APPROACH: Sensitive Detection of Exosomal Biomarkers by Aptamer-Mediated Proximity Ligation Assay and Time-Resolved Förster Resonance Energy Transfer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Lines and Cell Culture
2.3. Exosome Isolation and Characterization
2.4. Synthesis of CoraFluor-Like-NHS
2.5. Fluorescent Labeling of Oligonucleotides
2.6. Flow Cytometry Analysis of the Binding Affinities between Cho Primer/Apt-CD63 and Exosomes
2.7. PLA and RCA Reactions
2.8. Validation of RCA Products by Confocal Microscopy
2.9. Verification of RCA Products by Flow Cytometry
2.10. Exosomal Biomarker Analysis by APPROACH Strategy
3. Results and Discussion
3.1. Principles of APPROACH Strategy for Exosome Detection (Figure 1)
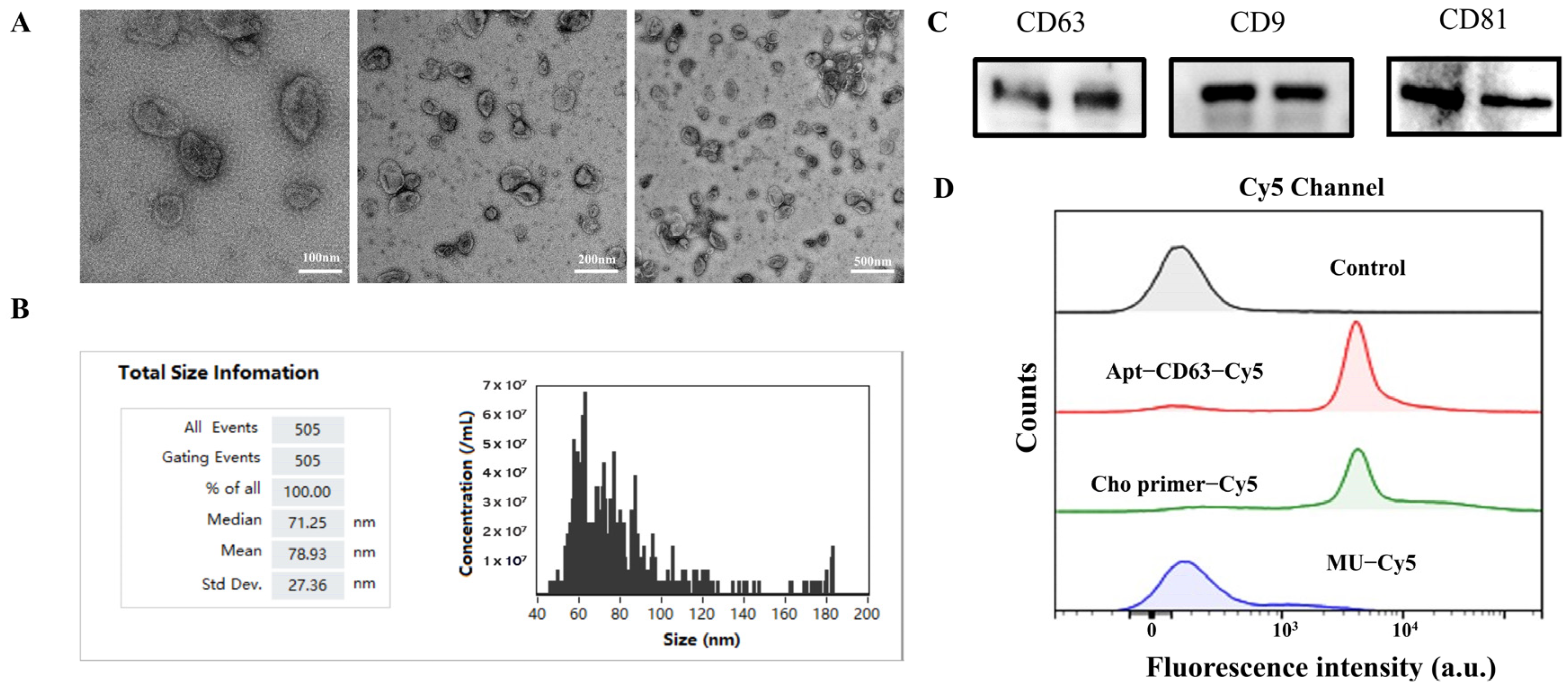
3.2. Characterization of Exosomes
3.3. Feasibility and Visualization Analyses of the APPROACH Strategy for Exosomal Biomarker Detection
3.4. Optimization of Experimental Conditions
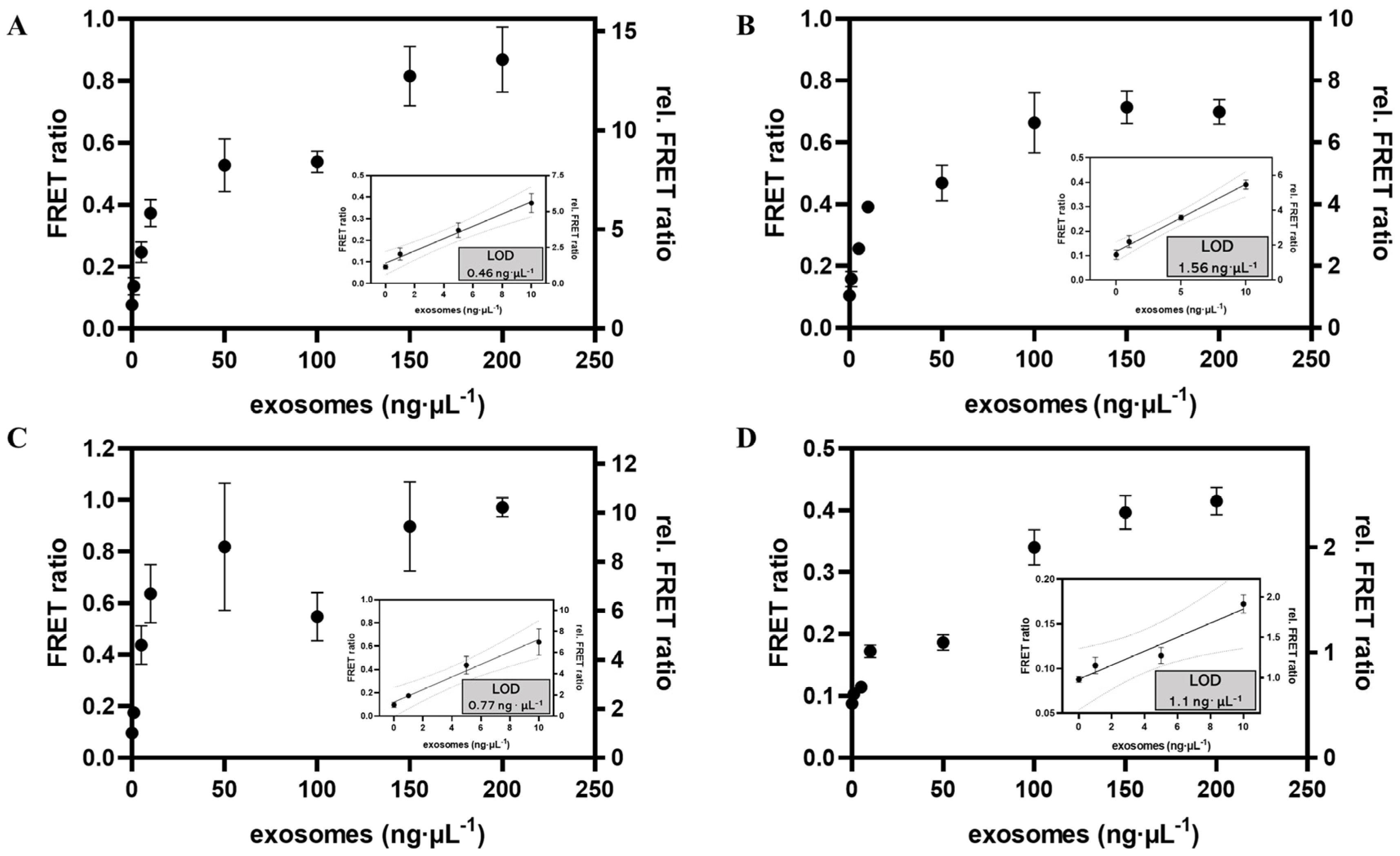
3.5. Analytical Performance of Exosomal Biomarkers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Bhatnagar, S. Exosome Function: From Tumor Immunology to Pathogen Biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef]
- Ni, Z.; Zhou, S.; Li, S.; Kuang, L.; Chen, H.; Luo, X.; Ouyang, J.; He, M.; Du, X.; Chen, L. Exosomes: Roles and therapeutic potential in osteoarthritis. Bone. Res. 2020, 8, 25. [Google Scholar] [CrossRef]
- Bu, H.; He, D.; He, X.; Wang, K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. ChemBioChem 2018, 20, 451–461. [Google Scholar] [CrossRef]
- Huda, M.N.; Nafiujjaman, M.; Deaguero, I.G.; Okonkwo, J.; Hill, M.L.; Kim, T.; Nurunnabi, M. Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 2021, 7, 2106–2149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Luo, C.; Mei, Q.; Zhang, H.; Zhang, W.; Su, D.; Fu, W.; Luo, Y. Aptamer-Cholesterol-Mediated Proximity Ligation Assay for Accurate Identification of Exosomes. Anal. Chem. 2020, 92, 5411–5418. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Liu, Z.; Li, J.; Yang, H.; Liu, Y.; Kong, J. Sensitive electrochemical detection of A549 exosomes based on DNA/ferrocene-modified single-walled carbon nanotube complex. Anal. Biochem. 2023, 660, 114971. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Garcia-Rico, E.; O’Loghlen, A.; Giannini, V.; Alvarez-Puebla, R.A. Surface-Enhanced Raman Scattering (SERS) Spectroscopy for Sensing and Characterization of Exosomes in Cancer Diagnosis. Cancers 2021, 13, 2179. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Bu, F.; Li, X.; Zhang, S.; Min, L. Mass spectrometry-based extracellular vesicle micromolecule detection in cancer biomarker discovery: An overview of metabolomics and lipidomics. View 2023, 4, 20220086. [Google Scholar] [CrossRef]
- Anoop, S.; Asha, S.; Aamir, A.; Sandeep, A.; Khosla, A. Recent advances in electrochemical biosensors: Applications, challenges, and future scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Leva-Bueno, J.; Peyman, S.A.; Millner, P.A. A review on impedimetric immunosensors for pathogen and biomarker detection. Med. Microbiol. Immunol. 2020, 209, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Abraham, P.R.; Mukhopadhyay, S. Aptamers: An Emerging Tool for Diagnosis and Therapeutics in Tuberculosis. Front. Cell. Infect. Microbiol. 2021, 11, 656421. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide Aptamers: New Tools for Targeted Cancer Therapy. Mol. Ther-Nucl. Acids 2014, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent progress of SELEX methods for screening nucleic acid aptamers. Talanta 2023, 266, 124998. [Google Scholar] [CrossRef] [PubMed]
- Manea, I.; Casian, M.; Hosu-Stancioiu, O.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Cristea, C. A review on magnetic beads-based SELEX technologies: Applications from small to large target molecules. Anal. Chim. Acta 2024, 1297, 342325. [Google Scholar] [CrossRef] [PubMed]
- Kohlberger, M.; Gadermaier, G. SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Biochem. 2022, 69, 1771–1792. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, C.; Wang, Y.; Chen, G. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol. Biol. Rep. 2022, 49, 7979–7993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, Z.; Liu, D.; Jiang, H.; Zhang, Z.K.; Lu, A.; Zhang, G. Structural biology for the molecular insight between aptamers and target proteins. Int. J. Mol. Sci. 2021, 22, 4093. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew. Chem. Int. Ed. 2021, 60, 16800–16823. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, C.; Song, M.; Cao, Y.; Huang, Q.; Lu, F. Optimization of Gonyautoxin1/4-Binding G-Quadruplex Aptamers by Label-Free Surface-Enhanced Raman Spectroscopy. Toxins 2022, 14, 622. [Google Scholar] [CrossRef]
- Chen, J.; Meng, H.-M.; An, Y.; Geng, X.; Zhao, K.; Qu, L.; Li, Z. Structure-switching aptamer triggering hybridization displacement reaction for label-free detection of exosomes. Talanta 2020, 209, 120510. [Google Scholar] [CrossRef]
- Bunka, D.H.J.; Stockley, P.G. Aptamers come of age—At last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Xu, L.; Lu, S.; Wang, H.; Xu, H.; Ye, B.-C. Dual-Recognition Triggered Proximity Ligation Combined with a Rolling Circle Amplification Strategy for Analysis of Exosomal Protein-Specific Glycosylation. Anal. Chem. 2023, 95, 15745–15754. [Google Scholar] [CrossRef]
- Fredriksson S, Gullberg M, Jarvius J, Protein detection using proximity-dependent DNA ligation assays. Nat. biotechnol. 2002, 20, 473–477. [CrossRef]
- Wu, J.; Lin, Z.; Zou, Z.; Liang, S.; Wu, M.; Hu, T.Y.; Zhang, Y. Identifying the Phenotypes of Tumor-Derived Extracellular Vesicles Using Size-Coded Affinity Microbeads. J. Am. Chem. Soc. 2022, 144, 23483–23491. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Gu, L.; Yan, W.; Liu, L.; Wang, S.; Zhang, X.; Lyu, M. Research Progress on Rolling Circle Amplification (RCA)-Based Biomedical Sensing. Pharmaceuticals 2018, 11, 35. [Google Scholar] [CrossRef]
- Kaur, A.; Kaur, P.; Ahuja, S. Förster resonance energy transfer (FRET) and applications thereof. Anal. Methods 2020, 12, 5532–5550. [Google Scholar] [CrossRef]
- Algar, W.R.; Hildebrandt, N.; Vogel, S.S.; Medintz, I.L. FRET as a biomolecular research tool—Understanding its potential while avoiding pitfalls. Nat. Methods. 2019, 16, 815–829. [Google Scholar] [CrossRef]
- Jares-Erijman, E.A.; Jovin, T.M. FRET imaging. Nat. Biotechnol. 2003, 21, 1387–1395. [Google Scholar] [CrossRef]
- Qiu, X.; Guo, J.; Xu, J.; Hildebrandt, N. Three-Dimensional FRET Multiplexing for DNA Quantification with Attomolar Detection Limits. J. Phys. Chem. Lett. 2018, 9, 4379–4384. [Google Scholar] [CrossRef]
- Padros, J.; Chatel, G.; Caron, M. Time-resolved Förster Resonance energy transfer assays for measurement of endogenous Phosphorylated STAT proteins in human cells. J. Vis. Exp. 2021, 2021, e62915. [Google Scholar] [CrossRef]
- Geiβler, D.; Hildebrandt, N. Lanthanide complexes in FRET applications. Curr. Inorg. Chem. (Discontin.) 2011, 1, 17–35. [Google Scholar] [CrossRef]
- Sahoo, H. Förster resonance energy transfer—A spectroscopic nanoruler: Principle and applications. J. Photochem. Photobiol. 2011, 12, 20–30. [Google Scholar] [CrossRef]
- Heffern, M.C.; Matosziuk, L.M.; Meade, T.J. Lanthanide Probes for Bioresponsive Imaging. Chem. Rev. 2013, 114, 4496–4539. [Google Scholar] [CrossRef]
- Payne, N.C.; Kalyakina, A.S.; Singh, K.; Tye, M.A.; Mazitschek, R. Bright and stable luminescent probes for target engagement profiling in live cells. Nat. Chem. Biol. 2021, 17, 1168–1177. [Google Scholar] [CrossRef]
- Rectenwald, J.M.; Hardy, P.B.; Norris-Drouin, J.L.; Cholensky, S.H.; James, L.I.; Frye, S.V.; Pearce, K.H. A general TR-FRET assay platform for high-throughput screening and characterizing inhibitors of methyl-lysine reader proteins. SLAS Discov. 2019, 24, 693–700. [Google Scholar] [CrossRef]
- Qiu, X.; Guittet, O.; Mingoes, C.; El Banna, N.; Huang, M.E.; Lepoivre, M.; Hildebrandt, N. Quantification of Cellular Deoxyribonucleoside Triphosphates by Rolling Circle Amplification and Forster Resonance Energy Transfer. Anal. Chem. 2019, 91, 14561–14568. [Google Scholar] [CrossRef]
- Zhang, F.C.; Sun, Z.Y.; Liao, L.P.; Zuo, Y.; Zhang, D.; Wang, J.; Luo, C. Discovery of novel CBP bromodomain inhibitors through TR-FRET-based high-throughput screening. Acta Pharmacol. Sin. 2020, 41, 286–292. [Google Scholar] [CrossRef]
- Qiu, X.; Xu, J.; Cardoso Dos Santos, M.; Hildebrandt, N. Multiplexed biosensing and bioimaging using lanthanide-based time-gated förster resonance energy transfer. Acc. Chem. Res. 2022, 55, 551–564. [Google Scholar] [CrossRef]
- Qiu, X.; Xu, J.; Guo, J.; Yahia-Ammar, A.; Kapetanakis, N.I.; Duroux-Richard, I.; Unterluggauer, J.J.; Golob-Schwarzl, N.; Regeard, C.; Uzan, C.; et al. Advanced microRNA-based cancer diagnostics using amplified time-gated FRET. Chem. Sci. 2018, 9, 8046–8055. [Google Scholar] [CrossRef] [PubMed]
- Manouchehri Doulabi, E.; Fredolini, C.; Gallini, R.; Lof, L.; Shen, Q.; Ikebuchi, R.; Dubois, L.; Azimi, A.; Loudig, O.; Gabrielsson, S.; et al. Surface protein profiling of prostate-derived extracellular vesicles by mass spectrometry and proximity assays. Commun. Biol. 2022, 5, 1402. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, H.; Hou, M.; He, J.; Jiang, J.-H. Dual Rolling Circle Amplification-Assisted Single-Particle Fluorescence Profiling of Exosome Heterogeneity for Discriminating Lung Adenocarcinoma from Pulmonary Nodules. CCS Chem. 2022, 5, 947–957. [Google Scholar] [CrossRef]
- Feng, Y.; Guo, Y.; Li, Y.; Tao, J.; Ding, L.; Wu, J.; Ju, H. Lectin-mediated in situ rolling circle amplification on exosomes for probing cancer-related glycan pattern. Anal. Chim. Acta 2018, 1039, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Nizamudeen, Z.; Markus, R.; Lodge, R.; Parmenter, C.; Platt, M.; Chakrabarti, L.; Sottile, V. Rapid and accurate analysis of stem cell-derived extracellular vesicles with super resolution microscopy and live imaging. BBA-Mol. Cell Res. 2018, 1865, 1891–1900. [Google Scholar] [CrossRef]
- Alenquer, M.; Amorim, M. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef] [PubMed]
- Daassi, D.; Mahoney, K.M.; Freeman, G.J. The importance of exosomal PD-L1 in tumour immune evasion. Nat. Rev. Immunol. 2020, 20, 209–215. [Google Scholar] [CrossRef]
- Shukla, H.D.; Vaitiekunas, P.; Cotter, R.J. Advances in membrane proteomics and cancer biomarker discovery: Current status and future perspective. Proteomics 2012, 12, 3085–3104. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Liu, X.; Xie, Y.; Chen, M.; Zheng, C.; Zhong, H.; Li, M. Integrated SERS-vertical flow biosensor enabling multiplexed quantitative profiling of serological exosomal proteins in patients for accurate breast cancer subtyping. ACS Nano 2023, 17, 4077–4088. [Google Scholar] [CrossRef]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef]
- Li, N.; Huang, Z.; Ye, Z.; Zhang, X.; Chen, L.; Xiao, Y. Total membrane lipid assay (MLA): Simple and practical quantification of exosomes based on efficient membrane-specific dyes unaffected by proteins. Mater. Chem. Front. 2018, 2, 2130–2139. [Google Scholar] [CrossRef]
- Hao, J.; Wang, J.; Dong, Y.; Yang, J.; Wang, Z.; Zhao, X.; Li, J. Homogeneous, simple, and direct analysis of exosomal PD-L1 via aptamer-bivalent-cholesterol-anchor assembly of DNAzyme (ABCzyme) for tumor immunotherapy. Anal. Chem. 2023, 95, 6854–6862. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.-C. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mao, Z.; Chen, Q.; Koh, K.; Hu, X.; Chen, H. Rapid and sensitive detection of PD-L1 exosomes using Cu-TCPP 2D MOF as a SPR sensitizer. Biosens. Bioelectron. 2022, 201, 113954. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Qiu, X.; Mei, Q.; Luo, Y.; Fu, W. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal. Bioanal. Chem. 2020, 412, 601–609. [Google Scholar] [CrossRef]



| Strategy | Identification Target | Linear Range | LOD | Reference |
|---|---|---|---|---|
| Size-coded affinity microbeads | PD-L1, EpCAM, EGFR | 0–60 ng∙μL−1 | --- | [25] |
| Electrochemical sandwich immunosensor | α-CD9 | 102–106 particles∙μL−1 | 200 particles∙μL−1 | [50] |
| Fluorescence detection | CD63 | 103–107 particles∙μL−1 | --- | [6] |
| Total membrane lipid assay | Bilipid layer | 0–200 ng∙μL−1 | 0.342 ng μL−1 | [51] |
| DNAzyme- based method | PD-L1 | 0–1000 ng∙μL−1 | 5.21 ng∙μL−1 | [52] |
| Magnetic-based microfluidic device | CD63 | 7.6 × (101–105) particles ∙μL−1 | 4.39 particles∙μL−1 | [53] |
| SERS-based method | MUC1, HER2, CEA | 1–3 × 107 particles∙mL−1 | 107–1012 particles∙mL−1 | [49] |
| SPR-based method | PD-L1 | 10–5000 particles ∙μL−1 | 0.0167 particles∙μL−1 | [54] |
| CRISPR/Cas12a | CD63 | 3 × 103–6 ×107 particles∙μL−1 | --- | [55] |
| APPROACH | CD63, PD-L1, HER2 | 0–200 ng∙μL−1 | 0.46 ng∙μL−1, 0.77 ng∙μL−1, 1.1 ng∙μL−1 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Qian, M.; Liu, Y.; Qiu, X. APPROACH: Sensitive Detection of Exosomal Biomarkers by Aptamer-Mediated Proximity Ligation Assay and Time-Resolved Förster Resonance Energy Transfer. Biosensors 2024, 14, 233. https://doi.org/10.3390/bios14050233
Li Y, Qian M, Liu Y, Qiu X. APPROACH: Sensitive Detection of Exosomal Biomarkers by Aptamer-Mediated Proximity Ligation Assay and Time-Resolved Förster Resonance Energy Transfer. Biosensors. 2024; 14(5):233. https://doi.org/10.3390/bios14050233
Chicago/Turabian StyleLi, Ying, Meiqi Qian, Yongpeng Liu, and Xue Qiu. 2024. "APPROACH: Sensitive Detection of Exosomal Biomarkers by Aptamer-Mediated Proximity Ligation Assay and Time-Resolved Förster Resonance Energy Transfer" Biosensors 14, no. 5: 233. https://doi.org/10.3390/bios14050233
APA StyleLi, Y., Qian, M., Liu, Y., & Qiu, X. (2024). APPROACH: Sensitive Detection of Exosomal Biomarkers by Aptamer-Mediated Proximity Ligation Assay and Time-Resolved Förster Resonance Energy Transfer. Biosensors, 14(5), 233. https://doi.org/10.3390/bios14050233






