MNAzyme-Assisted Nucleic Acid Lateral Flow Assay for Cost-Effective, On-Site Mercury Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Urea Polyacrylamide Gel Electrophoresis (Urea-PAGE) Analysis
2.3. Synthesis of Gold Nanoparticles (AuNPs)
2.4. Modification of AuNPs with DNA
2.5. Preparation of the NALFA Strip and Immobilization of Capture DNA
2.6. Hg2+ Detection Assay
2.7. Detection of Hg2+ in Tap Water
3. Results and Discussion
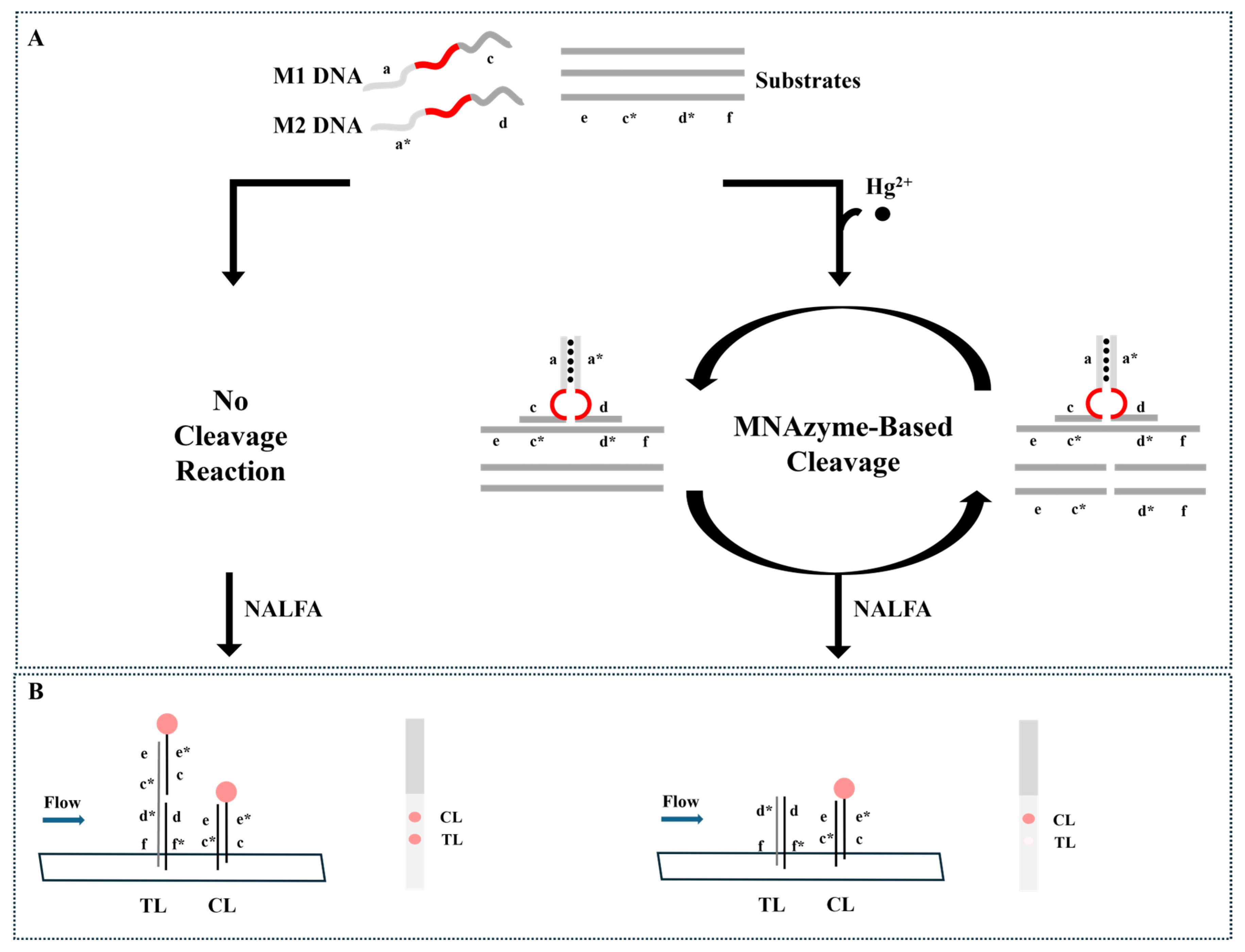
3.1. Hg2+ Detection Strategy
3.2. Detection Feasibility
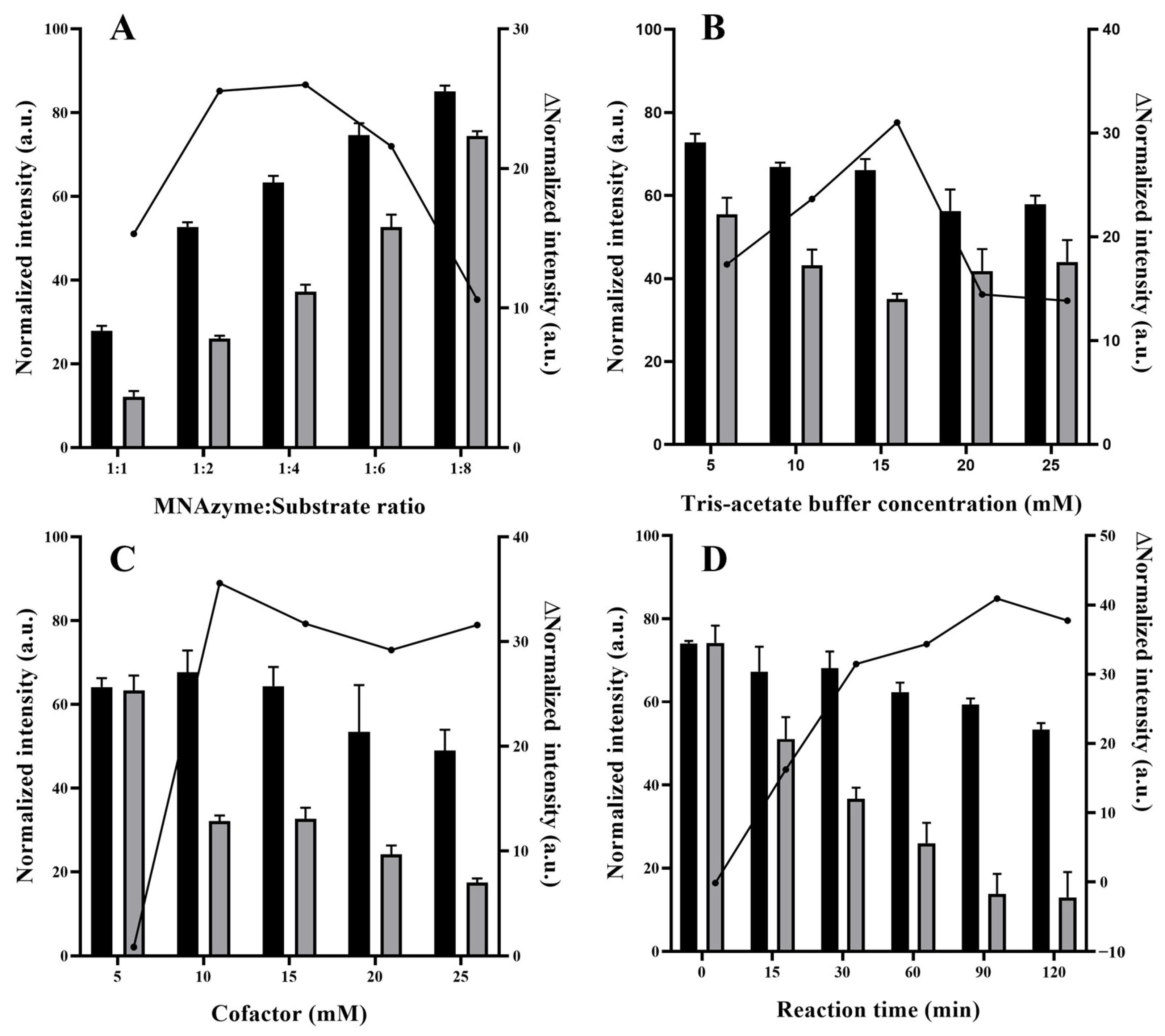
3.3. Optimization of the Hg2+ Detection Assay
3.4. Sensitivity and Specificity of the Hg2+ Detection Assay
3.5. Practical Application of the Hg2+ Detection Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.R.; Wang, J.J.; Zheng, Y.M.; Zhang, L.M.; He, J.Z. Patterns of Bacterial Diversity Along a Long-Term Mercury-Contaminated Gradient in the Paddy Soils. Microb. Ecol. 2014, 68, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chang, H.; Hirata, A.; Wu, H.; Xue, Q.K.; Chen, M. Nanoporous Gold Based Optical Sensor for Sub-Ppt Detection of Mercury Ions. ACS Nano 2013, 7, 4595–4600. [Google Scholar] [CrossRef] [PubMed]
- Selid, P.D.; Xu, H.; Collins, E.M.; Face-Collins, M.S.; Zhao, J.X. Sensing Mercury for Biomedical and Environmental Monitoring. Sensors 2009, 9, 5446–5459. [Google Scholar] [CrossRef]
- Li, Y.L.; Leng, Y.M.; Zhang, Y.J.; Li, T.H.; Shen, Z.Y.; Wu, A.G. A New Simple and Reliable Hg2+ Detection System Based on Anti-Aggregation of Unmodified Gold Nanoparticles in the Presence of O-Phenylenediamine. Sens. Actuators B Chem. 2014, 200, 140–146. [Google Scholar] [CrossRef]
- Chen, X.; Han, C.; Cheng, H.; Wang, Y.; Liu, J.; Xu, Z.; Hu, L. Rapid Speciation Analysis of Mercury in Seawater and Marine Fish by Cation Exchange Chromatography Hyphenated with Inductively Coupled Plasma Mass Spectrometry. J. Chromatogr. A 2013, 1314, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Shoaee, H.; Roshdi, M.; Khanlarzadeh, N.; Beiraghi, A. Simultaneous Preconcentration of Copper and Mercury in Water Samples by Cloud Point Extraction and Their Determination by Inductively Coupled Plasma Atomic Emission Spectrometry. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 98, 70–75. [Google Scholar] [CrossRef]
- Francisco, B.B.A.; Rocha, A.A.; Grinberg, P.; Sturgeon, R.E.; Cassella, R.J. Determination of Inorganic Mercury in Petroleum Production Water by Inductively Coupled Plasma Optical Emission Spectrometry Following Photochemical Vapor Generation. J. Anal. At. Spectrom. 2016, 31, 751–758. [Google Scholar] [CrossRef]
- Leermakers, M.; Baeyens, W.; Quevauviller, P.; Horvat, M. Mercury in Environmental Samples: Speciation, Artifacts and Validation. TrAC Trends Anal. Chem. 2005, 24, 383–393. [Google Scholar] [CrossRef]
- Ramalhosa, E.; Segade, S.R.; Pereira, E.; Vale, C.; Duarte, A. Microwave-Assisted Extraction for Methylmercury Determination in Sediments by High Performance Liquid Chromatography-Cold Vapour-Atomic Fluorescence Spectrometry. J. Anal. At. Spectrom. 2001, 16, 643–647. [Google Scholar] [CrossRef]
- Miyake, Y.; Togashi, H.; Tashiro, M.; Yamaguchi, H.; Oda, S.; Kudo, M.; Tanaka, Y.; Kondo, Y.; Sawa, R.; Fujimoto, T.; et al. MercuryII-Mediated Formation of Thymine-HgII-Thymine Base Pairs in DNA Duplexes. J. Am. Chem. Soc. 2006, 128, 2172–2173. [Google Scholar] [CrossRef]
- Ono, A.; Togashi, H. Highly Selective Oligonucleotide-Based Sensor for Mercury(II) in Aqueous Solutions. Angew. Chem. Int. Ed. 2004, 43, 4300–4302. [Google Scholar] [CrossRef] [PubMed]
- Torigoe, H.; Ono, A.; Kozasa, T. HgII Ion Specifically Binds with T:T Mismatched Base Pair in Duplex DNA. Chem. —A Eur. J. 2010, 16, 13218–13225. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. Mechanism and Utility of an RNA-Cleaving DNA Enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Y.; Huo, W.; Song, Y.; Chen, Y.; Zhang, J.; Yang, X.; Jin, Y.; Liang, X.J. RNA-Cleaving DNAzymes for Accurate Biosensing and Gene Therapy. Nanoscale 2023, 15, 11346–11365. [Google Scholar] [CrossRef] [PubMed]
- Mokany, E.; Bone, S.M.; Young, P.E.; Doan, T.B.; Todd, A.V. MNAzymes, a Versatile New Class of Nucleic Acid Enzymes That Can Function as Biosensors and Molecular Switches. J. Am. Chem. Soc. 2010, 132, 1051–1059. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, H.; Le, X.C. Split Locations and Secondary Structures of a DNAzyme Critical to Binding-Assembled Multicomponent Nucleic Acid Enzymes for Protein Detection. Anal. Chem. 2021, 93, 15712–15719. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.J.J.; Liu, J. Sensing Parts-per-Trillion Cd2+, Hg2+, and Pb2+ Collectively and Individually Using Phosphorothioate DNAzymes. Anal. Chem. 2014, 86, 5999–6005. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Rational Design of “Turn-On” Allosteric DNAzyme Catalytic Beacons for Aqueous Mercury Ions with Ultrahigh Sensitivity and Selectivity. Angew. Chem. Int. Ed. 2007, 46, 7587–7590. [Google Scholar] [CrossRef]
- Yun, W.; Li, F.; Liu, X.; Li, N.; Chen, L.; Yang, L. A Catalytic Cleavage Strategy for Fluorometric Determination of Hg(II) Based on the Use of a Mg(II)-Dependent Split DNAzyme and Hairpins Conjugated to Gold Nanoparticles. Microchim. Acta 2018, 185, 457. [Google Scholar] [CrossRef]
- Qi, L.; Zhao, Y.; Yuan, H.; Bai, K.; Zhao, Y.; Chen, F.; Dong, Y.; Wu, Y. Amplified Fluorescence Detection of Mercury(II) Ions (Hg2+) Using Target-Induced DNAzyme Cascade with Catalytic and Molecular Beacons. Analyst 2012, 137, 2799–2805. [Google Scholar] [CrossRef]
- Gong, L.; Du, B.; Pan, L.; Liu, Q.; Yang, K.; Wang, W.; Zhao, H.; Wu, L.; He, Y. Colorimetric Aggregation Assay for Arsenic(III) Using Gold Nanoparticles. Microchim. Acta 2017, 184, 1185–1190. [Google Scholar] [CrossRef]
- Huang, M.; Xiong, E.; Wang, Y.; Hu, M.; Yue, H.; Tian, T.; Zhu, D.; Liu, H.; Zhou, X. Fast Microwave Heating-Based One-Step Synthesis of DNA and RNA Modified Gold Nanoparticles. Nat. Commun. 2022, 13, 968. [Google Scholar] [CrossRef]
- Luka, G.S.; Nowak, E.; Toyata, Q.R.; Tasnim, N.; Najjaran, H.; Hoorfar, M. Portable On-Chip Colorimetric Biosensing Platform Integrated with a Smartphone for Label/PCR-Free Detection of Cryptosporidium RNA. Sci. Rep. 2021, 11, 23192. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, S.; Han, J.; Kim, J.H.; Park, K.S. Equipment-Free, Salt-Mediated Immobilization of Nucleic Acids for Nucleic Acid Lateral Flow Assays. Sens. Actuators B Chem. 2022, 351, 130975. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Kim, S.H.; Cha, B.S.; Lee, E.S.; Kim, S.; Park, K.S. Primer Exchange Reaction-Coupled Transcription Isothermal Amplification as a Sensitive Biomolecular Assay. Chem. Commun. 2024, 60, 4565–4568. [Google Scholar] [CrossRef] [PubMed]
- Cha, B.S.; Jang, Y.J.; Lee, E.S.; Kim, D.Y.; Woo, J.S.; Son, J.; Kim, S.; Shin, J.; Han, J.; Kim, S.; et al. Development of a Novel DNA Aptamer Targeting Colorectal Cancer Cell-Derived Small Extracellular Vesicles as a Potential Diagnostic and Therapeutic Agent. Adv. Healthc. Mater. 2023, 12, 2300854. [Google Scholar] [CrossRef]
- Lim, J.W.; Kim, T.Y.; Choi, S.W.; Woo, M.A. 3D-Printed Rolling Circle Amplification Chip for on-Site Colorimetric Detection of Inorganic Mercury in Drinking Water. Food Chem. 2019, 300, 125177. [Google Scholar] [CrossRef]
- Li, T.; Dong, S.; Wang, E. Label-Free Colorimetric Detection of Aqueous Mercury Ion (Hg2+) Using Hg2+-Modulated G-Quadruplex-Based Dnazymes. Anal. Chem. 2009, 81, 2144–2149. [Google Scholar] [CrossRef]
- Li, T.; Li, B.; Wang, E.; Dong, S. G-Quadruplex-Based DNAzyme for Sensitive Mercury Detection with the Naked Eye. Chem. Commun. 2009, 24, 3551–3553. [Google Scholar] [CrossRef]
- Zhang, D.; Deng, M.; Xu, L.; Zhou, Y.; Yuwen, J.; Zhou, X.; Zhang, D.; Deng, M.; Xu, L.; Zhou, Y.; et al. The Sensitive and Selective Optical Detection of Mercury(II) Ions by Using a Phosphorothioate DNAzyme Strategy. Chem.–Eur. J. 2009, 15, 8117–8120. [Google Scholar] [CrossRef]


| Sample | Added (ppb) | ICP-OES (ppb) | Found (ppb) a | SD b | CV (%) c | Recovery (%) d |
|---|---|---|---|---|---|---|
| Tap water | 10 | 12 | 11.07 | 0.30 | 2.69 | 92.23 |
| 15 | 15 | 16.59 | 0.35 | 2.12 | 110.61 | |
| 25 | 22 | 24.92 | 0.42 | 1.70 | 113.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Kim, Y.; Kim, S.; Lee, E.S.; Cha, B.S.; Park, K.S. MNAzyme-Assisted Nucleic Acid Lateral Flow Assay for Cost-Effective, On-Site Mercury Detection. Biosensors 2024, 14, 454. https://doi.org/10.3390/bios14100454
Kim SH, Kim Y, Kim S, Lee ES, Cha BS, Park KS. MNAzyme-Assisted Nucleic Acid Lateral Flow Assay for Cost-Effective, On-Site Mercury Detection. Biosensors. 2024; 14(10):454. https://doi.org/10.3390/bios14100454
Chicago/Turabian StyleKim, Seok Hyeon, Yujun Kim, Seokjoon Kim, Eun Sung Lee, Byung Seok Cha, and Ki Soo Park. 2024. "MNAzyme-Assisted Nucleic Acid Lateral Flow Assay for Cost-Effective, On-Site Mercury Detection" Biosensors 14, no. 10: 454. https://doi.org/10.3390/bios14100454
APA StyleKim, S. H., Kim, Y., Kim, S., Lee, E. S., Cha, B. S., & Park, K. S. (2024). MNAzyme-Assisted Nucleic Acid Lateral Flow Assay for Cost-Effective, On-Site Mercury Detection. Biosensors, 14(10), 454. https://doi.org/10.3390/bios14100454






