, R.; Xia, J.; Gu, J.; Hoare, T.; Soleymani, L.; Li, Y. A DNA Barcode-Based Aptasensor Enables Rapid Testing of Porcine Epidemic Diarrhea Viruses in Swine Saliva Using Electrochemical Readout. Angew. Chem. 2022, 134, e202204252. [Google Scholar] [CrossRef]
- Siavash Moakhar, R.; Mahimkar, R.; Khorrami Jahromi, A.; Mahshid, S.S.; del Real Mata, C.; Lu, Y.; Vasquez Camargo, F.; Dixon, B.; Gilleard, J.; J Da Silva, A.; et al. Aptamer-Based Electrochemical Microfluidic Biosensor for the Detection of Cryptosporidium parvum. ACS Sens. 2023, 8, 2149–2158. [Google Scholar] [CrossRef]
- Hou, H.; Jin, Y.; Wei, H.; Ji, W.; Xue, Y.; Hu, J.; Zhang, M.; Jiang, Y.; Mao, L. A generalizable and noncovalent strategy for interfacing aptamers with a microelectrode for the selective sensing of neurotransmitters in vivo. Angew. Chem. Int. Ed. 2020, 59, 18996–19000. [Google Scholar] [CrossRef]
- Bošković, F.; Zhu, J.; Tivony, R.; Ohmann, A.; Chen, K.; Alawami, M.F.; Đorđević, M.; Ermann, N.; Pereira-Dias, J.; Fairhead, M. Simultaneous identification of viruses and viral variants with programmable DNA nanobait. Nat. Nanotechnol. 2023, 18, 290–298. [Google Scholar] [CrossRef]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef]
- Li, Y.; Deng, J.; Han, Z.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.; Sun, J. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Dirks, R.M.; Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- An, J.E.; Kim, K.H.; Park, S.J.; Seo, S.E.; Kim, J.; Ha, S.; Bae, J.; Kwon, O.S. Wearable cortisol aptasensor for simple and rapid real-time monitoring. ACS Sens. 2022, 7, 99–108. [Google Scholar] [CrossRef]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef]
- Yang, X.; Fang, C.; Mei, H.; Chang, T.; Cao, Z.; Shangguan, D. Characterization of G-Quadruplex/Hemin Peroxidase: Substrate Specificity and Inactivation Kinetics. Chem. Eur. J. 2011, 17, 14475–14484. [Google Scholar] [CrossRef]
- Xiong, J.; Dong, C.; Zhang, J.; Fang, X.; Ni, J.; Gan, H.; Li, J.; Song, C. DNA walker-powered ratiometric SERS cytosensor of circulating tumor cells with single-cell sensitivity. Biosens. Bioelectron. 2022, 213, 114442. [Google Scholar] [CrossRef]
- Li, D.; Zhao, T.; Chen, J.; Shi, J.; Wang, J.; Yin, Y.; Chen, S.; Xu, S.; Luo, X. Spatiotemporally controlled ultrasensitive molecular imaging using a DNA computation-mediated DNAzyme platform. Anal. Chem. 2022, 94, 14467–14474. [Google Scholar] [CrossRef] [PubMed]
- Sfrazzetto, G.T.; Satriano, C.; Tomaselli, G.A.; Rizzarelli, E. Synthetic fluorescent probes to map metallostasis and intracellular fate of zinc and copper. Coord. Chem. Rev. 2016, 311, 125–167. [Google Scholar] [CrossRef]
- Hawtrey, T.; New, E.J. Molecular probes for fluorescent sensing of metal ions in non-mammalian organisms. Curr. Opin. Chem. Biol. 2023, 74, 102311. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J. Searching for harmony in transition-metal signaling. Nat. Chem. Biol. 2015, 11, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Zhao, H.; Zhao, J.; Li, L. Modular Engineering of DNAzyme-Based Sensors for Spatioselective Imaging of Metal Ions in Mitochondria. J. Am. Chem. Soc. 2022, 145, 1678–1685. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, W.; Brossard, F.; Zheng, S.; Moench, S.; Pavagada, S.; Owens, R.M.; Fruk, L. Activity-enhanced DNAzyme for design of label-free copper (ii) biosensor. Nanoscale 2023, 15, 10776–10782. [Google Scholar] [CrossRef]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, D.M.; Istok, J.D.; Watson, D.B.; Lu, Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef]
- He, X.; Zhou, X.; Liu, Y.; Wang, X. Ultrasensitive, recyclable and portable microfluidic surface-enhanced raman scattering (SERS) biosensor for uranyl ions detection. Sens. Actuators B Chem. 2020, 311, 127676. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.-I.; Sonoda, T. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Garo, L.P.; Ajay, A.K.; Fujiwara, M.; Gabriely, G.; Raheja, R.; Kuhn, C.; Kenyon, B.; Skillin, N.; Kadowaki-Saga, R.; Saxena, S. MicroRNA-146a limits tumorigenic inflammation in colorectal cancer. Nat. Commun. 2021, 12, 2419. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Scott, J.G.; Harris, A.L.; Buffa, F.M. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat. Commun. 2018, 9, 5228. [Google Scholar] [CrossRef]
- Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, A.; Shahba, S.; Malvandi, A.M.; Mohammadipour, A. Roles of the miR-155 in neuroinflammation and neurological disorders: A potent biological and therapeutic target. Cell. Mol. Neurobiol. 2023, 43, 455–467. [Google Scholar] [CrossRef]
- Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, A.; Shahba, S.; Malvandi, A.M.; Mohammadipour, A. MicroRNA-22: A novel and potent biological therapeutics in neurological disorders. Mol. Neurobiol. 2022, 59, 2694–2701. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, L.; Yang, Z.; Song, Z.; Zhang, Q.; He, Y. Graphene oxide-based qRT-PCR assay enables the sensitive and specific detection of miRNAs for the screening of ovarian cancer. Anal. Chim. Acta 2021, 1174, 338715. [Google Scholar] [CrossRef]
- Wei, J.; Wang, H.; Wu, Q.; Gong, X.; Ma, K.; Liu, X.; Wang, F. A smart, autocatalytic, DNAzyme biocircuit for in vivo, amplified, microRNA imaging. Angew. Chem. 2020, 132, 6021–6027. [Google Scholar] [CrossRef]
- Wang, J.; Yu, S.; Wu, Q.; Gong, X.; He, S.; Shang, J.; Liu, X.; Wang, F. A self-catabolic multifunctional DNAzyme nanosponge for programmable drug delivery and efficient gene silencing. Angew. Chem. Int. Ed. 2021, 60, 10766–10774. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, S.; Wu, C.; Li, Q.; Shen, Z.; Lu, Y.; Wu, Z.-S. Self-protected DNAzyme walker with a circular bulging DNA shield for amplified imaging of miRNAs in living cells and mice. ACS Nano 2021, 15, 19211–19224. [Google Scholar] [CrossRef]
- Yin, Y.; Chen, G.; Gong, L.; Ge, K.; Pan, W.; Li, N.; Machuki, J.; Yu, Y.; Geng, D.; Dong, H.; et al. DNAzyme-powered three-dimensional DNA walker nanoprobe for detection amyloid β-peptide oligomer in living cells and in vivo. Anal. Chem. 2020, 92, 9247–9256. [Google Scholar] [CrossRef]
- Zhao, J.; He, C.; Long, Y.; Lei, J.; Liu, H.; Hou, J.; Hou, C.; Huo, D. 3D DNAzyme walker based electrochemical biosensor for attomolar level microRNA-155 detection. Anal. Chim. Acta 2023, 1276, 341642. [Google Scholar] [CrossRef] [PubMed]
- Chorti, P.; Kazi, A.P.; Wiederoder, M.; Christodouleas, D.C. High-Throughput Flow-Through Direct Immunoassays for Targeted Bacteria Detection. Anal. Chem. 2021, 93, 14586–14592. [Google Scholar] [CrossRef]
- Guo, Q.; Han, J.-J.; Shan, S.; Liu, D.-F.; Wu, S.-S.; Xiong, Y.-H.; Lai, W.-H. DNA-based hybridization chain reaction and biotin–streptavidin signal amplification for sensitive detection of Escherichia coli O157: H7 through ELISA. Biosens. Bioelectron. 2016, 86, 990–995. [Google Scholar] [CrossRef]
- Yamashige, R.; Kimoto, M.; Okumura, R.; Hirao, I. Visual detection of amplified DNA by polymerase chain reaction using a genetic alphabet expansion system. J. Am. Chem. Soc. 2018, 140, 14038–14041. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ding, W.; Wang, C.; Wu, H.; Tian, X.; Lyu, M.; Wang, S. DNAzyme biosensors for the detection of pathogenic bacteria. Sens. Actuators B Chem. 2021, 331, 129422. [Google Scholar] [CrossRef]
- Xing, G.; Zhang, W.; Li, N.; Pu, Q.; Lin, J.-M. Recent progress on microfluidic biosensors for rapid detection of pathogenic bacteria. Chin. Chem. Lett. 2022, 33, 1743–1751. [Google Scholar] [CrossRef]
- Tram, K.; Kanda, P.; Salena, B.J.; Huan, S.; Li, Y. Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem. 2014, 126, 13013–13016. [Google Scholar] [CrossRef]
- Ali, M.M.; Wolfe, M.; Tram, K.; Gu, J.; Filipe, C.D.; Li, Y.; Brennan, J.D. A DNAzyme-based colorimetric paper sensor for Helicobacter pylori. Angew. Chem. 2019, 131, 10012–10016. [Google Scholar] [CrossRef]
- Pandey, R.; Chang, D.; Smieja, M.; Hoare, T.; Li, Y.; Soleymani, L. Integrating programmable DNAzymes with electrical readout for rapid and culture-free bacterial detection using a handheld platform. Nat. Chem. 2021, 13, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Balasubramanian, S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. 2003, 42, 5734–5736. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Guéron, M.; Leroy, J.-L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000, 299, 123–144. [Google Scholar] [CrossRef]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Nonin-Lecomte, S.; Leroy, J.L. Structure of a C-rich strand fragment of the human centromeric satellite III: A pH-dependent intercalation topology. J. Mol. Biol. 2001, 309, 491–506. [Google Scholar] [CrossRef]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef]
- Kaiser, C.E.; Van Ert, N.A.; Agrawal, P.; Chawla, R.; Yang, D.; Hurley, L.H. Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 2017, 139, 8522–8536. [Google Scholar] [CrossRef]
- Justus, C.R.; Dong, L.; Yang, L.V. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front. Physiol. 2013, 4, 354. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, D.; Li, C.; Yu, F.; Fan, L.; Lei, C.; Huang, Y.; Nie, Z.; Yao, S. Cell-surface-anchored ratiometric DNA tweezer for real-time monitoring of extracellular and apoplastic pH. Anal. Chem. 2018, 90, 13459–13466. [Google Scholar] [CrossRef]
- Yang, H.; Chen, J.; Liang, Y.; Zhang, Y.; Yin, W.; Xu, Y.; Liu, S.-Y.; Dai, Z.; Zou, X. A MOF-Shell-Confined I-Motif-Based pH Probe (MOFC-i) Strategy for Sensitive and Dynamic Imaging of Cell Surface pH. ACS Appl. Mater. Interfaces 2021, 13, 45291–45299. [Google Scholar] [CrossRef] [PubMed]
- Bus, T.; Traeger, A.; Schubert, U.S. The great escape: How cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B 2018, 6, 6904–6918. [Google Scholar] [CrossRef] [PubMed]
- Banushi, B.; Joseph, S.R.; Lum, B.; Lee, J.J.; Simpson, F. Endocytosis in cancer and cancer therapy. Nat. Rev. Cancer 2023, 23, 450–473. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, X.; Tian, T.; Zhang, T.; Gao, S.; Zhang, X.; Yao, Y.; Lin, Y.; Cai, X. A lysosome-activated tetrahedral Nanobox for encapsulated siRNA delivery. Adv. Mater. 2022, 34, 2201731. [Google Scholar] [CrossRef]
- He, S.; Liu, M.; Yin, F.; Liu, J.; Ge, Z.; Li, F.; Li, M.; Shi, J.; Wang, L.; Mao, X. Programming folding cooperativity of the dimeric i-motif with DNA frameworks for sensing small pH variations. Chem. Commun. 2021, 57, 3247–3250. [Google Scholar] [CrossRef]
- Nesterova, I.V.; Nesterov, E.E. Rational design of highly responsive pH sensors based on DNA i-motif. J. Am. Chem. Soc. 2014, 136, 8843–8846. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Qiao, Y.; Gu, D.; Qi, R.; Zhao, H.; Yin, Y.; Zhao, W.; Xi, R.; Meng, M. DNA-based pH nanosensor with adjustable FRET responses to track lysosomes and pH fluctuations. Anal. Chem. 2021, 93, 7250–7257. [Google Scholar] [CrossRef]
- Liu, J.; Jing, X.; Liu, M.; Li, F.; Li, M.; Li, Q.; Shi, J.; Li, J.; Wang, L.; Mao, X. Mechano-fluorescence actuation in single synaptic vesicles with a DNA framework nanomachine. Sci. Robot. 2022, 7, eabq5151. [Google Scholar] [CrossRef]
- Ghosal, G.; Muniyappa, K. Hoogsteen base-pairing revisited: Resolving a role in normal biological processes and human diseases. Biochem. Biophys. Res. Commun. 2006, 343, 1–7. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Georgiades, S.; Abd Karim, N.; Suntharalingam, K.; Vilar, R. Interaction of Metal Complexes with G-Quadruplex DNA. Angew. Chem. Int. Ed. 2009, 49, 4020–4034. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex-forming aptamers-characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. In The Alkali Metal Ions: Their Role for Life; Sigel, A., Sigel, H., Sigel, R., Eds.; Springer: Cham, Switzerland, 2016; Volume 16, pp. 203–258. [Google Scholar]
- Xu, J.; Jiang, R.; Feng, Y.; Liu, Z.; Huang, J.; Ma, C.; Wang, K. Functional nucleic acid-based fluorescent probes for metal ion detection. Coord. Chem. Rev. 2022, 459, 214453. [Google Scholar] [CrossRef]
- Pathak, P.; Yao, W.; Hook, K.D.; Vik, R.; Winnerdy, F.R.; Brown, J.Q.; Gibb, B.C.; Pursell, Z.F.; Phan, A.T.; Jayawickramarajah, J. Bright G-quadruplex nanostructures functionalized with porphyrin lanterns. J. Am. Chem. Soc. 2019, 141, 12582–12591. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Yett, A.; Lin, L.Y.; Beseiso, D.; Miao, J.; Yatsunyk, L.A. N-methyl mesoporphyrin IX as a highly selective light-up probe for G-quadruplex DNA. J. Porphyr. Phthalocyanines 2019, 23, 1195–1215. [Google Scholar] [CrossRef]
- Bhasikuttan, A.C.; Mohanty, J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: Promising fluorescence sensors. Chem. Commun. 2015, 51, 7581–7597. [Google Scholar] [CrossRef]
- Bhasikuttan, A.C.; Mohanty, J.; Pal, H. Interaction of malachite green with guanine-rich single-stranded DNA: Preferential binding to a G-quadruplex. Angew. Chem. Int. Ed. 2007, 46, 9305–9307. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Li, J.; Wu, H.; Yan, Y.; Yuan, T.; Shu, Y.; Gao, X.; Zhang, L.; Li, S.; Ding, S.; Cheng, W. Zippered G-quadruplex/hemin DNAzyme: Exceptional catalyst for universal bioanalytical applications. Nucleic Acids Res. 2021, 49, 13031–13044. [Google Scholar] [CrossRef]
- Yum, J.H.; Park, S.; Sugiyama, H. G-quadruplexes as versatile scaffolds for catalysis. Org. Biomol. Chem. 2019, 17, 9547–9561. [Google Scholar] [CrossRef]
- Alizadeh, N.; Salimi, A.; Hallaj, R. Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme: Principle and biosensing application. In Catalytically Active Nucleic Acids; Seitz, H., Stahl, F., Walter, J.G., Eds.; Springer: Cham, Switzerland, 2020; Volume 170, pp. 85–106. [Google Scholar]
- Mehta, N.; Benzerara, K.; Kocar, B.D.; Chapon, V. Sequestration of radionuclides radium-226 and strontium-90 by cyanobacteria forming intracellular calcium carbonates. Environ. Sci. Technol. 2019, 53, 12639–12647. [Google Scholar] [CrossRef]
- Newcombe, H. Magnitude of biological hazard from strontium-90. Science 1957, 126, 549–551. [Google Scholar] [CrossRef]
- Amano, H.; Sakamoto, H.; Shiga, N.; Suzuki, K. Method for rapid screening analysis of Sr-90 in edible plant samples collected near Fukushima, Japan. Appl. Radiat. Isot. 2016, 112, 131–135. [Google Scholar] [CrossRef]
- Kankia, B.I.; Marky, L.A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: Stability, heat, and hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. [Google Scholar] [CrossRef]
- Leung, K.-H.; Ma, V.P.-Y.; He, H.-Z.; Chan, D.S.-H.; Yang, H.; Leung, C.-H.; Ma, D.-L. A highly selective G-quadruplex-based luminescent switch-on probe for the detection of nanomolar strontium(II) ions in sea water. RSC Adv. 2012, 2, 8273–8276. [Google Scholar] [CrossRef]
- Feng, L.; Wang, H.; Liu, T.; Feng, T.; Cao, M.; Zhang, J.; Liu, T.; Guo, Z.; Galiotis, C.; Yuan, Y. Ultrasensitive and highly selective detection of strontium ions. Nat. Sustain. 2023, 6, 789–796. [Google Scholar] [CrossRef]
- He, W.; Li, G.; Ma, X.; Wang, H.; Huang, J.; Xu, M.; Huang, C. WEEE recovery strategies and the WEEE treatment status in China. J. Hazard. Mater. 2006, 136, 502–512. [Google Scholar] [CrossRef]
- Recknagel, S.; Radant, H.; Kohlmeyer, R. Survey of mercury, cadmium and lead content of household batteries. Waste Manag. 2014, 34, 156–161. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Liu, L.; Xing, Y.; Zhou, X. Duplex functional G-quadruplex/NMM fluorescent probe for label-free detection of lead (II) and mercury (II) ions. J. Hazard. Mater. 2018, 355, 50–55. [Google Scholar] [CrossRef]
- Chen, F.; Lu, Q.; Huang, L.; Liu, B.; Liu, M.; Zhang, Y.; Liu, J. DNA triplex and quadruplex assembled nanosensors for correlating K+ and pH in lysosomes. Angew. Chem. Int. Ed. 2021, 60, 5453–5458. [Google Scholar] [CrossRef]
- Sun, J.; Lu, Y.; He, L.; Pang, J.; Yang, F.; Liu, Y. Colorimetric sensor array based on gold nanoparticles: Design principles and recent advances. Trends Anal. Chem. 2020, 122, 115754. [Google Scholar] [CrossRef]
- Yang, X.; Yuan, L.; Xu, Y.; He, B. Target-catalyzed self-assembled spherical G-quadruplex/hemin DNAzymes for highly sensitive colorimetric detection of microRNA in serum. Anal. Chim. Acta 2023, 1247, 340879. [Google Scholar] [CrossRef] [PubMed]
- Zahra, Q.U.A.; Luo, Z.; Ali, R.; Khan, M.I.; Li, F.; Qiu, B. Advances in gold nanoparticles-based colorimetric aptasensors for the detection of antibiotics: An overview of the past decade. Nanomaterials 2021, 11, 840. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yu, Y.-L.; Wang, J.-H. Inner filter effect-based fluorescent sensing systems: A review. Anal. Chim. Acta 2018, 999, 13–26. [Google Scholar] [CrossRef]
- Ge, J.; Hu, Y.; Deng, R.; Li, Z.; Zhang, K.; Shi, M.; Yang, D.; Cai, R.; Tan, W. Highly sensitive microRNA detection by coupling nicking-enhanced rolling circle amplification with MoS2 quantum dots. Anal. Chem. 2020, 92, 13588–13594. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Carbon Dots and Graphene Quantum Dots in Electrochemical Biosensing. Nanomaterials 2019, 9, 634. [Google Scholar] [CrossRef]
- Lu, J.; Wang, J.; Hu, X.; Gyimah, E.; Yakubu, S.; Wang, K.; Wu, X.; Zhang, Z. Electrochemical Biosensor Based on Tetrahedral DNA Nanostructures and G-Quadruplexb. Anal. Chem. 2019, 91, 7353–7359. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, G.; Szurszewski, J.H. Carbon Monoxide, Hydrogen Sulfide, and Nitric Oxide as Signaling Molecules in the Gastrointestinal Tract. Gastroenterology 2014, 147, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Luo, X.; Lu, X.; Xie, S.; Deng, L.; Kang, W.; He, F.; Zhang, J.; Lei, C.; Lin, B.; et al. Engineering of Nucleic Acids and Synthetic Cofactors as Holo Sensors for Probing Signaling Molecules in the Cellular Membrane Microenvironment. Angew. Chem. Int. Ed. 2019, 58, 6590–6594. [Google Scholar] [CrossRef]
- Bours, M.J.L.; Swennen, E.L.R.; Di Virgilio, F.; Cronstein, B.N.; Dagnelie, P.C. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol. Ther. 2006, 112, 358–404. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, Q.; Shi, L.; Peng, P.; Shi, L.; Li, T. Logic-Gated Proximity Aptasensing for Cell-Surface Real-Time Monitoring of Apoptosis. Angew. Chem. Int. Ed. 2021, 60, 20858–20864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X. Endocrine and metabolism. In Clinical Molecular Diagnostics; Pan, S., Tang, J., Eds.; Springer: Singapore, 2021; pp. 155–166. [Google Scholar]
- Del Prato, S.; Marchetti, P.; Bonadonna, R.C. Phasic Insulin Release and Metabolic Regulation in Type 2 Diabetes. Diabetes 2002, 51, 109–116. [Google Scholar] [CrossRef]
- Wu, Y.; Midinov, B.; White, R.J. Electrochemical Aptamer-Based Sensor for Real-Time Monitoring of Insulin. ACS Sens. 2019, 4, 498–503. [Google Scholar] [CrossRef]
- Raveendran, M.; Lee, A.J.; Sharma, R.; Wälti, C.; Actis, P. Rational design of DNA nanostructures for single molecule biosensing. Nat. Commun. 2020, 11, 4384. [Google Scholar] [CrossRef]
- Seferos, D.S.; Giljohann, D.A.; Hill, H.D.; Prigodich, A.E.; Mirkin, C.A. Nano-flares: Probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [Google Scholar] [CrossRef]
- Prigodich, A.E.; Alhasan, A.H.; Mirkin, C.A. Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J. Am. Chem. Soc. 2011, 133, 2120–2123. [Google Scholar] [CrossRef]
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical nucleic acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef]
- Kelich, P.; Jeong, S.; Navarro, N.; Adams, J.; Sun, X.; Zhao, H.; Landry, M.P.; Vukovic, L. Discovery of DNA–Carbon Nanotube Sensors for Serotonin with Machine Learning and Near-infrared Fluorescence Spectroscopy. ACS Nano 2021, 16, 736–745. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Wu, Y.; Guo, M.; Gu, C.; Dai, C.; Kong, D.; Wang, Y.; Zhang, C.; Qu, D.; et al. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat. Biomed. Eng. 2022, 6, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Hao, C.; Xu, L.; Xu, C.; Kuang, H. Spiny Nanorod and Upconversion Nanoparticle Satellite Assemblies for Ultrasensitive Detection of Messenger RNA in Living Cells. Anal. Chem. 2018, 90, 5414–5421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Peng, R.; Liu, W.; Donovan, M.J.; Wang, L.; Ismail, I.; Li, J.; Li, J.; Qu, F.; Tan, W. Engineering DNA on the surface of upconversion nanoparticles for bioanalysis and therapeutics. ACS Nano 2021, 15, 17257–17274. [Google Scholar] [CrossRef]
- Chen, T.; Wu, C.S.; Jimenez, E.; Zhu, Z.; Dajac, J.G.; You, M.; Han, D.; Zhang, X.; Tan, W. DNA Micelle Flares for Intracellular mRNA Imaging and Gene Therapy. Angew. Chem. Int. Ed. 2013, 52, 2012–2016. [Google Scholar] [CrossRef]
- Cao, L.; Li, C.M.; Zhen, S.J.; Huang, C.Z. A General Signal Amplifier of Self-Assembled DNA Micelles for Sensitive Quantification of Biomarkers. Anal. Chem. 2023, 95, 1794–1800. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Xu, X.; Xu, R.; Li, H.; Teng, X.; Du, Y.; Miao, Y.; Lin, H.C.; Han, D. Cancer diagnosis with DNA molecular computation. Nat. Nanotechnol. 2020, 15, 709–715. [Google Scholar] [CrossRef]
- Vicente, A.M.; Ballensiefen, W.; Jönsson, J.-I. How personalised medicine will transform healthcare by 2030: The ICPerMed vision. J. Transl. Med. 2020, 18, 180. [Google Scholar] [CrossRef]
- Wang, A.G.; Dong, T.; Mansour, H.; Matamoros, G.; Sanchez, A.L.; Li, F. Based DNA reader for visualized quantification of soil-transmitted helminth infections. ACS Sens. 2018, 3, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, A.; Chang, D.; Das, J.; Gomis, S.; Foroutan, F.; Chen, J.; Pandey, L.; Flynn, C.; Yousefi, H.; Geraili, A.; et al. Monitoring Cardiac Biomarkers with Aptamer-Based Molecular Pendulum Sensors. Angew. Chem. 2023, 135, e202213567. [Google Scholar] [CrossRef]
- Williamson, P.; Piskunen, P.; Ijäs, H.; Butterworth, A.; Linko, V.; Corrigan, D.K. Signal amplification in electrochemical DNA biosensors using target-capturing DNA origami tiles. ACS Sens. 2023, 8, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, S.; Liu, Z.; Zhu, B.; Zhou, Z.; Li, G.; Meana, J.J.; González-Maeso, J.; Lu, C. Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes. Nat. Commun. 2023, 14, 4672. [Google Scholar] [CrossRef] [PubMed]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.; He, T.; Wang, Q.; Cui, C. Unraveling the Possibilities: Recent Progress in DNA Biosensing. Biosensors 2023, 13, 889. https://doi.org/10.3390/bios13090889
Yu M, He T, Wang Q, Cui C. Unraveling the Possibilities: Recent Progress in DNA Biosensing. Biosensors. 2023; 13(9):889. https://doi.org/10.3390/bios13090889
Chicago/Turabian StyleYu, Meng, Tingli He, Qianqian Wang, and Cheng Cui. 2023. "Unraveling the Possibilities: Recent Progress in DNA Biosensing" Biosensors 13, no. 9: 889. https://doi.org/10.3390/bios13090889




