Competitive Immunoassay in a Microfluidic Biochip for In-Field Detection of Abscisic Acid in Grapes
Abstract
:1. Introduction
2. Materials and Methods
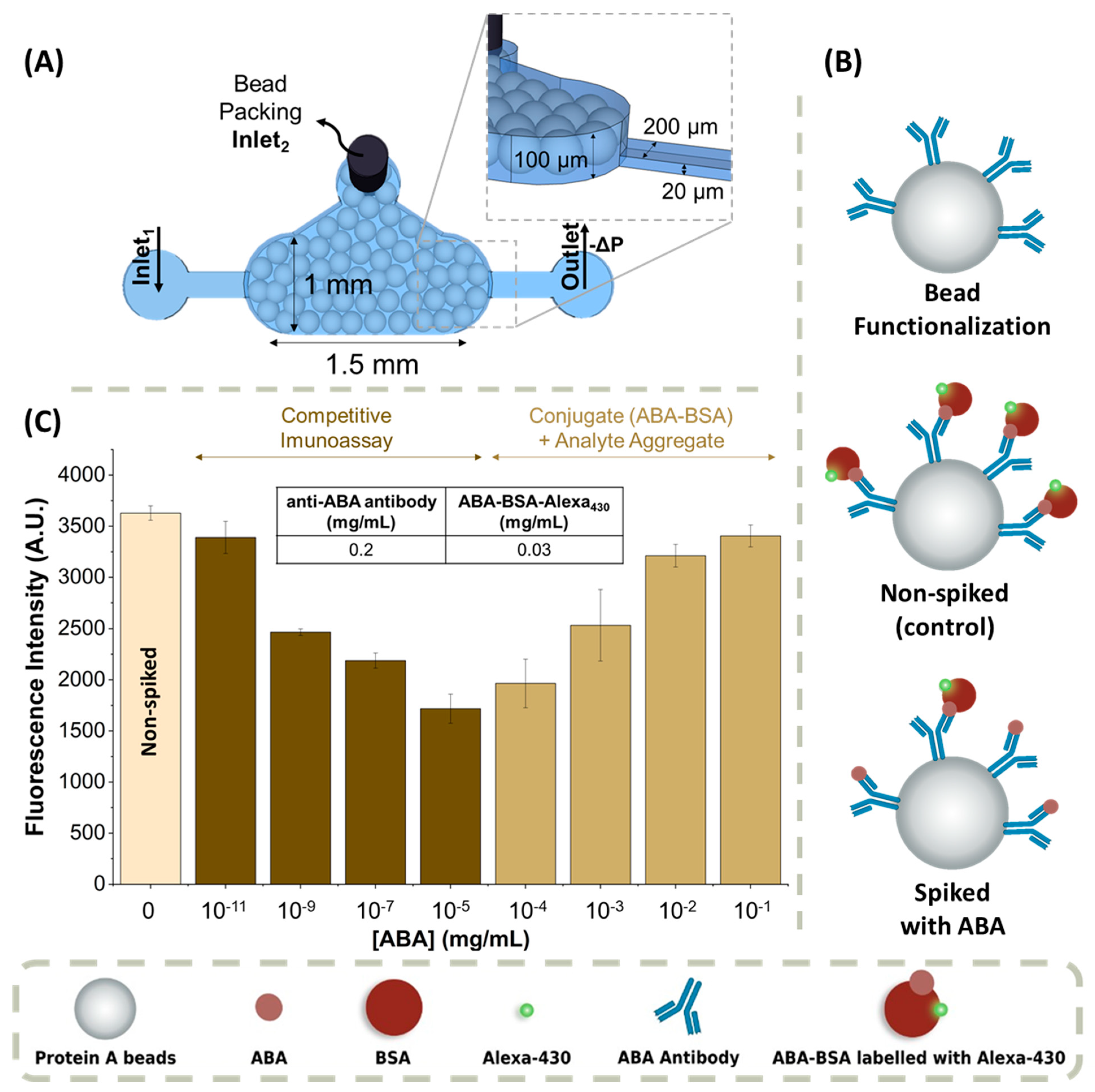
2.1. Fabrication of the Microfluidic Devices
2.2. Competitive Immunoassay for ABA Detection
2.2.1. Reagents
2.2.2. Functionalization of the Microbeads
2.2.3. Packing the Biomarker Detection Chamber
2.2.4. ABA-BSA Conjugate Labelling
2.2.5. ABA Detection Immunoassay
2.2.6. Image Acquisition and Processing
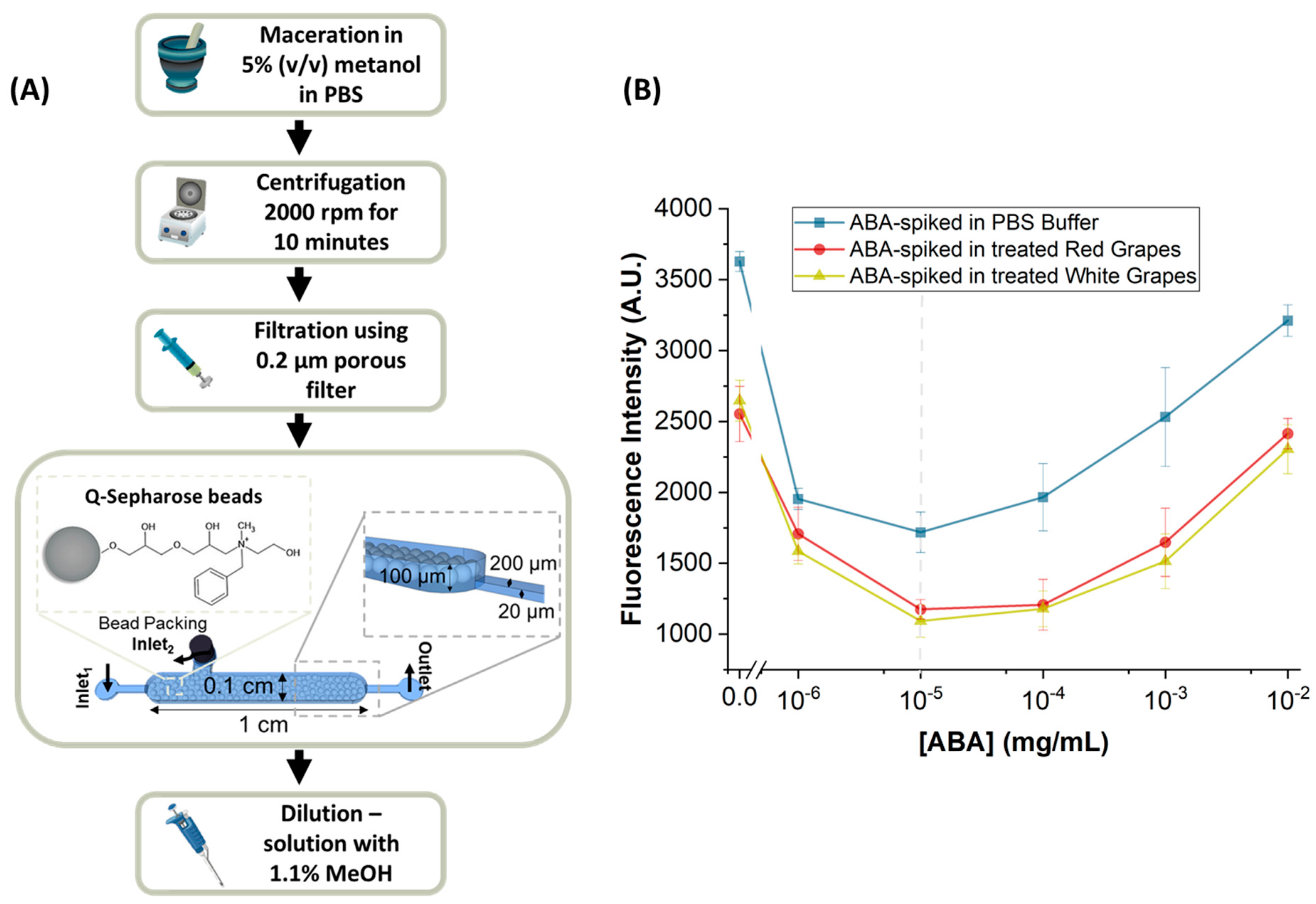
2.3. Sample Collection and Treatment for ABA Detection
Packing of the Sample Treatment Chamber
2.4. Reverse-Phase HPLC Detection of ABA
3. Results and Discussion
3.1. ABA-Spiked in PBS 1× Buffer
3.2. ABA-Spiked in Treated Table Grape Samples
3.3. ABA in Real Wine Grape Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Causas Das Alterações Climáticas. Available online: https://climate.ec.europa.eu/climate-change/causes-climate-change_pt (accessed on 9 November 2023).
- Lo Presti, D.; Di Tocco, J.; Massaroni, C.; Cimini, S.; De Gara, L.; Singh, S.; Raucci, A.; Manganiello, G.; Woo, S.L.; Schena, E.; et al. Current understanding, challenges and perspective on portable systems applied to plant monitoring and precision agriculture. Biosens. Bioelectron. 2023, 222, 115005. [Google Scholar] [CrossRef]
- OIV 2019 Statistical Report on World Vitiviniculture. Available online: https://www.oiv.int/public/medias/6782/oiv-2019-statistical-report-on-world-vitiviniculture.pdf (accessed on 30 July 2023).
- Li, J.; Li, C.; Smith, S.M. Hormone Metabolism and Signaling in Plants; Acdemic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van Der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Shah, J. Plants under attack: Systemic signals in defence. Curr. Opin. Plant Biol. 2009, 12, 459–464. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Raikhy, G.; Kumar, D. Chemical elicitors of systemic acquired resistance—Salicylic acid and its functional analogs. Curr. Plant Biol. 2019, 17, 48–59. [Google Scholar] [CrossRef]
- Checker, V.G.; Kushwaha, H.R.; Kumari, P.; Yadav, S. Role of Phytohormones in Plant Defense: Signaling and Cross Talk BT—Molecular Aspects of Plant-Pathogen Interaction. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 159–184. [Google Scholar] [CrossRef]
- Haynes, W.M. CRC Handbook of Chemistry and Physics; CRC press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Liaqat, S.; Saffeullah, P.; Umar, S.; Siddiqi, T.O. The molecular mechanism of brassinosteroids in mediating the abiotic stress responses of plants. In Frontiers in Plant-Soil Interaction; Academic Press: Cambridge, MA, USA, 2021; pp. 547–566. [Google Scholar] [CrossRef]
- Shatil-Cohen, A.; Attia, Z.; Moshelion, M. Bundle-sheath cell regulation of xylem-mesophyll water transport via aquaporins under drought stress: A target of xylem-borne ABA? Plant J. 2011, 67, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Coelho, J.; Almeida-Trapp, M.; Pimentel, D.; Soares, F.; Reis, P.; Rego, C.; Mithöfer, A.; Fortes, A.M. The study of hormonal metabolism of Trincadeira and Syrah cultivars indicates new roles of salicylic acid, jasmonates, ABA and IAA during grape ripening and upon infection with Botrytis cinerea. Plant Sci. 2019, 283, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Fortes, A.M.; Teixeira, R.T.; Agudelo-Romero, P. Complex interplay of hormonal signals during grape berry ripening. Molecules 2015, 20, 9326–9343. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Merrium, S.; Habib-ur-Rahman, M.; Hakeem, S.; Saddique, M.A.B.; Sher, M.A. Wetting mechanism and morphological adaptation; leaf rolling enhancing atmospheric water acquisition in wheat crop—A review. Environ. Sci. Pollut. Res. 2022, 29, 30967–30985. [Google Scholar] [CrossRef]
- Simonneau, T.; Lebon, E.; Coupel-Ledru, A.; Marguerit, E.; Rossdeutsch, L.; Ollat, N. Adapting plant material to face water stress in vineyards: Which physiological targets for an optimal control of plant water status? Oeno One 2017, 51, 167–179. [Google Scholar] [CrossRef]
- Feng, S.; Chen, C.; Song, C.; Ding, X.; Wang, W.; Que, L. Optical Aptamer-Based Sensors for Detecting Plant Hormones. IEEE Sens. J. 2021, 21, 5743–5750. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Zhang, P.; Hasi, G.; Huang, Y.; Lu, J.; Zhang, Y.-L. Response of phytohormones and correlation of SAR signal pathway genes to the different resistance levels of grapevine against Plasmopara viticola infection. Plant Physiol. Biochem. 2016, 107, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Owen, S.J.; Lafond, M.D.; Bowen, P.; Bogdanoff, C.; Usher, K.; Abrams, S.R. Profiles of abscisic acid and its catabolites in developing Merlot grape (Vitis vinifera) berries. Am. J. Enol. Vitic. 2009, 60, 277–284. [Google Scholar] [CrossRef]
- Fernandes, P.; Carvalho, F.; Marques, M.P.C. Miniaturization in Biotechnology: Speeding up the Development of Bioprocesses. Recent Pat. Biotechnol. 2011, 5, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Elvira, K.S.; Solvas, X.C.I.; Wootton, R.C.R.; Demello, A.J. The past, present and potential for microfluidic reactor technology in chemical synthesis. Nat. Chem. 2013, 5, 905–915. [Google Scholar] [CrossRef]
- Lim, C.T.; Zhang, Y. Bead-based microfluidic immunoassays: The next generation. Biosens. Bioelectron. 2007, 22, 1197–1204. [Google Scholar] [CrossRef]
- Brás, E.J.S.; Fortes, A.M.; Chu, V.; Fernandes, P.; Conde, J.P. Microfluidic device for the point of need detection of a pathogen infection biomarker in grapes. Analyst 2019, 22, 1197–1204. [Google Scholar] [CrossRef]
- Domingues, C.; Fontes, P.M.; Condelipes, P.G.M.; Marques, V.; Afonso, M.B.; Chu, V.; Rodrigues, C.M.P.; Conde, J.P. Microfluidic Detection of Adenylate Kinase as a Cell Damage Biomarker. Chemosensors 2023, 11, 220. [Google Scholar] [CrossRef]
- Mata, A.; Fleischman, A.J.; Roy, S. Characterization of Polydimethylsiloxane (PDMS) Properties for Biomedical Micro/Nanosystems. Biomed. Microdevices 2005, 7, 281–293. [Google Scholar] [CrossRef]
- Schneider, W.; Rasband, S.; Eliceiri, K.W. Nih image to imagej: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chen, C.; Che, X.; Wang, W.; Que, L. Detection of plant hormone abscisic acid (ABA) using an optical aptamer-based sensor with a microfluidics capillary interface. In Proceedings of the 2017 IEEE 30th International Conference on Micro Electro Mechanical Systems, Las Vegas, NV, USA, 22–26 January 2017; pp. 370–373. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domingues, C.; Meirinho, R.; Rodrigues, R.G.; Fortes, A.M.; Chu, V.; Conde, J.P. Competitive Immunoassay in a Microfluidic Biochip for In-Field Detection of Abscisic Acid in Grapes. Biosensors 2024, 14, 123. https://doi.org/10.3390/bios14030123
Domingues C, Meirinho R, Rodrigues RG, Fortes AM, Chu V, Conde JP. Competitive Immunoassay in a Microfluidic Biochip for In-Field Detection of Abscisic Acid in Grapes. Biosensors. 2024; 14(3):123. https://doi.org/10.3390/bios14030123
Chicago/Turabian StyleDomingues, Cristiana, Rui Meirinho, Rodolfo G. Rodrigues, Ana Margarida Fortes, Virginia Chu, and João Pedro Conde. 2024. "Competitive Immunoassay in a Microfluidic Biochip for In-Field Detection of Abscisic Acid in Grapes" Biosensors 14, no. 3: 123. https://doi.org/10.3390/bios14030123
APA StyleDomingues, C., Meirinho, R., Rodrigues, R. G., Fortes, A. M., Chu, V., & Conde, J. P. (2024). Competitive Immunoassay in a Microfluidic Biochip for In-Field Detection of Abscisic Acid in Grapes. Biosensors, 14(3), 123. https://doi.org/10.3390/bios14030123




