Advancing Brain Research through Surface-Enhanced Raman Spectroscopy (SERS): Current Applications and Future Prospects
Abstract
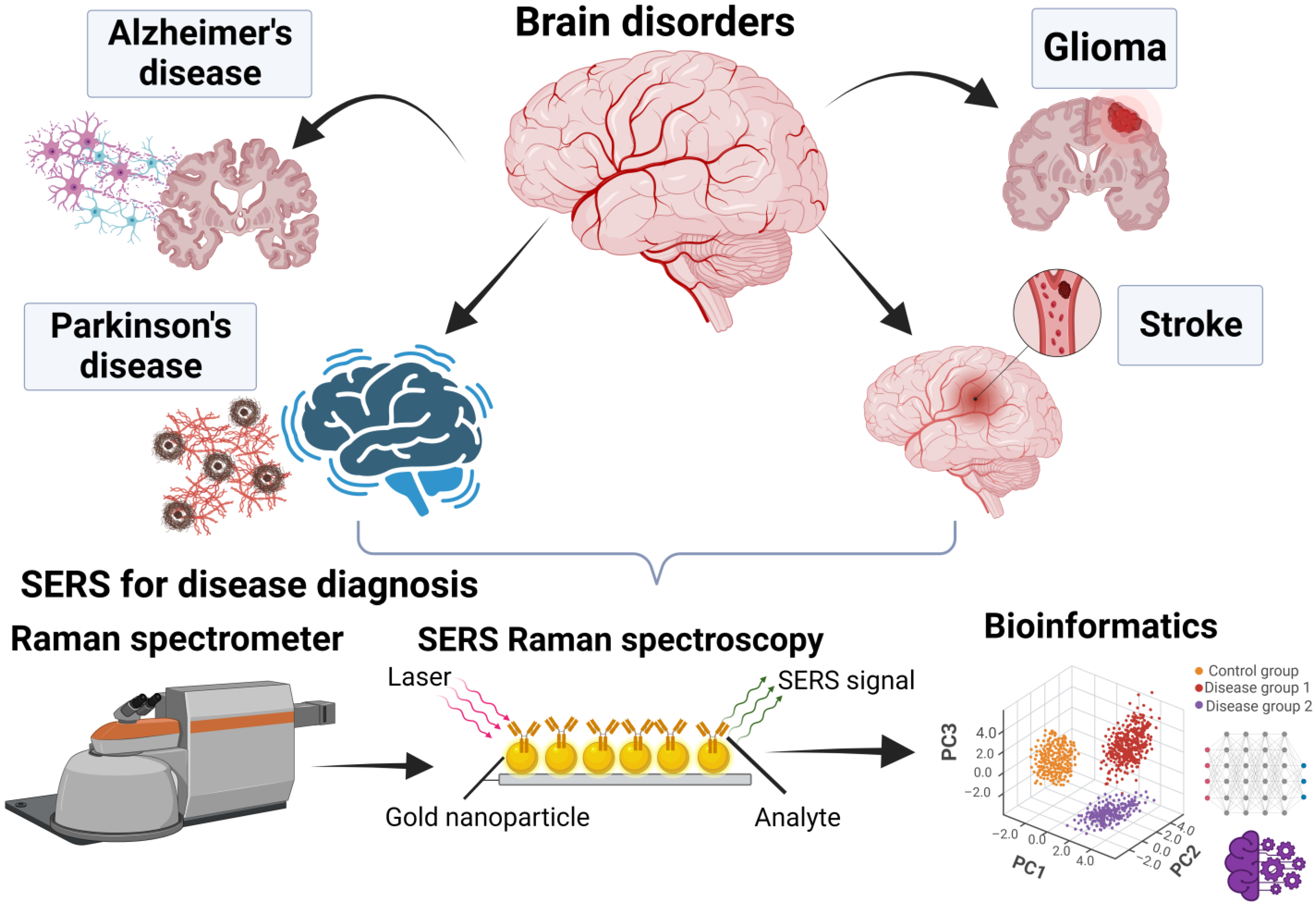
:1. Introduction
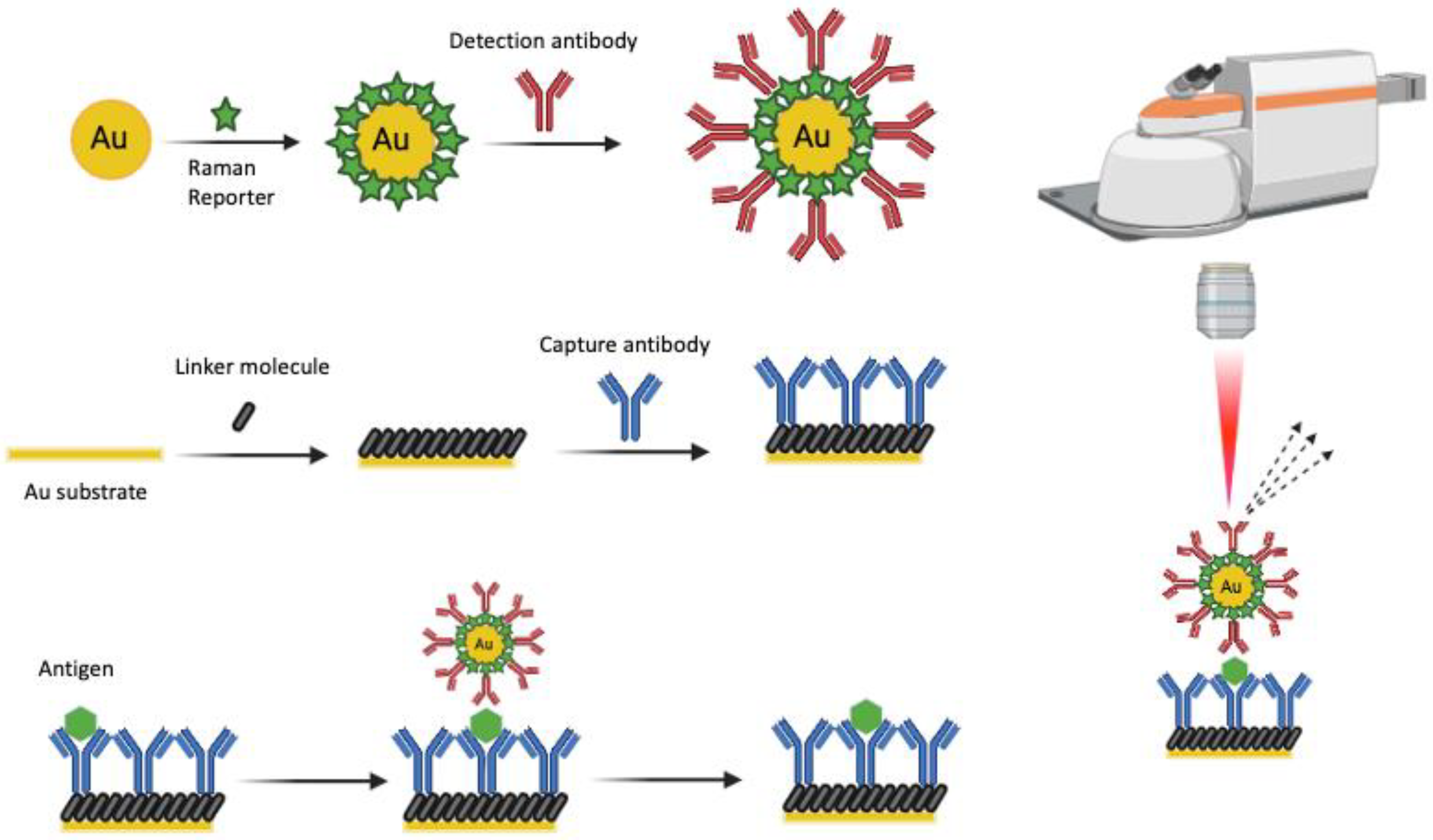
2. Fundamentals of SERS
3. Technological Advancements in SERS for Brain Research
4. Applications of SERS in Brain Research
4.1. The Use of SERS in Glioma Research
4.2. The Use of SERS in Alzheimer’s Disease Research
4.3. The Use of SERS in Parkinson’s Disease Research
4.4. The Use of SERS in Stroke Research
4.5. The Use of SERS in Neurotransmitter Detection
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2016 Neurology Collaborators. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.; Ali, G.C.; Guerchet, M.; Prina, A.M.; Albanese, E.; Wu, Y.T. Recent global trends in the prevalence and incidence of dementia, and survival with dementia. Alzheimer’s Res. Ther. 2016, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Francis, S.S.; Barnholtz-Sloan, J.S. Epidemiology of Brain and Other CNS Tumors. Curr. Neurol. Neurosci. Rep. 2021, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.Q.; Gowani, Z.S.; O’Connor, M.; Pence, I.J.; Nguyen, T.Q.; Holt, G.E.; Schwartz, H.S.; Halpern, J.L.; Mahadevan-Jansen, A. Intraoperative Raman spectroscopy of soft tissue sarcomas. Lasers Surg. Med. 2016, 48, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Picardi, G.; Spalloni, A.; Generosi, A.; Paci, B.; Mercuri, N.B.; Luce, M.; Longone, P.; Cricenti, A. Tissue degeneration in ALS affected spinal cord evaluated by Raman spectroscopy. Sci. Rep. 2018, 8, 13110. [Google Scholar] [CrossRef]
- Surmacki, J.M.; Ansel-Bollepalli, L.; Pischiutta, F.; Zanier, E.R.; Ercole, A.; Bohndiek, S.E. Label-free monitoring of tissue biochemistry following traumatic brain injury using Raman spectroscopy. Analyst 2016, 142, 132–139. [Google Scholar] [CrossRef]
- Yan, D.; Xiong, C.; Zhong, Q.; Yao, Y.; Chen, S.; Mei, X.; Zhu, S. Identification of late-life depression and mild cognitive impairment via serum surface-enhanced Raman spectroscopy and multivariate statistical analysis. Biomed. Opt. Express 2023, 14, 2920–2933. [Google Scholar] [CrossRef]
- Kralova, K.; Kral, M.; Vrtelka, O.; Setnicka, V. Comparative study of Raman spectroscopy techniques in blood plasma-based clinical diagnostics: A demonstration on Alzheimer’s disease. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 304, 123392. [Google Scholar] [CrossRef]
- Ralbovsky, N.M.; Halamkova, L.; Wall, K.; Anderson-Hanley, C.; Lednev, I.K. Screening for Alzheimer’s Disease Using Saliva: A New Approach Based on Machine Learning and Raman Hyperspectroscopy. J. Alzheimer’s Dis. 2019, 71, 1351–1359. [Google Scholar] [CrossRef]
- Zhang, L.; Su, Y.; Liang, X.; Cao, K.; Luo, Q.; Luo, H. Ultrasensitive and point-of-care detection of plasma phosphorylated tau in Alzheimer’s disease using colorimetric and surface-enhanced Raman scattering dual-readout lateral flow assay. Nano Res. 2023, 16, 7459–7469. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Halliwell, D.E.; Mann, D.M.A.; Allsop, D.; Martin-Hirsch, P.L.; Martin, F.L. Raman Spectroscopy to Diagnose Alzheimer’s Disease and Dementia with Lewy Bodies in Blood. ACS Chem. Neurosci. 2018, 9, 2786–2794. [Google Scholar] [CrossRef] [PubMed]
- Huefner, A.; Kuan, W.L.; Mason, S.L.; Mahajan, S.; Barker, R.A. Serum Raman spectroscopy as a diagnostic tool in patients with Huntington’s disease. Chem. Sci. 2020, 11, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhao, Y.; Hou, Y.; Li, H.; Yang, M.; Wang, Y.; Sun, B. Multiplexed electrochemical and SERS dual-mode detection of stroke biomarkers: Rapid screening with high sensitivity. New J. Chem. 2019, 43, 13381–133874. [Google Scholar] [CrossRef]
- Auner, G.W.; Koya, S.K.; Huang, C.; Broadbent, B.; Trexler, M.; Auner, Z.; Elias, A.; Mehne, K.C.; Brusatori, M.A. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018, 37, 691–717. [Google Scholar] [CrossRef] [PubMed]
- Crow, P.; Stone, N.; Kendall, C.A.; Uff, J.S.; Farmer, J.A.M.; Barr, H.; Wright, M.P.J. The use of Raman spectroscopy to identify and grade prostatic adenocarcinoma in vitro. Br. J. Cancer 2003, 89, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Quesnel, A.; Coles, N.; Angione, C.; Dey, P.; Polvikoski, T.M.; Outeiro, T.F.; Islam, M.; Khundakar, A.A.; Filippou, P.S. Glycosylation spectral signatures for glioma grade discrimination using Raman spectroscopy. BMC Cancer 2023, 23, 174. [Google Scholar] [CrossRef] [PubMed]
- Hanna, K.; Krzoska, E.; Shaaban, A.M.; Muirhead, D.; Abu-Eid, R.; Speirs, V. Raman spectroscopy: Current applications in breast cancer diagnosis, challenges and future prospects. Br. J. Cancer 2022, 126, 1125–1139. [Google Scholar] [CrossRef]
- Duckworth, J.; Krasnoslobodtsev, A.V. Modular Micro Raman Reader Instrument for Fast SERS-Based Detection of Biomarkers. Micromachines 2022, 13, 1570. [Google Scholar] [CrossRef]
- Conti, F.; D’Acunto, M.; Caudai, C.; Colantonio, S.; Gaeta, R.; Moroni, D.; Pascali, M.A. Raman spectroscopy and topological machine learning for cancer grading. Sci. Rep. 2023, 13, 7282. [Google Scholar] [CrossRef]
- Ryzhikova, E.; Ralbovsky, N.M.; Sikirzhytski, V.; Kazakov, O.; Halamkova, L.; Quinn, J.; Zimmerman, E.A.; Lednev, I.K. Raman spectroscopy and machine learning for biomedical applications: Alzheimer’s disease diagnosis based on the analysis of cerebrospinal fluid. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 248, 119188. [Google Scholar] [CrossRef]
- Wang, W.; Ma, P.; Song, D. Applications of surface-enhanced Raman spectroscopy based on portable Raman spectrometers: A review of recent developments. Luminescence 2022, 37, 1822–1835. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-enhanced Raman spectroscopy: Benefits, trade-offs and future developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef] [PubMed]
- Chisanga, M.; Williams, H.; Boudreau, D.; Pelletier, J.N.; Trottier, S.; Masson, J.F. Label-Free SERS for Rapid Differentiation of SARS-CoV-2-Induced Serum Metabolic Profiles in Non-Hospitalized Adults. Anal. Chem. 2023, 95, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, M.; Hadjigeorgiou, K.; Abalde-Cela, S.; Andreou, C. Label-Free Sensing with Metal Nanostructure-Based Surface-Enhanced Raman Spectroscopy for Cancer Diagnosis. ACS Appl. Nano Mater. 2022, 5, 12276–12299. [Google Scholar] [CrossRef] [PubMed]
- Badillo-Ramirez, I.; Janssen, S.A.J.; Soufi, G.; Slipets, R.; Zor, K.; Boisen, A. Label-free SERS assay combined with multivariate spectral data analysis for lamotrigine quantification in human serum. Mikrochim. Acta 2023, 190, 495. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Wei, Y.; Xu, Y.; Zheng, M.; Wang, C.; Zhang, S.; Xie, X.; Ye, C.; Mi, X. Microfluidic magnetic detection system combined with a DNA framework-mediated immune-sandwich assay for rapid and sensitive detection of tumor-derived exosomes. Microsyst. Nanoeng. 2023, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Irudayaraj, J. Surface-enhanced Raman spectroscopy at single-molecule scale and its implications in biology. Philos. Trans. R. Soc. Lond B Biol. Sci. 2013, 368, 20120026. [Google Scholar] [CrossRef]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef]
- Unser, S.; Bruzas, I.; He, J.; Sagle, L. Localized Surface Plasmon Resonance Biosensing: Current Challenges and Approaches. Sensors 2015, 15, 15684–15716. [Google Scholar] [CrossRef]
- Kim, J.; Jang, Y.; Kim, N.J.; Kim, H.; Yi, G.C.; Shin, Y.; Kim, M.H.; Yoon, S. Study of Chemical Enhancement Mechanism in Non-plasmonic Surface Enhanced Raman Spectroscopy (SERS). Front. Chem. 2019, 7, 582. [Google Scholar] [CrossRef]
- Szaniawska, A.; Kudelski, A. Applications of Surface-Enhanced Raman Scattering in Biochemical and Medical Analysis. Front. Chem. 2021, 9, 664134. [Google Scholar] [CrossRef]
- Desroches, J.; Jermyn, M.; Mok, K.; Lemieux-Leduc, C.; Mercier, J.; St-Arnaud, K.; Urmey, K.; Guiot, M.C.; Marple, E.; Petrecca, K.; et al. Characterization of a Raman spectroscopy probe system for intraoperative brain tissue classification. Biomed. Opt. Express 2015, 6, 2380–2397. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.C.; Petrecca, K.; Leblond, F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 2015, 7, 274ra219. [Google Scholar] [CrossRef] [PubMed]
- Badillo-Ramírez, I.; Landeros-Rivera, B.; Saniger, J.M.; Popp, J.; Cialla-May, D. SERS-based detection of 5-S-cysteinyl-dopamine as a novel biomarker of Parkinson’s disease in artificial biofluids. Analyst 2023, 148, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ge, S.; Chen, M.; Mao, H.; Wang, Y. LoC-SERS Platform Integrated with the Signal Amplification Strategy toward Parkinson’s Disease Diagnosis. ACS Appl. Mater. Interfaces 2023, 15, 21830–21842. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, G.; Montorio, D.; Morra, V.B.; Criscuolo, C.; Lanzillo, R.; Salvatore, E.; Camerlingo, C.; Lisitskiy, M.; Delfino, I.; Portaccio, M.; et al. Surface-enhanced Raman spectroscopy of tears: Toward a diagnostic tool for neurodegenerative disease identification. J. Biomed. Opt. 2020, 25, 1–12. [Google Scholar] [CrossRef]
- Moisoiu, V.; Iancu, S.D.; Stefancu, A.; Moisoiu, T.; Pardini, B.; Dragomir, M.P.; Crisan, N.; Avram, L.; Crisan, D.; Andras, I.; et al. SERS liquid biopsy: An emerging tool for medical diagnosis. Colloids Surf. B Biointerfaces 2021, 208, 112064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mi, X.; Tan, X.; Xiang, R. Recent Progress on Liquid Biopsy Analysis using Surface-Enhanced Raman Spectroscopy. Theranostics 2019, 9, 491–525. [Google Scholar] [CrossRef]
- Avci, E.; Yilmaz, H.; Sahiner, N.; Tuna, B.G.; Cicekdal, M.B.; Eser, M.; Basak, K.; Altintoprak, F.; Zengin, I.; Dogan, S.; et al. Label-Free Surface Enhanced Raman Spectroscopy for Cancer Detection. Cancers 2022, 14, 5021. [Google Scholar] [CrossRef]
- Kaminska, A.; Szymborski, T.; Witkowska, E.; Kijenska-Gawronska, E.; Swieszkowski, W.; Nicinski, K.; Trzcinska-Danielewicz, J.; Girstun, A. Detection of Circulating Tumor Cells Using Membrane-Based SERS Platform: A New Diagnostic Approach for ‘Liquid Biopsy’. Nanomaterials 2019, 9, 366. [Google Scholar] [CrossRef]
- Avram, L.; Stefancu, A.; Crisan, D.; Leopold, N.; Donca, V.; Buzdugan, E.; Craciun, R.; Andras, D.; Coman, I. Recent advances in surface-enhanced Raman spectroscopy based liquid biopsy for colorectal cancer (Review). Exp. Ther. Med. 2020, 20, 213. [Google Scholar] [CrossRef]
- Ge, S.; Chen, G.; Deng, J.; Gu, Y.; Mao, Y.; Zhou, X.; Li, G. Multiplex signal amplification strategy-based early-stage diagnosis of Parkinson’s disease on a SERS-enabled LoC system. Anal. Chim. Acta 2023, 1247, 340890. [Google Scholar] [CrossRef]
- Kasera, S.; Herrmann, L.O.; del Barrio, J.; Baumberg, J.J.; Scherman, O.A. Quantitative multiplexing with nano-self-assemblies in SERS. Sci. Rep. 2014, 4, 6785. [Google Scholar] [CrossRef]
- Nicolson, F.; Jamieson, L.E.; Mabbott, S.; Plakas, K.; Shand, N.C.; Detty, M.R.; Graham, D.; Faulds, K. Multiplex imaging of live breast cancer tumour models through tissue using handheld surface enhanced spatially offset resonance Raman spectroscopy (SESORRS). Chem. Commun. 2018, 54, 8530–8533. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Su, E.; Chen, H.-Y.; Gu, Z.; Zhao, X. Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuators B Chem. 2018, 277, 502–509. [Google Scholar] [CrossRef]
- Li, J.; Wuethrich, A.; Zhang, Z.; Wang, J.; Lin, L.L.; Behren, A.; Wang, Y.; Trau, M. SERS Multiplex Profiling of Melanoma Circulating Tumor Cells for Predicting the Response to Immune Checkpoint Blockade Therapy. Anal. Chem. 2022, 94, 14573–14582. [Google Scholar] [CrossRef]
- Verdin, A.; Malherbe, C.; Muller, W.H.; Bertrand, V.; Eppe, G. Multiplex micro-SERS imaging of cancer-related markers in cells and tissues using poly(allylamine)-coated Au@Ag nanoprobes. Anal. Bioanal. Chem. 2020, 412, 7739–7755. [Google Scholar] [CrossRef]
- Lai, H.; Yu, Z.; Li, G.; Zhang, Z. Advanced sample preparation techniques for rapid surface-enhanced Raman spectroscopy analysis of complex samples. J. Chromatogr. A 2022, 1675, 463181. [Google Scholar] [CrossRef]
- Demers, J.L.; Esmonde-White, F.W.; Esmonde-White, K.A.; Morris, M.D.; Pogue, B.W. Next-generation Raman tomography instrument for non-invasive in vivo bone imaging. Biomed. Opt. Express 2015, 6, 793–806. [Google Scholar] [CrossRef]
- Harmsen, S.; Huang, R.; Wall, M.A.; Karabeber, H.; Samii, J.M.; Spaliviero, M.; White, J.R.; Monette, S.; O’Connor, R.; Pitter, K.L.; et al. Surface-enhanced resonance Raman scattering nanostars for high-precision cancer imaging. Sci. Transl. Med. 2015, 7, 271ra277. [Google Scholar] [CrossRef]
- Keren, S.; Zavaleta, C.; Cheng, Z.; de la Zerda, A.; Gheysens, O.; Gambhir, S.S. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2008, 105, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.R.; Andreou, C.; Oseledchyk, A.; Marcus, W.D.; Wong, H.C.; Massague, J.; Kircher, M.F. Tissue factor-specific ultra-bright SERRS nanostars for Raman detection of pulmonary micrometastases. Nanoscale 2017, 9, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, F.; Andreiuk, B.; Andreou, C.; Hsu, H.T.; Rudder, S.; Kircher, M.F. Non-invasive In Vivo Imaging of Cancer Using Surface-Enhanced Spatially Offset Raman Spectroscopy (SESORS). Theranostics 2019, 9, 5899–5913. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Peng, X.H.; Ansari, D.O.; Yin-Goen, Q.; Chen, G.Z.; Shin, D.M.; Yang, L.; Young, A.N.; Wang, M.D.; Nie, S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008, 26, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Solis, D.M.; Taboada, J.M.; Obelleiro, F.; Liz-Marzan, L.M.; Garcia de Abajo, F.J. Optimization of Nanoparticle-Based SERS Substrates through Large-Scale Realistic Simulations. ACS Photonics 2017, 4, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.E.; McCabe, S.M.; Sloan-Dennison, S.; Laing, S.; Shand, N.C.; Graham, D.; Faulds, K. Tomographic Imaging and Localization of Nanoparticles in Tissue Using Surface-Enhanced Spatially Offset Raman Spectroscopy. ACS Appl. Mater. Interfaces 2022, 14, 31613–31624. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Ma, K.; Glucksberg, M.R.; Van Duyne, R.P. Seeing through bone with surface-enhanced spatially offset Raman spectroscopy. J. Am. Chem. Soc. 2013, 135, 17290–17293. [Google Scholar] [CrossRef]
- Odion, R.A.; Strobbia, P.; Crawford, B.M.; Vo-Dinh, T. Inverse surface-enhanced spatially offset Ramanspectroscopy (SESORS) through a monkey skull. J. Raman Spectrosc. 2018, 49, 1452–1460. [Google Scholar] [CrossRef]
- Nicolson, F.; Jamieson, L.E.; Mabbott, S.; Plakas, K.; Shand, N.C.; Detty, M.R.; Graham, D.; Faulds, K. Through tissue imaging of a live breast cancer tumour model using handheld surface enhanced spatially offset resonance Raman spectroscopy (SESORRS). Chem. Sci. 2018, 9, 3788–3792. [Google Scholar] [CrossRef]
- Iakab, S.A.; Baquer, G.; Lafuente, M.; Pina, M.P.; Ramirez, J.L.; Rafols, P.; Correig-Blanchar, X.; Garcia-Altares, M. SALDI-MS and SERS Multimodal Imaging: One Nanostructured Substrate to Rule Them Both. Anal. Chem. 2022, 94, 2785–2793. [Google Scholar] [CrossRef]
- Karabeber, H.; Huang, R.; Iacono, P.; Samii, J.M.; Pitter, K.; Holland, E.C.; Kircher, M.F. Guiding brain tumor resection using surface-enhanced Raman scattering nanoparticles and a hand-held Raman scanner. ACS Nano 2014, 8, 9755–9766. [Google Scholar] [CrossRef]
- Kircher, M.F.; de la Zerda, A.; Jokerst, J.V.; Zavaleta, C.L.; Kempen, P.J.; Mittra, E.; Pitter, K.; Huang, R.; Campos, C.; Habte, F.; et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat. Med. 2012, 18, 829–834. [Google Scholar] [CrossRef]
- Han, L.; Duan, W.; Li, X.; Wang, C.; Jin, Z.; Zhai, Y.; Cao, C.; Chen, L.; Xu, W.; Liu, Y.; et al. Surface-Enhanced Resonance Raman Scattering-Guided Brain Tumor Surgery Showing Prognostic Benefit in Rat Models. ACS Appl. Mater. Interfaces 2019, 11, 15241–15250. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Orringer, D.A.; Freudiger, C.W.; Ramkissoon, S.; Liu, X.; Lau, D.; Golby, A.J.; Norton, I.; Hayashi, M.; Agar, N.Y.R.; et al. Rapid, Label-Free Detection of Brain Tumors with Stimulated Raman Scattering Microscopy. Sci. Transl. Med. 2013, 5, 201ra119. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Lee, J.U.; Jeon, M.J.; Sim, S.J. Highly sensitive surface-enhanced Raman scattering-based immunosensor incorporating half antibody-fragment for quantitative detection of Alzheimer’s disease biomarker in blood. Anal. Chim. Acta 2022, 1195, 339445. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, C.; Cazzaniga, F.A.; Bistaffa, E.; Barucci, A.; de Angelis, M.; Banchelli, M.; Farnesi, E.; Polykretis, P.; Marzi, C.; Indaco, A.; et al. Impact of seed amplification assay and surface-enhanced Raman spectroscopy combined approach on the clinical diagnosis of Alzheimer’s disease. Transl. Neurodegener. 2023, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Schwartzbaum, J.A.; Fisher, J.L.; Aldape, K.D.; Wrensch, M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006, 2, 494–503. [Google Scholar] [CrossRef]
- Pitter, K.L.; Tamagno, I.; Alikhanyan, K.; Hosni-Ahmed, A.; Pattwell, S.S.; Donnola, S.; Dai, C.; Ozawa, T.; Chang, M.; Chan, T.A.; et al. Corticosteroids compromise survival in glioblastoma. Brain 2016, 139, 1458–1471. [Google Scholar] [CrossRef]
- Chen, C.; Wu, W.; Chen, C.; Chen, F.; Dong, X.; Ma, M.; Yan, Z.; Lv, X.; Ma, Y.; Zhu, M. Rapid diagnosis of lung cancer and glioma based on serum Raman spectroscopy combined with deep learning. J. Raman Spectrosc. 2021, 52, 1798–1809. [Google Scholar] [CrossRef]
- Li, Q.; Shen, J.; Zhou, Y. Diagnosis of Glioma Using Raman Spectroscopy and the Entropy Weight Fuzzy-Rough Nearest Neighbor (EFRNN) Algorithm on Fresh Tissue. Anal. Lett. 2023, 56, 895–905. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, Y.; Song, W.; Lu, L. Delineating the tumor margin with intraoperative surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2019, 411, 3993–4006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Li, Y.; Xu, H.; Yang, L.; Shan, P.; Du, Y.; Yan, X.; Chen, X. Raman spectroscopy: A prospective intraoperative visualization technique for gliomas. Front. Oncol. 2022, 12, 1086643. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Yue, Q.; Duan, W.; Sui, A.; Zhao, B.; Deng, Y.; Zhai, Y.; Zhang, Y.; Sun, T.; Zhang, G.P.; et al. Intelligent SERS Navigation System Guiding Brain Tumor Surgery by Intraoperatively Delineating the Metabolic Acidosis. Adv. Sci. 2022, 9, e2104935. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, K.; Qu, X.; Xu, W.; Xu, S. Ratiometric pH-responsive SERS strategy for glioma boundary determination. Talanta 2022, 250, 123750. [Google Scholar] [CrossRef] [PubMed]
- Strobbia, P.; Cupil-Garcia, V.; Crawford, B.M.; Fales, A.M.; Pfefer, T.J.; Liu, Y.; Maiwald, M.; Sumpf, B.; Vo-Dinh, T. Accurate in vivo tumor detection using plasmonic-enhanced shifted-excitation Raman difference spectroscopy (SERDS). Theranostics 2021, 11, 4090–4102. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Primers 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404. [Google Scholar] [CrossRef]
- Thal, D.R.; Rub, U.; Orantes, M.; Braak, H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology 2002, 58, 1791–1800. [Google Scholar] [CrossRef]
- Gao, F.; Li, F.; Wang, J.; Yu, H.; Li, X.; Chen, H.; Wang, J.; Qin, D.; Li, Y.; Liu, S.; et al. SERS-Based Optical Nanobiosensors for the Detection of Alzheimer’s Disease. Biosensors 2023, 13, 880. [Google Scholar] [CrossRef]
- Yu, D.; Yin, Q.; Wang, J.; Yang, J.; Chen, Z.; Gao, Z.; Huang, Q.; Li, S. SERS-Based Immunoassay Enhanced with Silver Probe for Selective Separation and Detection of Alzheimer’s Disease Biomarkers. Int. J. Nanomed. 2021, 16, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Fei, R.; Lu, Y.; Wan, Y.; Wu, X.; Dong, J.; Meng, D.; Ge, Q.; Zhao, X. Ultrasensitive detection of multiple Alzheimer’s disease biomarkers by SERS-LFA. Analyst 2022, 147, 4124–4131. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, C.; Cabinio, M.; Picciolini, S.; Gualerzi, A.; Baglio, F.; Bedoni, M. SERS-based biosensor for Alzheimer disease evaluation through the fast analysis of human serum. J. Biophotonics 2020, 13, e201960033. [Google Scholar] [CrossRef] [PubMed]
- Alexander, G.C.; Emerson, S.; Kesselheim, A.S. Evaluation of Aducanumab for Alzheimer Disease: Scientific Evidence and Regulatory Review Involving Efficacy, Safety, and Futility. JAMA 2021, 325, 1717–1718. [Google Scholar] [CrossRef] [PubMed]
- Soderberg, L.; Johannesson, M.; Nygren, P.; Laudon, H.; Eriksson, F.; Osswald, G.; Moller, C.; Lannfelt, L. Lecanemab, Aducanumab, and Gantenerumab—Binding Profiles to Different Forms of Amyloid-Beta Might Explain Efficacy and Side Effects in Clinical Trials for Alzheimer’s Disease. Neurotherapeutics 2023, 20, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R.B.; Berg, D.; Adler, C.H.; Bloem, B.R.; Chan, P.; Deuschl, G.; Gasser, T.; Goetz, C.G.; Halliday, G.; Joseph, L.; et al. The new definition and diagnostic criteria of Parkinson’s disease. Lancet Neurol. 2016, 15, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Morato Torres, C.A.; Wassouf, Z.; Zafar, F.; Sastre, D.; Outeiro, T.F.; Schule, B. The Role of Alpha-Synuclein and Other Parkinson’s Genes in Neurodevelopmental and Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 5724. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. Neurodegeneration and the ordered assembly of alpha-synuclein. Cell Tissue Res. 2018, 373, 137–148. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Brundin, P.; Melki, R. Prying into the Prion Hypothesis for Parkinson’s Disease. J. Neurosci. 2017, 37, 9808–9818. [Google Scholar] [CrossRef]
- Fairfoul, G.; McGuire, L.I.; Pal, S.; Ironside, J.W.; Neumann, J.; Christie, S.; Joachim, C.; Esiri, M.; Evetts, S.G.; Rolinski, M.; et al. Alpha-synuclein RT-QuIC in the CSF of patients with alpha-synucleinopathies. Ann. Clin. Transl. Neurol. 2016, 3, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Okuzumi, A.; Hatano, T.; Matsumoto, G.; Nojiri, S.; Ueno, S.I.; Imamichi-Tatano, Y.; Kimura, H.; Kakuta, S.; Kondo, A.; Fukuhara, T.; et al. Propagative alpha-synuclein seeds as serum biomarkers for synucleinopathies. Nat. Med. 2023, 29, 1448–1455. [Google Scholar] [CrossRef]
- Feigin, V.L.; Brainin, M.; Norrving, B.; Martins, S.; Sacco, R.L.; Hacke, W.; Fisher, M.; Pandian, J.; Lindsay, P. World Stroke Organization (WSO): Global Stroke Fact Sheet 2022. Int. J. Stroke 2022, 17, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, W.; Tseng, M.-C.; Sandercock, P. Blood Biomarkers in the Diagnosis of Ischemic Stroke. Stroke 2008, 39, 2902–2909. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-B.; Li, J.-J.; Weng, G.-J.; Zhu, J.; Guo, Y.-B.; Zhao, J.-W. An anisotropic nanobox based core-shell-satellite nanoassembly of multiple SERS enhancement with heterogeneous interface for stroke marker determination. J. Colloid Interface Sci. 2023, 647, 81–92. [Google Scholar] [CrossRef]
- Schildkraut, J.J. The catecholamine hypothesis of affective disorders: A review of supporting evidence. Am. J. Psychiatry 1965, 122, 509–522. [Google Scholar] [CrossRef]
- Farley, I.J.; Shannak, K.S.; Hornykiewicz, O. Brain monoamine changes in chronic paranoid schizophrenia and their possible relation to increased dopamine receptor sensitivity. Adv. Biochem. Psychopharmacol. 1980, 21, 427–433. [Google Scholar]
- Brown, W.C.; Schiffman, D.O.; Swinyard, E.A.; Goodman, L.S. Comparative assay of an antiepileptic drugs by psychomotor seizure test and minimal electroshock threshold test. J. Pharmacol. Exp. Ther. 1953, 107, 273–283. [Google Scholar]
- Hornykiewicz, O. The metabolism of brain dopamine in human parkinsonism. Riv. Patol. Nerv. Ment. 1970, 91, 281–286. [Google Scholar]
- Perry, E.K.; Perry, R.H.; Tomlinson, B.E.; Blessed, G.; Gibson, P.H. Coenzyme A-acetylating enzymes in Alzheimer’s disease: Possible cholinergic ‘compartment’ of pyruvate dehydrogenase. Neurosci. Lett. 1980, 18, 105–110. [Google Scholar] [CrossRef]
- Lussier, F.; Brule, T.; Bourque, M.J.; Ducrot, C.; Trudeau, L.E.; Masson, J.F. Dynamic SERS nanosensor for neurotransmitter sensing near neurons. Faraday Discuss 2017, 205, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jeon, C.S.; Kim, K.B.; Kim, H.J.; Pyun, S.H.; Park, Y.M. Quantitative detection of dopamine in human serum with surface-enhanced Raman scattering (SERS) of constrained vibrational mode. Talanta 2023, 260, 124590. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Pisano, F.; Collard, L.; Balena, A.; Pisanello, M.; Spagnolo, B.; Mach-Batlle, R.; Tantussi, F.; Carbone, L.; De Angelis, F.; et al. Toward Plasmonic Neural Probes: SERS Detection of Neurotransmitters through Gold-Nanoislands-Decorated Tapered Optical Fibers with Sub-10 nm Gaps. Adv. Mater. 2023, 35, e2200902. [Google Scholar] [CrossRef] [PubMed]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef]
- Moody, A.S.; Sharma, B. Multi-metal, Multi-wavelength Surface-Enhanced Raman Spectroscopy Detection of Neurotransmitters. ACS Chem. Neurosci. 2018, 9, 1380–1387. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsheikh, S.; Coles, N.P.; Achadu, O.J.; Filippou, P.S.; Khundakar, A.A. Advancing Brain Research through Surface-Enhanced Raman Spectroscopy (SERS): Current Applications and Future Prospects. Biosensors 2024, 14, 33. https://doi.org/10.3390/bios14010033
Elsheikh S, Coles NP, Achadu OJ, Filippou PS, Khundakar AA. Advancing Brain Research through Surface-Enhanced Raman Spectroscopy (SERS): Current Applications and Future Prospects. Biosensors. 2024; 14(1):33. https://doi.org/10.3390/bios14010033
Chicago/Turabian StyleElsheikh, Suzan, Nathan P. Coles, Ojodomo J. Achadu, Panagiota S. Filippou, and Ahmad A. Khundakar. 2024. "Advancing Brain Research through Surface-Enhanced Raman Spectroscopy (SERS): Current Applications and Future Prospects" Biosensors 14, no. 1: 33. https://doi.org/10.3390/bios14010033
APA StyleElsheikh, S., Coles, N. P., Achadu, O. J., Filippou, P. S., & Khundakar, A. A. (2024). Advancing Brain Research through Surface-Enhanced Raman Spectroscopy (SERS): Current Applications and Future Prospects. Biosensors, 14(1), 33. https://doi.org/10.3390/bios14010033





