Recent Advance in Single-Molecule Fluorescent Biosensors for Tumor Biomarker Detection
Abstract
1. Introduction
2. Single-Molecule Fluorescent Biosensors
3. Biosensing Applications
3.1. Detection of DNAs and DNA Modifications
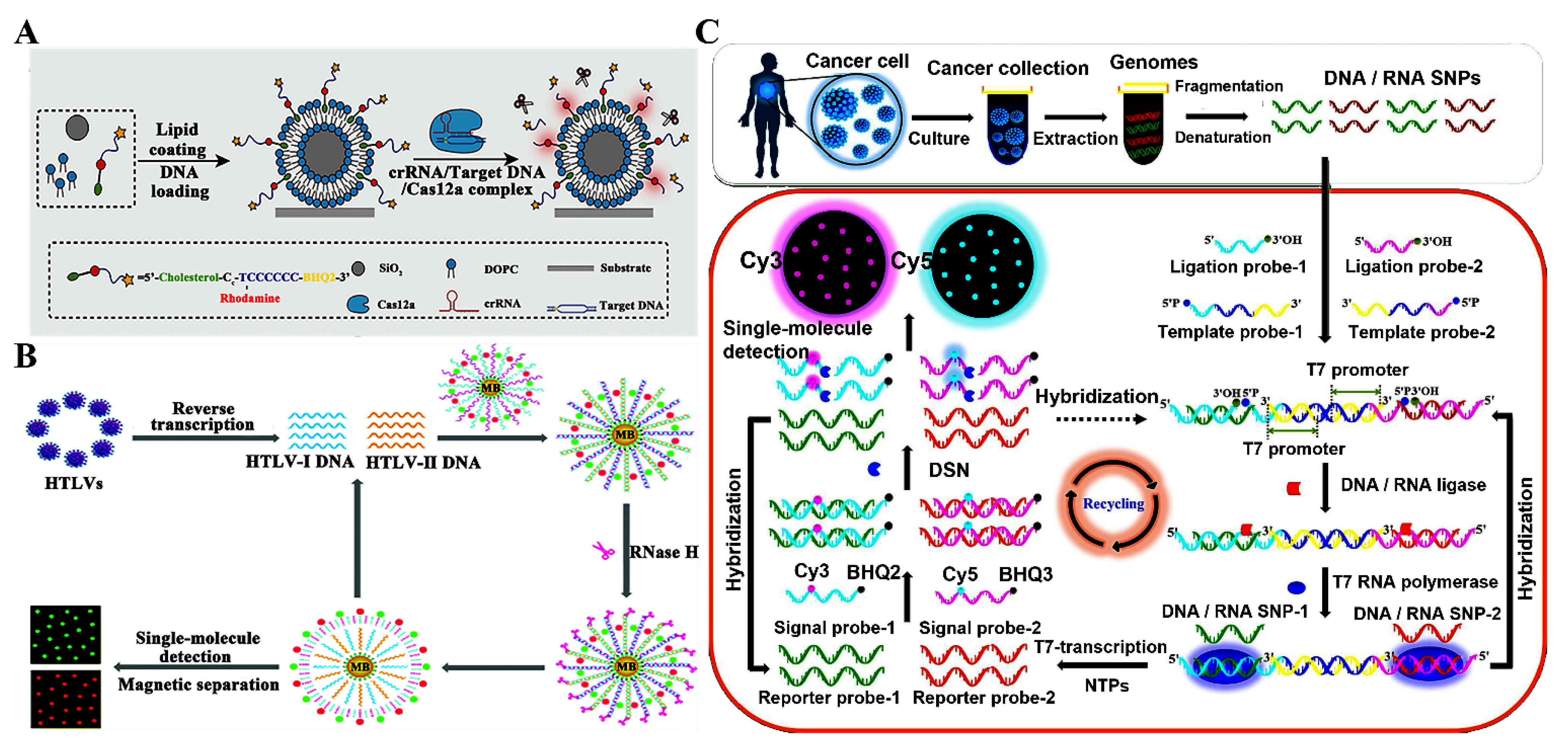
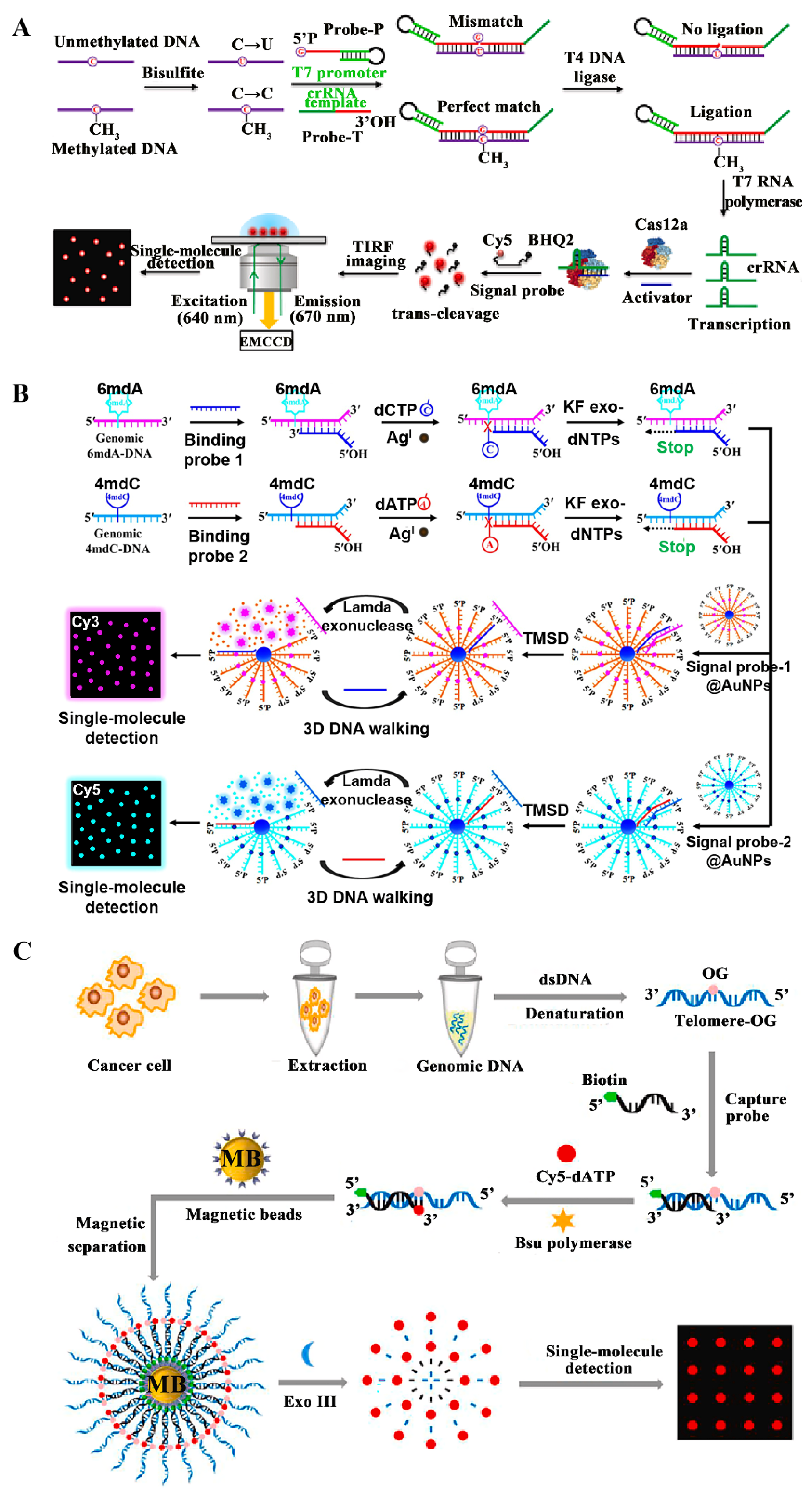
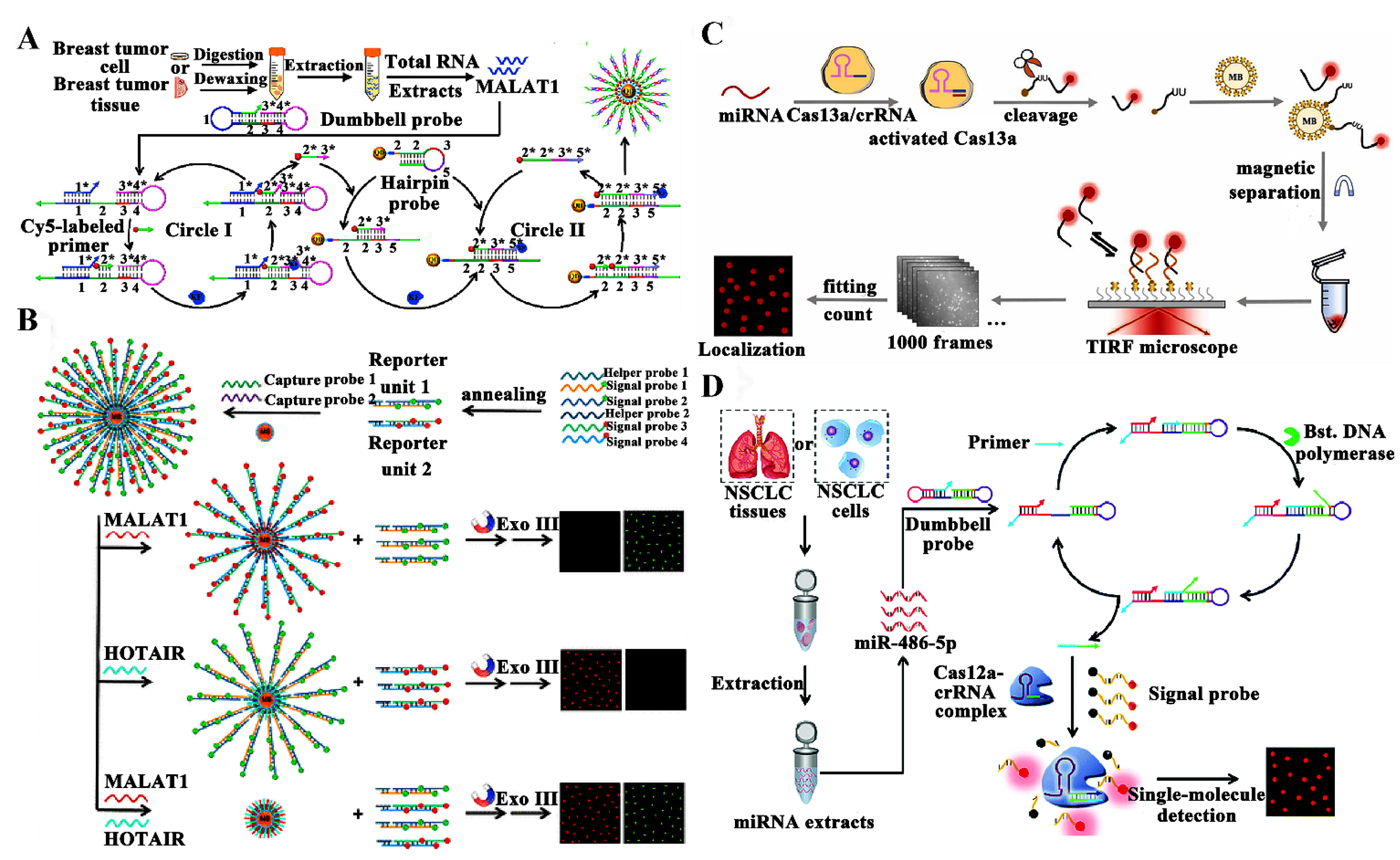
3.2. Detection of RNAs

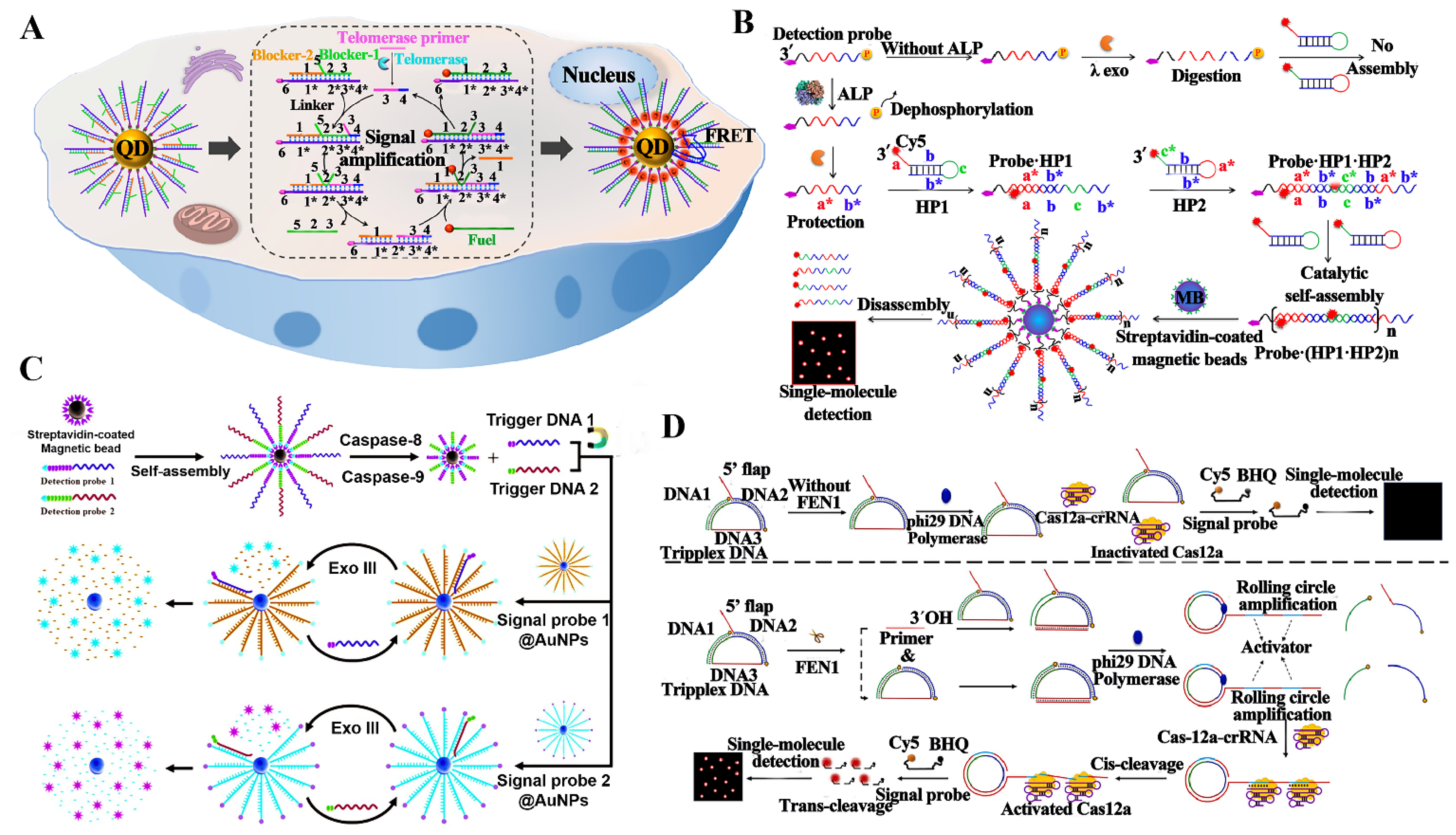
3.3. Detection of Enzymes

4. Conclusions
Funding
Conflicts of Interest
References
- Zhang, P.; Wang, R. Label-Free Biosensor. Biosensors 2023, 13, 556. [Google Scholar] [CrossRef]
- Yao, B.; Yao, J.T.; Fan, Z.Q.; Zhao, J.F.; Zhang, K.; Huang, W. Rapid Advances of Versatile MXenes for Electrochemical Enzyme-Based Biosensors, Immunosensors, and Nucleic Acid-Based Biosensors. ChemElectroChem 2022, 9, 00103. [Google Scholar] [CrossRef]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2016, 88, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Spanos, A.; Sedgwick, S.G.; Yarranton, G.T.; Hübscher, U.; Banks, G.R. Detection of the catalytic activities of DNA polymerases and their associated exonucleases following SDS-polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981, 9, 1825–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Z.; Wang, S.; Cheng, F.; Chen, L. Iodine-Mediated Etching of Gold Nanorods for Plasmonic ELISA Based on Colorimetric Detection of Alkaline Phosphatase. ACS Appl. Mater. Interfaces 2015, 7, 27639–27645. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Yin, B.C.; Wang, S.; Xu, Z.; Ye, B.C. Improved ligation-mediated PCR method coupled with T7 RNA polymerase for sensitive DNA detection. Anal. Chem. 2014, 86, 7214–7218. [Google Scholar] [CrossRef] [PubMed]
- Harrer, S.; Kim, S.C.; Schieber, C.; Kannam, S.; Gunn, N.; Moore, S.; Scott, D.; Bathgate, R.; Skafidas, S.; Wagner, J.M. Label-free screening of single biomolecules through resistive pulse sensing technology for precision medicine applications. Nanotechnology 2015, 26, 182502. [Google Scholar] [CrossRef]
- Zhang, C.; Hu, H.; Ma, C.; Li, Y.; Wang, X.; Li, D.; Movsesyan, A.; Wang, Z.; Govorov, A.; Gan, Q.; et al. Quantum plasmonics pushes chiral sensing limit to single molecules: A paradigm for chiral biodetections. Nat. Commun. 2024, 15, 2. [Google Scholar] [CrossRef]
- Herkert, E.K.; Bermeo Alvaro, D.R.; Recchia, M.; Langbein, W.; Borri, P.; Garcia-Parajo, M.F. Hybrid Plasmonic Nanostructures for Enhanced Single-Molecule Detection Sensitivity. ACS Nano 2023, 17, 8453–8464. [Google Scholar] [CrossRef]
- Hupfel, M.; Yu Kobitski, A.; Zhang, W.; Nienhaus, G.U. Wavelet-based background and noise subtraction for fluorescence microscopy images. Biomed. Opt. Express 2021, 12, 969–980. [Google Scholar] [CrossRef]
- Kang, S.; Nieuwenhuis, A.F.; Mathwig, K.; Mampallil, D.; Lemay, S.G. Electrochemical single-molecule detection in aqueous solution using self-aligned nanogap transducers. ACS Nano 2013, 7, 10931–10937. [Google Scholar] [CrossRef] [PubMed]
- Mia, A.K.; Meyyappan, M.; Giri, P.K. Two-Dimensional Transition Metal Dichalcogenide Based Biosensors: From Fundamentals to Healthcare Applications. Biosensors 2023, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Y.; Zhang, K.; Ji, J.; Liu, J.; Liu, B. Single Molecule Fluorescent Colocalization of Split Aptamers for Ultrasensitive Detection of Biomolecules. Anal. Chem. 2018, 90, 9315–9321. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Yi, N.; Jiang, D. Advances in single molecule arrays (SIMOA) for ultra-sensitive detection of biomolecules. Talanta 2024, 270, 125529. [Google Scholar] [CrossRef]
- Stabile, F.; Shaheen, C.; Scott, S.; Berard, D.; Levens, D.; Benham, C.; Leslie, S. Single Molecule Imaging of DNA Structure: CLIC Microscopy Powers Mechanistic Insights for Drug Development. Biophys. J. 2019, 116, 475a. [Google Scholar] [CrossRef]
- Firman, K.; Evans, L.; Youell, J. A Synthetic Biology Project—Developing a single-molecule device for screening drug-target interactions. FEBS Lett. 2012, 586, 2157–2163. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Chen, P.F.; He, S.W.X.; Jiang, B.W.; Wang, Y.; Cheng, Y.H.; Peng, J.; Verpoort, F.; Wang, J.H.; Kou, Z.K. Single-atom metal-nitrogen-carbon catalysts energize single molecule detection for biosensing. InfoMat 2023, 5, e12421. [Google Scholar] [CrossRef]
- Omair, Z.; Talukder, M.A. Sensitivity Analysis of Gold Nanorod Biosensors for Single Molecule Detection. Plasmonics 2019, 14, 1611–1619. [Google Scholar] [CrossRef]
- Ochmann, S.E.; Vietz, C.; Trofymchuk, K.; Acuna, G.P.; Lalkens, B.; Tinnefeld, P. Optical Nanoantenna for Single Molecule-Based Detection of Zika Virus Nucleic Acids without Molecular Multiplication. Anal. Chem. 2017, 89, 13000–13007. [Google Scholar] [CrossRef]
- Kim, H.; Ha, T. Single-molecule nanometry for biological physics. Rep. Prog. Phys. 2013, 76, 016601. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, X.; Li, D.L.; Wang, T.T.; Ma, F.; Zhang, C.Y. Construction of a gold nanoparticle-based single-molecule biosensor for simple and sensitive detection of Argonaute 2 activity. J. Mater. Chem. B 2022, 10, 5594–5601. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Meng, Y.R.; Hu, J.; Qiu, J.G.; Zhang, C.Y. Development of a single-molecule biosensor with an ultra-low background for the simultaneous detection of multiple retroviral DNAs. J. Mater. Chem. B 2022, 10, 5465–5472. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, C.C.; Zhang, C.Y. Nucleic acid amplification-integrated single-molecule fluorescence imaging for in vitro and in vivo biosensing. Chem. Commun. 2021, 57, 13415–13428. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Lee, G.; Kang, S.H. Simultaneous quantification of thyroid hormones using an ultrasensitive single-molecule fourplex nanoimmunosensor in an evanescent field. Biosens. Bioelectron. 2023, 220, 114894. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.A.; Kang, S.H. Wavelength-dependent three-dimensional single-molecule superlocalization imaging for yoctomole detection of thyroid-stimulating hormone on a quantum dot nanobiosensor. Chin. Chem. Lett. 2023, 34, 108383. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Zhang, S.; Li, C.; Ma, J. Amplification-Free, Single-Microbead-Based Cas12a Assay for One-Step DNA Detection at the Single-Molecule Level. Anal. Chem. 2022, 94, 13076–13083. [Google Scholar] [CrossRef]
- Chen, S.; Sun, Y.; Xia, Y.; Lv, K.; Man, B.; Yang, C. Donor effect dominated molybdenum disulfide/graphene nanostructure-based field-effect transistor for ultrasensitive DNA detection. Biosens. Bioelectron. 2020, 156, 112128. [Google Scholar] [CrossRef]
- Suhailin, F.H.; Alwahib, A.A.; Chee, H.Y.; Mustafa, F.H.; Hamzah, A.S.A.; Zainuddin, N.H.; Omar, M.F.; Razak, A.A.A.; Mahdi, M.A. Fiber-Based Surface Plasmon Resonance Biosensor for DNA Detection. IEEE Sens. J. 2022, 22, 20516–20523. [Google Scholar] [CrossRef]
- Cha, B.H.; Lee, S.-M.; Park, J.C.; Hwang, K.S.; Kim, S.K.; Lee, Y.-S.; Ju, B.-K.; Kim, T.S. Detection of Hepatitis B Virus (HBV) DNA at femtomolar concentrations using a silica nanoparticle-enhanced microcantilever sensor. Biosens. Bioelectron. 2009, 25, 130–135. [Google Scholar] [CrossRef]
- Lu, Y.; Peng, S.; Luo, D.; Lal, A. Low-concentration mechanical biosensor based on a photonic crystal nanowire array. Nat. Commun. 2011, 2, 578. [Google Scholar] [CrossRef]
- Song, W.; Guo, X.; Sun, W.; Yin, W.; He, P.; Yang, X.; Zhang, X. Target-triggering multiple-cycle signal amplification strategy for ultrasensitive detection of DNA based on QCM and SPR. Anal. Biochem. 2018, 553, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Grimm, J.B.; Lavis, L.D. Caveat fluorophore: An insiders’ guide to small-molecule fluorescent labels. Nat. Methods 2021, 19, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Schneckenburger, H. Förster resonance energy transfer-what can we learn and how can we use it? Methods Appl. Fluoresc. 2019, 8, 013001. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, K.; Rai, H.; Mondal, S. Quantum dots: An overview of synthesis, properties, and applications. Mater. Res. Express 2023, 10, 062001. [Google Scholar] [CrossRef]
- Park, J.; Lee, H.J.; Han, Y.K.; Kang, K.; Yi, J.M. Identification of DNA methylation biomarkers for evaluating cardiovascular disease risk from epigenome profiles altered by low-dose ionizing radiation. Clin. Epigenetics 2024, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Barros, B.; Oliveira, M.; Morais, S. Firefighters’ occupational exposure: Contribution from biomarkers of effect to assess health risks. Environ. Int. 2021, 156, 106704. [Google Scholar] [CrossRef]
- Gutteridge, A.; Rathbone, V.M.; Gibbons, R.; Bi, M.; Archard, N.; Davies, K.E.J.; Brown, J.; Plagnol, V.; Pillay, N.; Amary, F.; et al. Digital PCR analysis of circulating tumor DNA: A biomarker for chondrosarcoma diagnosis, prognostication, and residual disease detection. Cancer Med. 2017, 6, 2194–2202. [Google Scholar] [CrossRef]
- Groeneweg, J.W.; Roze, J.F.; Peters, E.D.J.; Sereno, F.; Brink, A.G.J.; Paijens, S.T.; Nijman, H.W.; van Meurs, H.S.; van Lonkhuijzen, L.; Piek, J.M.J.; et al. FOXL2 and TERT promoter mutation detection in circulating tumor DNA of adult granulosa cell tumors as biomarker for disease monitoring. Gynecol. Oncol. 2021, 162, 413–420. [Google Scholar] [CrossRef]
- Taksinwarajarn, T.; Sobhonslidsuk, A.; Kantachuvesiri, S.; Thongprayoon, C.; Cheungpasitporn, W.; Bruminhent, J.; Ramathibodi Transplant Infectious Diseases (RTID) group. Role of highly sensitive nucleic acid amplification testing for plasma cytomegalovirus DNA load in diagnosis of cytomegalovirus gastrointestinal disease among kidney transplant recipients. Transpl. Infect. Dis. 2021, 23, e13635. [Google Scholar] [CrossRef]
- Li, S.; Zeng, W.; Ni, X.; Liu, Q.; Li, W.; Stackpole, M.L.; Zhou, Y.; Gower, A.; Krysan, K.; Ahuja, P.; et al. Comprehensive tissue deconvolution of cell-free DNA by deep learning for disease diagnosis and monitoring. Proc. Natl. Acad. Sci. USA 2023, 120, e2305236120. [Google Scholar] [CrossRef]
- Li, P.; Liu, S.; Du, L.; Mohseni, G.; Zhang, Y.; Wang, C. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin. Epigenetics 2022, 14, 118. [Google Scholar] [CrossRef] [PubMed]
- Labugger, I.; Heyckendorf, J.; Dees, S.; Haussinger, E.; Herzmann, C.; Kohl, T.A.; Richter, E.; Rivera-Milla, E.; Lange, C. Detection of transrenal DNA for the diagnosis of pulmonary tuberculosis and treatment monitoring. Infection 2017, 45, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, I.R.; Sebastian, S. Novel viral vectors in infectious diseases. Immunology 2018, 153, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Smith, J.O. Infectious diseases: Predictions of virus spillover across species. Nature 2017, 546, 603–604. [Google Scholar] [CrossRef] [PubMed]
- Brites, C.; Miranda, F.; Luz, E.; Netto, E.M. Early and Successful Combination Antiretroviral Therapy Normalizes Survival Time in Patients Coinfected With Human Immunodeficiency Virus and Human T-cell Lymphotrophic Virus Type 1. Clin. Infect. Dis. 2020, 71, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.L.; Lee, T.H.; Chafets, D.; Nass, C.C.; Wang, B.; Loughlin, K.; Smith, D.; HTLV Outcomes Study Investigators. Higher human T lymphotropic virus (HTLV) provirus load is associated with HTLV-I versus HTLV-II, with HTLV-II subtype A versus B, and with male sex and a history of blood transfusion. J. Infect. Dis. 2004, 190, 504–510. [Google Scholar] [CrossRef]
- Ohmoto, A.; Fuji, S.; Kohmo, S.; Katsura, K. HTLV-I associated bronchioloalveolar disorder (HABA): Disease concept and differential diagnosis of an unsolved disease entity. Expert Rev. Anti-Infect. Ther. 2023, 21, 57–63. [Google Scholar] [CrossRef]
- Xu, T.; Sun, J.; Lv, J.; Watanabe, H.K.; Li, T.Q.; Zou, W.W.; Rouse, G.W.; Wang, S.; Qian, P.Y.; Bao, Z.M.; et al. Genome-wide discovery of single nucleotide polymorphisms (SNPs) and single nucleotide variants (SNVs) in deep-sea mussels: Potential use in population genomics and cross-species application. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 2017, 137, 318–326. [Google Scholar] [CrossRef]
- Cubizolles, N.; Rey, E.; Choulet, F.; Rimbert, H.; Laugier, C.; Balfourier, F.; Bordes, J.; Poncet, C.; Jack, P.; James, C.; et al. Exploiting the Repetitive Fraction of the Wheat Genome for High-Throughput Single-Nucleotide Polymorphism Discovery and Genotyping. Plant Genome 2016, 9, 78. [Google Scholar] [CrossRef]
- Tesfaye, M.; Feyissa, T.; Hailesilassie, T.; Kanagarajan, S.; Zhu, L.H. Genetic Diversity and Population Structure in Ethiopian Mustard (Brassica carinata A. Braun) as Revealed by Single Nucleotide Polymorphism Markers. Genes 2023, 14, 1757. [Google Scholar] [CrossRef]
- Fransen, N.L.; Crusius, J.B.A.; Smolders, J.; Mizee, M.R.; van Eden, C.G.; Luchetti, S.; Remmerswaal, E.B.M.; Hamann, J.; Mason, M.R.J.; Huitinga, I. Post-mortem multiple sclerosis lesion pathology is influenced by single nucleotide polymorphisms. Brain Pathol. 2020, 30, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Hull, B.P.; Jiramongkolchai, P.; Turner, J.H.; Olson, L.; Chandra, R.K. Single nucleotide polymorphisms related to cystic fibrosis in chronic rhinositus-a pilot study. Int. Forum Allergy Rhinol. 2017, 7, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Ershov, N.I.; Markel, A.L.; Redina, O.E. Strain-Specific Single-Nucleotide Polymorphisms in Hypertensive ISIAH Rats. Biochemistry 2017, 82, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Laxmi; Golmei, P.; Srivastava, S.; Kumar, S. Single nucleotide polymorphism-based biomarker in primary hypertension. Eur. J. Pharmacol. 2024, 972, 176584. [Google Scholar] [CrossRef]
- Meng, L.; Song, Z.; Liu, A.; Dahmen, U.; Yang, X.; Fang, H. Effects of Lipopolysaccharide-Binding Protein (LBP) Single Nucleotide Polymorphism (SNP) in Infections, Inflammatory Diseases, Metabolic Disorders and Cancers. Front. Immunol. 2021, 12, 681810. [Google Scholar] [CrossRef]
- Hua, T.; Zhang, C.; Fu, Y.; Qin, N.; Liu, S.; Chen, C.; Gong, L.; Ma, H.; Ding, Y.; Wei, X.; et al. Integrative analyses of N6-methyladenosine-associated single-nucleotide polymorphisms (m6A-SNPs) identify tumor suppressor gene AK9 in lung cancer. Mol. Carcinog. 2024, 63, 538–548. [Google Scholar] [CrossRef]
- Saikia, S.; Postwala, H.; Athilingam, V.P.; Anandan, A.; Padma, V.V.; Kalita, P.P.; Chorawala, M.; Prajapati, B. Single Nucleotide Polymorphisms (SNPs) in the Shadows: Uncovering their Function in Non-Coding Region of Esophageal Cancer. Curr. Pharm. Biotechnol. 2024, 25, 1915–1938. [Google Scholar] [CrossRef] [PubMed]
- Allemailem, K.S.; Almatroudi, A.; Alrumaihi, F.; Makki Almansour, N.; Aldakheel, F.M.; Rather, R.A.; Afroze, D.; Rah, B. Single nucleotide polymorphisms (SNPs) in prostate cancer: Its implications in diagnostics and therapeutics. Am. J. Transl. Res. 2021, 13, 3868. [Google Scholar]
- Wang, L.J.; Liu, X.W.; Zhang, X.Y.; Han, Y.; Liu, Q.; Wang, Z.Y.; Zhang, C.Y. Construction of a multiple ligation-driven exponentially symmetric T7-transcription machinery for single-molecule monitoring of diverse single-nucleotide polymorphisms in human cancers. Chem. Eng. J. 2024, 480, 148251. [Google Scholar] [CrossRef]
- Zhang, W.; Duan, N.; Zhang, Q.; Song, T.; Li, Z.; Zhang, C.; Chen, X.; Wang, K. DNA Methylation Mediated Down-Regulation of miR-370 Regulates Cell Growth through Activation of the Wnt/beta-Catenin Signaling Pathway in Human Osteosarcoma Cells. Int. J. Biol. Sci. 2017, 13, 561–573. [Google Scholar] [CrossRef]
- He, L.; Huang, H.; Bradai, M.; Zhao, C.; You, Y.; Ma, J.; Zhao, L.; Lozano-Duran, R.; Zhu, J.K. DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 2022, 13, 1335. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.A.; Stockwell, P.A.; Rodger, E.J.; Parry, M.F.; Eccles, M.R.; Stayner, C.; Chatterjee, A. Extensive Inter-Cyst DNA Methylation Variation in Autosomal Dominant Polycystic Kidney Disease Revealed by Genome Scale Sequencing. Front. Genet. 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Mazloumi, Z.; Farahzadi, R.; Rafat, A.; Dizaji Asl, K.; Karimipour, M.; Montazer, M.; Movassaghpour, A.A.; Dehnad, A.; Nozad Charoudeh, H. Effect of aberrant DNA methylation on cancer stem cell properties. Exp. Mol. Pathol. 2022, 125, 104757. [Google Scholar] [CrossRef] [PubMed]
- Zappe, K.; Cichna-Markl, M. Aberrant DNA Methylation of ABC Transporters in Cancer. Cells 2020, 9, 2281. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.W.; Ma, F.; Zhang, C.Y. Methylation-sensitive transcription-enhanced single-molecule biosensing of DNA methylation in cancer cells and tissues. Anal. Chim. Acta 2023, 1251, 340996. [Google Scholar] [CrossRef]
- Lv, H.; Dao, F.Y.; Zhang, D.; Yang, H.; Lin, H. Advances in mapping the epigenetic modifications of 5-methylcytosine (5mC), N6-methyladenine (6mA), and N4-methylcytosine (4mC). Biotechnol. Bioeng. 2021, 118, 4204–4216. [Google Scholar] [CrossRef]
- Ye, P.; Luan, Y.; Chen, K.; Liu, Y.; Xiao, C.; Xie, Z. MethSMRT: An integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2017, 45, D85–D89. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Xing, J.F.; Chen, W.; Luan, M.W.; Xie, R.; Huang, J.; Xie, S.Q.; Xiao, C.L. MDR: An integrative DNA N6-methyladenine and N4-methylcytosine modification database for Rosaceae. Hortic. Res. 2019, 6, 78. [Google Scholar] [CrossRef]
- Liang, D.; Wang, H.; Song, W.; Xiong, X.; Zhang, X.; Hu, Z.; Guo, H.; Yang, Z.; Zhai, S.; Zhang, L.H.; et al. The decreased N(6)-methyladenine DNA modification in cancer cells. Biochem. Biophys. Res. Commun. 2016, 480, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Wang, L.J.; Liu, Q.; Lu, Y.Y.; Liang, L.; Zhang, C.Y. Silver-Coordinated Watson-Crick Pairing-Driven Three-Dimensional DNA Walker for Locus-Specific Detection of Genomic N(6)-Methyladenine and N(4)-Methylcytosine at the Single-Molecule Level. Anal. Chem. 2024, 96, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, A.V.; Gudkov, S.V.; Usacheva, A.M.; Bruskov, V.I. Exogenous 8-Oxo-7,8-dihydro-2’-deoxyguanosine: Biomedical Properties, Mechanisms of Action, and Therapeutic Potential. Biochemistry 2017, 82, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Radak, Z.; Boldogh, I. 8-Oxo-7,8-dihydroguanine: Links to gene expression, aging, and defense against oxidative stress. Free Radic. Biol. Med. 2010, 49, 587–596. [Google Scholar] [CrossRef]
- Sassa, A.; Odagiri, M. Understanding the sequence and structural context effects in oxidative DNA damage repair. DNA Repair 2020, 93, 102906. [Google Scholar] [CrossRef] [PubMed]
- Ibuki, Y.; Warashina, T.; Noro, T.; Goto, R. Coexposure to benzo[a]pyrene plus ultraviolet A induces 8-oxo-7,8-dihydro-2’-deoxyguanosine formation in human skin fibroblasts: Preventive effects of anti-oxidant agents. Environ. Toxicol. Pharmacol. 2002, 12, 37–42. [Google Scholar] [CrossRef]
- Gonzalez-Rivera, J.C.; Baldridge, K.C.; Wang, D.S.; Patel, K.; Chuvalo-Abraham, J.C.L.; Hildebrandt Ruiz, L.; Contreras, L.M. Post-transcriptional air pollution oxidation to the cholesterol biosynthesis pathway promotes pulmonary stress phenotypes. Commun. Biol. 2020, 3, 392. [Google Scholar] [CrossRef]
- Kamiya, H.; Makino, T.; Suzuki, T.; Kobayashi, M.; Matsuoka, I. Mutations induced by 8-oxo-7,8-dihydroguanine in WRN- and DNA polymerase lambda-double knockdown cells. Mutagenesis 2018, 33, 301–310. [Google Scholar] [CrossRef]
- Liang, Y.D.; Liu, Q.; Du, M.H.; Liu, Z.; Yao, S.M.; Zheng, P.P.; Wan, Y.H.; Sun, N.; Li, Y.Y.; Liu, J.P.; et al. Urinary 8-oxo-7,8-dihydroguanosine as a potential biomarker of frailty for elderly patients with cardiovascular disease. Free Radic. Biol. Med. 2020, 152, 248–254. [Google Scholar] [CrossRef]
- European Standards Committee on Urinary (DNA) Lesion Analysis; Evans, M.D.; Olinski, R.; Loft, S.; Cooke, M.S. Toward consensus in the analysis of urinary 8-oxo-7,8-dihydro-2’-deoxyguanosine as a noninvasive biomarker of oxidative stress. FASEB J. 2010, 24, 1249–1260. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Z.; Wang, C.C.; Gao, X.; Li, C.B.; Wang, M.; Wang, Q.; Cai, J.P. Increased production of 8-oxo-7,8-dihydroguanine in human urine, a novel biomarker of osteoporosis. Free Radic. Res. 2022, 56, 358–365. [Google Scholar] [CrossRef]
- Li, P.; Wang, Z.Y.; Yueying, L.; Liu, L.Z.; Qiu, J.G.; Zhang, C.Y. Bsu polymerase-mediated fluorescence coding for rapid and sensitive detection of 8-oxo-7,8-dihydroguanine in telomeres of cancer cells. Talanta 2022, 243, 123340. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.M. On the Origin of lncRNAs: Missing Link Found. Trends Genet. 2017, 33, 660–662. [Google Scholar] [CrossRef] [PubMed]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef]
- Lin, C.; Wang, Y.; Wang, Y.; Zhang, S.; Yu, L.; Guo, C.; Xu, H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene 2017, 36, 5392–5406. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.X.; Liu, Y.; Wang, J.; Xie, Y.; Li, R.Y.; Ma, Q.; Tu, Q.; Melhem, N.A.; Couldwell, S.; El-Araby, R.E.; et al. A novel lncRNA-mediated epigenetic regulatory mechanism in periodontitis. Int. J. Biol. Sci. 2023, 19, 5187–5203. [Google Scholar] [CrossRef]
- Chen, Z.-l.; Wei, L.-L.; Shi, L.-Y.; Li, M.; Jiang, T.-T.; Chen, J.; Liu, C.-M.; Yang, S.; Tu, H.-h.; Hu, Y.-t.; et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci. Rep. 2017, 7, 16751. [Google Scholar] [CrossRef]
- Huo, X.; Han, S.; Wu, G.; Latchoumanin, O.; Zhou, G.; Hebbard, L.; George, J.; Qiao, L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: Implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol. Cancer 2017, 16, 165. [Google Scholar] [CrossRef]
- Hernandez-Aguilar, A.I.; Luciano-Villa, C.A.; Tello-Flores, V.A.; Beltran-Anaya, F.O.; Zubillaga-Guerrero, M.I.; Flores-Alfaro, E. Dysregulation of lncRNA-H19 in cardiometabolic diseases and the molecular mechanism involved: A systematic review. Expert Rev. Mol. Diagn. 2021, 21, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, C.; Zou, X.; Tian, X.; Hu, J.; Zhang, C.Y. Simultaneous Enzyme-Free Detection of Multiple Long Noncoding RNAs in Cancer Cells at Single-Molecule/Particle Level. Nano Lett. 2021, 21, 4193–4201. [Google Scholar] [CrossRef]
- Ma, F.; Lu, G.A.; Chen, Q.; Ruan, Y.; Li, X.; Lu, X.; Li, C. Dynamic global analysis of transcription reveals the role of miRNAs in synergistic stabilization of gene expression. Sci. Bull. 2020, 65, 2130–2140. [Google Scholar] [CrossRef] [PubMed]
- Gurtan, A.M.; Sharp, P.A. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013, 425, 3582–3600. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Raia, T.; Orticello, M.; Lucarelli, M. The complex interplay between DNA methylation and miRNAs in gene expression regulation. Biochimie 2020, 173, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Satish, D.; Mukherjee, S.K.; Gupta, D. PAmiRDB: A web resource for plant miRNAs targeting viruses. Sci. Rep. 2019, 9, 4627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, X.; Zhou, K.; Ling, X.; Zhang, J.; Wu, P.; Zhang, T.; Xie, K.; Dai, G. miRNA-10a-5p Targeting the BCL6 Gene Regulates Proliferation, Differentiation and Apoptosis of Chicken Myoblasts. Int. J. Mol. Sci. 2022, 23, 9545. [Google Scholar] [CrossRef]
- Yang, M.; Gao, X.; Hu, C.; Wang, S.; Sheng, H.; Ma, Y. Bta-miR-484 Targets SFRP1 and Affects Preadipocytes Proliferation, Differentiation, and Apoptosis. Int. J. Mol. Sci. 2023, 24, 12710. [Google Scholar] [CrossRef]
- Yan, H.; Cai, H.; Guan, Q.; He, J.; Zhang, J.; Guo, Y.; Huang, H.; Li, X.; Li, Y.; Gu, Y.; et al. Individualized analysis of differentially expressed miRNAs with application to the identification of miRNAs deregulated commonly in lung cancer tissues. Brief. Bioinform. 2018, 19, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.; Samowitz, W.S.; Wolff, R.K.; Stevens, J.R.; Herrick, J.S. The NF-kappaB signalling pathway in colorectal cancer: Associations between dysregulated gene and miRNA expression. J. Cancer Res. Clin. Oncol. 2018, 144, 269–283. [Google Scholar] [CrossRef]
- Apps, J. RARE-16. Differential expression of miRNAs in adamantinomatous craniopharynngioma reveals dysregulation of pathogenic pathways. Neuro Oncol. 2022, 24, i13. [Google Scholar] [CrossRef]
- Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. Dysregulation of miRNA in Leukemia: Exploiting miRNA Expression Profiles as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7156. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Guan, L.W.; Peng, J.W.; Huang, S.Y.; Liu, T.; Xiong, T.; Yang, Y.F.; Wang, X.L.; Hao, X. Ultra-sensitive and specificity quantitative detection of miRNA using a combined CRISPR/Cas13a and DNA-PAINT. Sens. Actuator B-Chem. 2023, 395, 134451. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Y.; Li, C.-C.; Zou, X.; Ma, F.; Zhang, C.-Y. Construction of a dual-functional dumbbell probe-based fluorescent biosensor for cascade amplification detection of miRNAs in lung cancer cells and tissues. Chem. Commun. 2022, 58, 5538–5541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, R.; Zhang, Y.; Zou, X.; Zhang, C.-Y. One-step self-assembly of quantum dot-based spherical nucleic acid nanostructure for accurate monitoring of long noncoding RNA MALAT1 in living cells and tissues. Chem. Eng. J. 2023, 469, 144021. [Google Scholar] [CrossRef]
- Zemora, G.; Handl, S.; Waldsich, C. Human telomerase reverse transcriptase binds to a pre-organized hTR in vivo exposing its template. Nucleic Acids Res. 2016, 44, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tran, H.; Mathahs, M.M.; Fink, B.D.; Albert, J.A.; Moninger, T.O.; Meier, J.L.; Li, M.; Schmidt, W.N. Zinc protoporphyrin binding to telomerase complexes and inhibition of telomerase activity. Pharmacol. Res. Perspect. 2021, 9, e00882. [Google Scholar] [CrossRef] [PubMed]
- Park, T.W.; Riethdorf, S.; Riethdorf, L.; Loning, T.; Janicke, F. Differential telomerase activity, expression of the telomerase catalytic sub-unit and telomerase-RNA in ovarian tumors. Int. J. Cancer 1999, 84, 426–431. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H.; Xu, Q.; Liu, H.; Han, Y.; Li, D.L.; Ma, F.; Zhang, C.Y. Construction of a 3D Quantum Dot Nanoassembly with Two-Step FRET for One-Step Sensing of Human Telomerase RNA in Breast Cancer Cells and Tissues. Anal. Chem. 2024, 96, 7738–7746. [Google Scholar] [CrossRef]
- Janszky, N.; Susal, C. Circulating and urinary microRNAs as possible biomarkers in kidney transplantation. Transplant. Rev. 2018, 32, 110–118. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Tao, R.; He, G.; Liu, J.; Li, C.; Hou, Y. The potential use of Piwi-interacting RNA biomarkers in forensic body fluid identification: A proof-of-principle study. Forensic Sci. Int.-Genet. 2019, 39, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Lai, Y.C.; Husna, A.A.; Chen, H.W.; Tanaka, Y.; Kawaguchi, H.; Hatai, H.; Miyoshi, N.; Nakagawa, T.; Fukushima, R.; et al. Aberrantly expressed snoRNA, snRNA, piRNA and tRFs in canine melanoma. Vet. Comp. Oncol. 2020, 18, 353–361. [Google Scholar] [CrossRef]
- Yin, J.; Qi, W.; Ji, C.G.; Zhang, D.X.; Xie, X.L.; Ding, Q.; Jiang, X.Y.; Han, J.; Jiang, H.Q. Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol. Rep. 2019, 42, 263–272. [Google Scholar] [CrossRef]
- Ge, Q.Q.; Han, Q.; Han, Y.; Ma, F.; Li, C.Z.; Zhang, C.Y. A multi-cycle signal amplification-mediated single quantum dot nanosensor for PIWI-interacting RNA detection. Chem. Commun. 2024, 60, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Saaoud, F.; Drummer, I.V.C.; Shao, Y.; Sun, Y.; Lu, Y.; Xu, K.; Ni, D.; Jiang, X.; Wang, H.; Yang, X. Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol. Ther. 2021, 220, 107715. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, J. Mammalian circular RNAs result largely from splicing errors. Cell Rep. 2021, 36, 109439. [Google Scholar] [CrossRef] [PubMed]
- Di Timoteo, G.; Rossi, F.; Bozzoni, I. Circular RNAs in cell differentiation and development. Development 2020, 147, dev182725. [Google Scholar] [CrossRef]
- Liu, C.; Wu, X.; Gokulnath, P.; Li, G.; Xiao, J. The Functions and Mechanisms of Translatable Circular RNAs. J. Pharmacol. Exp. Ther. 2023, 384, 52–60. [Google Scholar] [CrossRef]
- Shen, H.; Liu, B.; Xu, J.; Zhang, B.; Wang, Y.; Shi, L.; Cai, X. Circular RNAs: Characteristics, biogenesis, mechanisms and functions in liver cancer. J. Hematol. Oncol. 2021, 14, 134. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, S.; Wang, X.; Zhu, X.; Han, S. Progress in research on the role of circular RNAs in lung cancer. World J. Surg. Oncol. 2018, 16, 215. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S. Roles of circular RNAs in colorectal cancer. Oncol. Lett. 2021, 22, 602. [Google Scholar] [CrossRef]
- Jahani, S.; Nazeri, E.; Majidzadeh, A.K.; Jahani, M.; Esmaeili, R. Circular RNA; a new biomarker for breast cancer: A systematic review. J. Cell. Physiol. 2020, 235, 5501–5510. [Google Scholar] [CrossRef]
- Liu, W.-J.; Song, R.; Yang, D.; Zhao, S.; Zhang, C.-Y. A programmable quantum dot nanosensor guided by three-way junction skeleton-mediated cascade signal amplification for sensitive detection of circRNAs in breast cancer. Chem. Eng. J. 2024, 484, 149788. [Google Scholar] [CrossRef]
- Zuidhof, H.R.; Calkhoven, C.F. Oncogenic and Tumor-Suppressive Functions of the RNA Demethylase FTO. Cancer Res. 2022, 82, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, G.; Xu, H.; Dong, W.; Dong, Z.; Qiu, Z.; Zhang, Z.; Li, F.; Huang, Y.; Li, Y.; et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021, 33, 1221–1233.e1211. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yu, X.; Yang, L.; Liu, X.; Gao, B.; Huang, B.; Dou, X.; Liu, J.; Zou, Z.; Cui, X.L.; et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science 2022, 376, 968–973. [Google Scholar] [CrossRef]
- Chang, R.; Huang, Z.; Zhao, S.; Zou, J.; Li, Y.; Tan, S. Emerging Roles of FTO in Neuropsychiatric Disorders. Biomed. Res. Int. 2022, 2022, 2677312. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, Y.; Liao, X.; Tian, D.; Xu, Y.; Zhou, C.; Liu, J.; Li, S.; Zhou, J.; Nie, Y.; et al. FTO: A critical role in obesity and obesity-related diseases. Br. J. Nutr. 2023, 130, 1657–1664. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Jing, X.; Xiong, X.D. Emerging Role and Mechanism of the FTO Gene in Cardiovascular Diseases. Biomolecules 2023, 13, 850. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.N.; Liu, Y.Z.; Zhang, L.; Liu, W.; Zou, X.; Xu, Q.; Zhang, C.Y. Construction of Multiple DNAzymes Driven by Single Base Elongation and Ligation for Single-Molecule Monitoring of FTO in Cancer Tissues. Anal. Chem. 2023, 95, 12974–12981. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, D.; Hu, J.P.; Wang, L.J.; Qiu, J.G.; Zhang, C.Y. Construction of bidirectional strand displacement-driven three-dimensional DNA walkers for single-molecule monitoring of multiple DNA glycosylases. Sens. Actuator B-Chem. 2023, 380, 133357. [Google Scholar] [CrossRef]
- Shi, R.; Mullins, E.A.; Shen, X.X.; Lay, K.T.; Yuen, P.K.; David, S.S.; Rokas, A.; Eichman, B.F. Selective base excision repair of DNA damage by the non-base-flipping DNA glycosylase AlkC. EMBO J. 2018, 37, 63–74. [Google Scholar] [CrossRef]
- Mullins, E.A.; Rodriguez, A.A.; Bradley, N.P.; Eichman, B.F. Emerging Roles of DNA Glycosylases and the Base Excision Repair Pathway. Trends Biochem. Sci. 2019, 44, 765–781. [Google Scholar] [CrossRef]
- Kaur, R.; Nikkel, D.J.; Wetmore, S.D. Computational studies of DNA repair: Insights into the function of monofunctional DNA glycosylases in the base excision repair pathway. Wiley Interdiscip. Rev.-Comput. Mol. Sci. 2020, 10, e1471. [Google Scholar] [CrossRef]
- Olmon, E.D.; Delaney, S. Differential Ability of Five DNA Glycosylases to Recognize and Repair Damage on Nucleosomal DNA. ACS Chem. Biol. 2017, 12, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat. Res.-Rev. Mutat. Res. 2017, 771, 99–127. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Murakami, K.; Tada, H.; Uehara, Y.; Nogami, J.; Maehara, K.; Ohkawa, Y.; Saitoh, H.; Nishitani, H.; Ono, T.; et al. Thymine DNA glycosylase modulates DNA damage response and gene expression by base excision repair-dependent and independent mechanisms. Genes Cells 2017, 22, 392–405. [Google Scholar] [CrossRef]
- Hans, F.; Senarisoy, M.; Bhaskar Naidu, C.; Timmins, J. Focus on DNA Glycosylases-A Set of Tightly Regulated Enzymes with a High Potential as Anticancer Drug Targets. Int. J. Mol. Sci. 2020, 21, 9226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, X.; Ma, F.; Zhang, C.-Y. Advances in quantum dot-based biosensors for DNA-modifying enzymes assay. Coord. Chem. Rev. 2022, 469, 214674. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, J.; Li, D.-L.; Qiu, J.-G.; Jiang, B.-H.; Zhang, C.-Y. Construction of single-molecule counting-based biosensors for DNA-modifying enzymes: A review. Anal. Chim. Acta 2024, 1298, 342395. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, C.C.; Ma, F.; Luo, X.; Zhang, C.Y. Catalytic single-molecule Forster resonance energy transfer biosensor for uracil-DNA glycosylase detection and cellular imaging. Biosens. Bioelectron. 2022, 213, 114447. [Google Scholar] [CrossRef]
- Silvas, T.V.; Schiffer, C.A. APOBEC3s: DNA-editing human cytidine deaminases. Protein Sci. 2019, 28, 1552–1566. [Google Scholar] [CrossRef]
- Kawale, A.S.; Ran, X.; Patel, P.S.; Saxena, S.; Lawrence, M.S.; Zou, L. APOBEC3A induces DNA gaps through PRIMPOL and confers gap-associated therapeutic vulnerability. Sci. Adv. 2024, 10, eadk2771. [Google Scholar] [CrossRef]
- Duan, Y.; Du, Y.; Gu, Z.; Zheng, X.; Wang, C. Prognostic value, immune signature and molecular mechanisms of the APOBEC family members APOBEC1, APOBEC3A, APOBEC3G and APOBEC3H in pancreatic adenocarcinoma. Front. Mol. Biosci. 2022, 9, 1036287. [Google Scholar] [CrossRef] [PubMed]
- Langenbucher, A.; Bowen, D.; Sakhtemani, R.; Bournique, E.; Wise, J.F.; Zou, L.; Bhagwat, A.S.; Buisson, R.; Lawrence, M.S. An extended APOBEC3A mutation signature in cancer. Nat. Commun. 2021, 12, 1602. [Google Scholar] [CrossRef] [PubMed]
- Gansmo, L.B.; Romundstad, P.; Hveem, K.; Vatten, L.; Nik-Zainal, S.; Lonning, P.E.; Knappskog, S. APOBEC3A/B deletion polymorphism and cancer risk. Carcinogenesis 2018, 39, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Song, R.; Zou, X.R.; Li, D.L.; Xu, Q.; Zhang, C.Y. Enzymatic DNA repairing amplification-powered construction of an Au nanoparticle-based nanosensor for single-molecule monitoring of cytosine deaminase activity in cancer cells. Anal. Chim. Acta 2023, 1281, 341895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yuan, Z.-Z.; Zhang, X.; Zhang, Y.; Zou, X.; Ma, F.; Zhang, C.-Y. Entropy-Driven Self-Assembly of Single Quantum Dot Sensor for Catalytic Imaging of Telomerase in Living Cells. Anal. Chem. 2022, 94, 18092–18098. [Google Scholar] [CrossRef] [PubMed]
- Kupiec, M. Biology of telomeres: Lessons from budding yeast. Fems Microbiol. Rev. 2014, 38, 144–171. [Google Scholar] [CrossRef]
- Matthe, D.M.; Thoma, O.M.; Sperka, T.; Neurath, M.F.; Waldner, M.J. Telomerase deficiency reflects age-associated changes in CD4+ T cells. Immun. Ageing 2022, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Kuru, G.; Üner, G.; Bedir, E. Is telomerase a hidden player? Therapeutic potential of natural telomerase activators against age-related diseases. Phytochem. Rev. 2022, 22, 35–72. [Google Scholar] [CrossRef]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the Human Telomerase Gene TERT by Telomere Position Effect-Over Long Distances (TPE-OLD): Implications for Aging and Cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef]
- Han, W.H.; Zou, C.; Qian, L.X.; Wang, C.; Wang, X.W.; Liu, Y.Q.; Wang, X.R. Functional Analysis of Alkaline Phosphatase in Whitefly Bemisia tabaci (Middle East Asia Minor 1 and Mediterranean) on Different Host Plants. Genes 2021, 12, 497. [Google Scholar] [CrossRef]
- Lopez-Delgado, L.; Riancho-Zarrabeitia, L.; Garcia-Unzueta, M.T.; Tenorio, J.A.; Garcia-Hoyos, M.; Lapunzina, P.; Valero, C.; Riancho, J.A. Abnormal bone turnover in individuals with low serum alkaline phosphatase. Osteoporosis Int. 2018, 29, 2147–2150. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.; Costa Ramos, L.F.; Dos Santos Seckler, H.; Mendonca Gomes, F.; Reis Cortines, J.; Ramos, I.; Dinis Anobom, C.; de Alcantara Machado, E.; Perpetua de Oliveira, D.M. Biochemical characterization of digestive membrane-associated alkaline phosphatase from the velvet bean caterpillar Anticarsia gemmatalis. Arch. Insect Biochem. Physiol. 2019, 102, e21591. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Irie, M.; Iwata, K.; Nakane, H.; Yoshikane, M.; Koyama, Y.; Uehara, Y.; Takeyama, Y.; Kitamura, Y.; Sohda, T.; et al. Altered expression of alkaline phosphatase (ALP) in the liver of primary biliary cirrhosis (PBC) patients. Hepatol. Res. 2006, 35, 37–44. [Google Scholar] [CrossRef]
- Whyte, M.P.; Ma, N.S.; Mumm, S.; Gottesman, G.S.; McAlister, W.H.; Nenninger, A.R.; Bijanki, V.N.; Ericson, K.L.; Magnusson, P. Persistent idiopathic hyperphosphatasemia from bone alkaline phosphatase in a healthy boy. Bone 2020, 138, 115459. [Google Scholar] [CrossRef]
- Huang, C.W.; Wu, T.H.; Hsu, H.Y.; Pan, K.T.; Lee, C.W.; Chong, S.W.; Huang, S.F.; Lin, S.E.; Yu, M.C.; Chen, S.M. Reappraisal of the Role of Alkaline Phosphatase in Hepatocellular Carcinoma. J. Pers. Med. 2022, 12, 518. [Google Scholar] [CrossRef]
- Ma, F.; Zhao, N.N.; Liu, M.; Xu, Q.; Zhang, C.Y. Single-Molecule Biosensing of Alkaline Phosphatase in Cells and Serum Based on Dephosphorylation-Triggered Catalytic Assembly and Disassembly of the Fluorescent DNA Chain. Anal. Chem. 2022, 94, 6004–6010. [Google Scholar] [CrossRef] [PubMed]
- Groborz, K.; Gonzalez Ramirez, M.L.; Snipas, S.J.; Salvesen, G.S.; Drag, M.; Poreba, M. Exploring the prime site in caspases as a novel chemical strategy for understanding the mechanisms of cell death: A proof of concept study on necroptosis in cancer cells. Cell Death Differ. 2020, 27, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Gheyas, R.; Menko, A.S. The involvement of caspases in the process of nuclear removal during lens fiber cell differentiation. Cell Death Discov. 2023, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Tang, H.; Wang, C.; Zhang, H.; Zheng, C.; Zhang, N.; Chen, B.; Sun, L. Baohuoside I Inhibits the Proliferation of Pancreatic Cancer Cells via mTOR/S6K1-Caspases/Bcl2/Bax Apoptotic Signaling. Cancer Manag. Res. 2019, 11, 10609–10621. [Google Scholar] [CrossRef]
- Ross, C.; Chan, A.H.; von Pein, J.B.; Maddugoda, M.P.; Boucher, D.; Schroder, K. Inflammatory Caspases: Toward a Unified Model for Caspase Activation by Inflammasomes. Annu. Rev. Immunol. 2022, 40, 249–269. [Google Scholar] [CrossRef]
- Gao, L.; Li, Q.; Zhang, Z.; Ge, P.; Sun, J.; Qiao, X.; Wang, L.; Song, L. Species-specific CgCaspase-Cg-5 in the pacific oyster induces haemocyte apoptosis by regulating the mRNA expression of apoptosis-related genes in the early stage of immune response. Fish Shellfish Immunol. 2023, 138, 108856. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yi, Y.; Lv, Q.; Zhang, J.; Wu, K.; Wu, W.; Zhang, W. Novel 1,3,5-triazine derivatives exert potent anti-cervical cancer effects by modulating Bax, Bcl2 and Caspases expression. Chem. Biol. Drug Des. 2018, 91, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Arechaga-Ocampo, E.; Pereira-Suarez, A.L.; del Moral-Hernandez, O.; Cedillo-Barron, L.; Rodriguez-Sastre, M.A.; Castillo-Alvarez, A.; Lopez-Bayghen, E.; Villegas-Sepulveda, N. HPV+ cervical carcinomas and cell lines display altered expression of caspases. Gynecol. Oncol. 2008, 108, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zhang, C.W.; Mao, Y.; Li, L.; Gao, N.; Lim, K.L.; Xu, Q.H.; Yao, S.Q. Two-Photon Enzymatic Probes Visualizing Sub-cellular/Deep-brain Caspase Activities in Neurodegenerative Models. Sci. Rep. 2016, 6, 26385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, J.; Wang, B.; Zhang, C.F.; Xu, X.H.; Zhang, M. A C(21)-Steroidal Glycoside from Cynanchum atratum Attenuates Concanavalin A-Induced Liver Injury in Mice. Molecules 2019, 24, 1087. [Google Scholar] [CrossRef]
- Liu, M.; Xu, R.; Liu, W.; Qiu, J.G.; Wang, Y.; Ma, F.; Zhang, C.Y. Integration of exonuclease III-powered three-dimensional DNA walker with single-molecule detection for multiple initiator caspases assay. Chem. Sci. 2021, 12, 15645–15654. [Google Scholar] [CrossRef]
- Zhang, X.; Dan, S.; Pan, X.; Li, J.; Wei, Q.; Huang, L.; Kang, B.; Chen, C. Identification of VPS34-PI(3)P-FEN1-mediated DNA repair pathway as a potential drug target to overcome chemoresistance. Biochem. Biophys. Res. Commun. 2023, 674, 27–35. [Google Scholar] [CrossRef]
- Schilling, E.M.; Scherer, M.; Rothemund, F.; Stamminger, T. Functional regulation of the structure-specific endonuclease FEN1 by the human cytomegalovirus protein IE1 suggests a role for the re-initiation of stalled viral replication forks. PLoS Pathog. 2021, 17, e1009460. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, S.; Zhu, J.K.; Gong, Z. Requirement for flap endonuclease 1 (FEN1) to maintain genomic stability and transcriptional gene silencing in Arabidopsis. Plant J. 2016, 87, 629–640. [Google Scholar] [CrossRef]
- Shi, R.; Wang, Y.; Gao, Y.; Xu, X.; Mao, S.; Xiao, Y.; Song, S.; Wang, L.; Tian, B.; Zhao, Y.; et al. Succinylation at a key residue of FEN1 is involved in the DNA damage response to maintain genome stability. Am. J. Physiol.-Cell Physiol. 2020, 319, C657–C666. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Liu, L.; Chen, J.; Hu, Q.; Shen, S.; Zhou, Y.; Chen, S.; Xue, C.; Cui, G.; et al. Upregulation of FEN1 Is Associated with the Tumor Progression and Prognosis of Hepatocellular Carcinoma. Dis. Markers 2020, 2020, 2514090. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Luo, S. miR-140-5p inhibits cervical cancer cell phenotypes via downregulating FEN1 to halt the cell cycle. Mol. Med. Rep. 2020, 22, 4919–4930. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yu, X.-D.; Liu, W.; Liu, H.; Xu, Q.; Zhang, C.-Y. Dual signal amplification-integrated single-molecule biosensing of flap endonuclease 1 in breast cancer tissues. Sens. Actuator B-Chem. 2023, 394, 134383. [Google Scholar] [CrossRef]
- Diekmann, R.; Helle, Ø.I.; Øie, C.I.; McCourt, P.; Huser, T.R.; Schüttpelz, M.; Ahluwalia, B.S. Chip-based wide field-of-view nanoscopy. Nat. Photonics 2017, 11, 322–328. [Google Scholar] [CrossRef]
| Methods | Analyte Type | Linear Range | LOD | Sample Volume | Ref. |
|---|---|---|---|---|---|
| Single-molecule fluorescent-based biosensor | HPV16 DNA | 1 aM–1 fM | 3.0 aM | 1 uL | [26] |
| MoS2/Graphene nanostructure field effect transistor-based detection | DNA | 10 aM–100 pM | 10 aM | N/A | [27] |
| Fiber-based surface plasmon resonance biosensor | secY DNA | 1.0 fM–1.0 pM | 1.0 fM | N/A | [28] |
| silica nanoparticle-enhanced microcantilever sensor | Hepatitis B Virus DNA | 23.1 fM–2.3 nM | 2.3 fM | 20 uL | [29] |
| nanomechanical resonator biosensor based on a photonic crystal nanowire array | DNA | 500 aM–10 pM | 500 aM | 1 mL | [30] |
| quartz crystal microbalance-based detection | DNA | 1.0 fM–10 pM | 0.7 fM | 30 uL | [31] |
| Target | Combined Amplification Strategy | Assay Time | Linear Range | LOD | Real Sample | Ref. |
|---|---|---|---|---|---|---|
| DNA | no | 10 min | 1 aM–1 fM | 5.00 aM | vaginal secretions; MRSA-spiked milk samples | [26] |
| Retroviral DNA | no | 70 min | 100 aM–1 nM | 66.1 aM, 82.8 aM | HuT-78 cells; U266B1 cells | [22] |
| SNP | T7 transcription | 1 h | 0.1 aM–100 pM | 0.0724 × 10 aM; 0.0372 × 10 aM | PANC-1 cells; HCT-116 cells; HL-7702 cells; H358 cells; HLF-1 cells; and A549 cells | [59] |
| DNA methylation | T7 transcription | 4.5 h | 1 fM–10 nM | 337 aM | A549 cells; HepG2 cells; HT-29 cells; HBE cells; H358 cell; breast cancer patient tissue; healthy individual tissue. | [65] |
| 6mdA-DNA; 4mdC-DNA | TMSD | 1 h | 10 fM–100 nM | 9.80 fM; 9.97 fM | HepG2 cells; E. coli plasmid cloning vector (pUC19) | [71] |
| 8-oxo-7, 8-dihydroguanine | 70 min | 0.5 aM–5 pM | 2.45 aM | A549 cells; HeLa cells; HL-7702 cells | [81] | |
| lncRNA | PER; SDA | 40 min | 100 aM–1 nM | 65.25 aM | MCF-7 cells; HeLa cells; A549 cells; HBE cells; breast cancer patient tissues; healthy individual tissues | [102] |
| MALAT1; HOTAIR | No | 1 h | 0.1 aM–1 pM | 0.10 aM; 0.031 aM | SW480 cells; MCF-7 cells; A549 cells; HBE cells | [89] |
| microRNA | No | ~3 h | 1 fM–100 pM | 1.12 fM | Serum | [100] |
| microRNA | PER | 1 fM–100 pM | 0.45 fM | H460 cells; H292 cells; H358 cells; A549 cells; and H1975 cells; NSCLC patient tissue and healthy individual tissue | [101] | |
| Human telomerase RNA | No | 1 h | 10 fM–1 nM | 2.10 fM | MCF-7 cells; HL-7702 cells; A549 cells; HeLa cells; breast cancer patient tissue; healthy individual tissue | [106] |
| piRNA | Loop connection | ~5 h | 1 fM–10 nM | 0.104 fM | MCF-7 cells; A549 cells; HepG2 cells; HeLa cells; HCT-116 cells; breast cancer patient tissue; healthy individual tissue | [111] |
| circRNAs | EXPAR, RCA | ~3 h | 100 aM–10 pM | 41.3 aM | MCF-7 cells; HepG2 cells; HeLa cells; HL-7702 cells; A549 cells; breast cancer patient tissue; healthy individual tissue | [120] |
| UDG | SDA | ~2.5 h | 0.0005–0.5 U/mL | 0.00029 U/mL | HeLa cells; HL-7702 cells | [138] |
| hSMUG1; hAAG | SP-SDA | ~4 h | 1.00 × 10−9–0.02 U/μL | 8.14 × 10−10 U/μL; 4.50 × 10−9 U/μL | A549 cells; HEK-293 cells; HeLa cells | [128] |
| FTO | RCA | ~5 h | 1 fM–1 nM | 0.596 fM | MDA-MB-231 cells; MCF-10A cells; MCF-7 cells; A549 cells; HeLa cells; breast cancer patient tissue; healthy individual tissue | [121] |
| APOBEC3A | ERA | ~3 h | 1 fM–50 pM | 0.855 aM | A549 cells; HEK-293 cells; HeLa cells; MCF-7 cells | [144] |
| telomerase | EDC | 100 min | no | no | MCF-7 cells; HeLa cells; HL-7702 cells A549 cells; | [145] |
| ALP | HCR | ~2.5 h | 1 × 10−5–1 × 10−2 U/mL | 2.61× 10−6 U/mL | HeLa cells; MCF-7 cells; HEK cells; clinical serum samples | [156] |
| caspase-8; caspase-9 | Exo III-mediated amplification | ~2.5 h | 2.50 × 10−6 –2.50 × 10−3 U/μL | 2.08 × 10−6 U/μL; 1.71 × 10−6 U/μL | HeLa cells; MCF-7 cells; Jurkat cells | [166] |
| FEN1 | RCA | 2.5 h | 0–0.5 U/μL | 2.24 × 10−5 U/μL | A549 cells; HeLa cells; MCF-7 cells; HepG2 cells; HL-7702 cells; HEK-293 cells; breast cancer patient tissue; healthy individual tissue | [173] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, J.; Qiao, L.; Zhang, Q.; Hu, J.; Zhang, C.-y. Recent Advance in Single-Molecule Fluorescent Biosensors for Tumor Biomarker Detection. Biosensors 2024, 14, 540. https://doi.org/10.3390/bios14110540
Zhang J, Liu J, Qiao L, Zhang Q, Hu J, Zhang C-y. Recent Advance in Single-Molecule Fluorescent Biosensors for Tumor Biomarker Detection. Biosensors. 2024; 14(11):540. https://doi.org/10.3390/bios14110540
Chicago/Turabian StyleZhang, Jie, Jiawen Liu, Lixue Qiao, Qian Zhang, Juan Hu, and Chun-yang Zhang. 2024. "Recent Advance in Single-Molecule Fluorescent Biosensors for Tumor Biomarker Detection" Biosensors 14, no. 11: 540. https://doi.org/10.3390/bios14110540
APA StyleZhang, J., Liu, J., Qiao, L., Zhang, Q., Hu, J., & Zhang, C.-y. (2024). Recent Advance in Single-Molecule Fluorescent Biosensors for Tumor Biomarker Detection. Biosensors, 14(11), 540. https://doi.org/10.3390/bios14110540





