DNA-Based Molecular Machines: Controlling Mechanisms and Biosensing Applications
Abstract
1. Introduction
2. The Controlling Mechanisms of DMMs
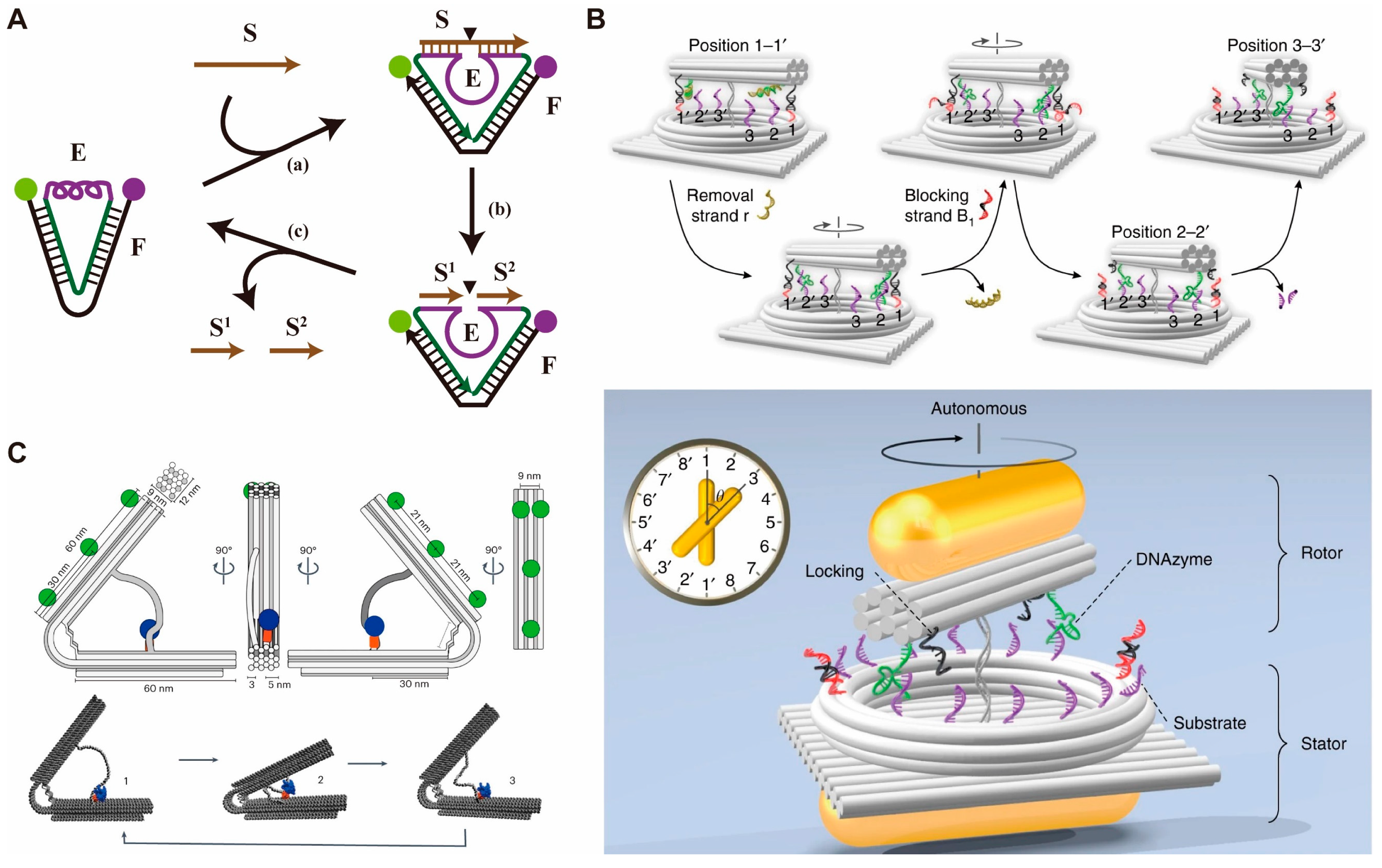
2.1. DMMs Driven by DNA Fuels

2.2. DMMs Driven by Enzymes/DNAzymes
2.3. DMMs Driven by Electric Field
2.4. DMMs Driven by Light
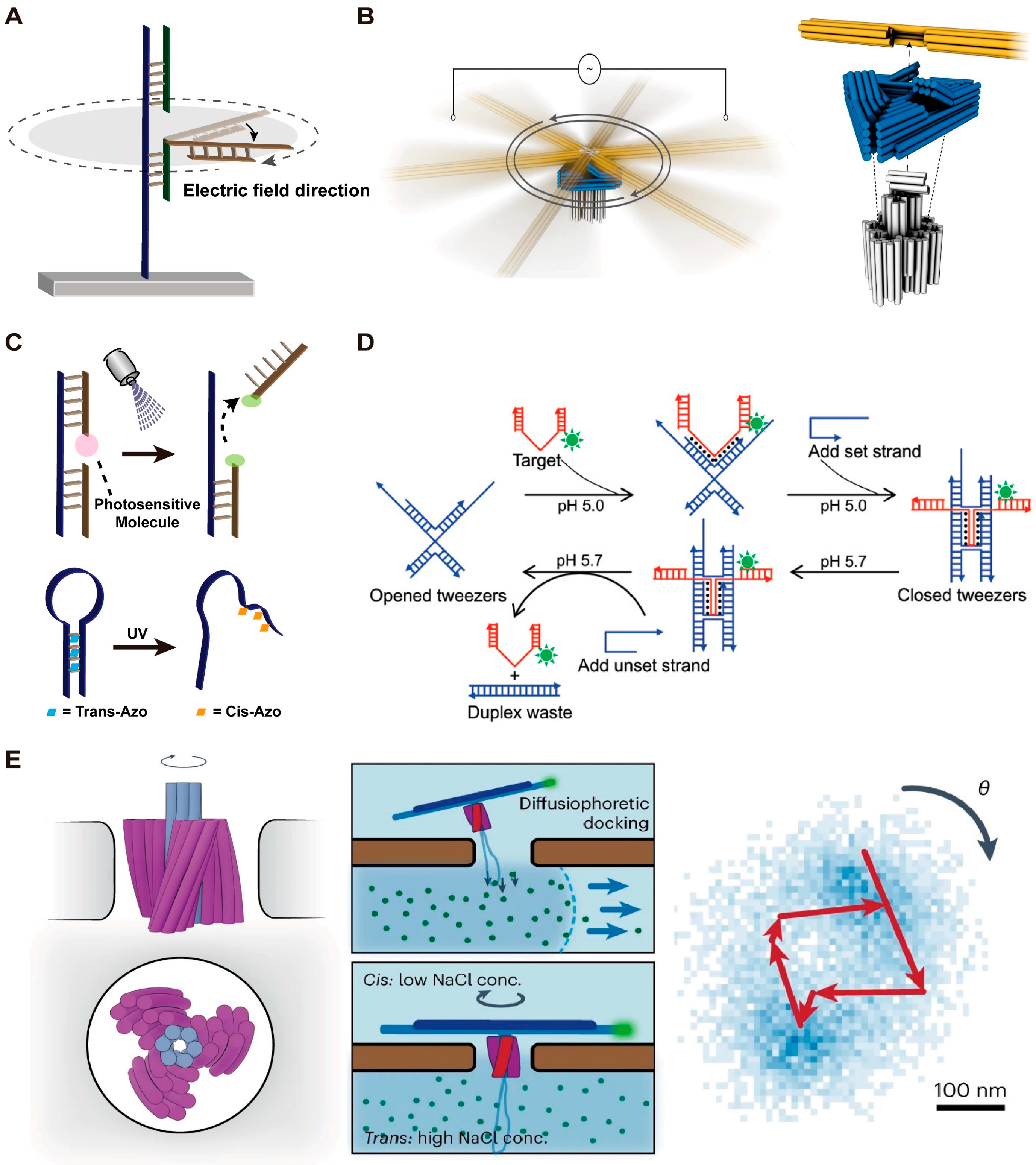
2.5. DMMs Driven by Other Stimuli
3. Biosensing Applications of DMMs
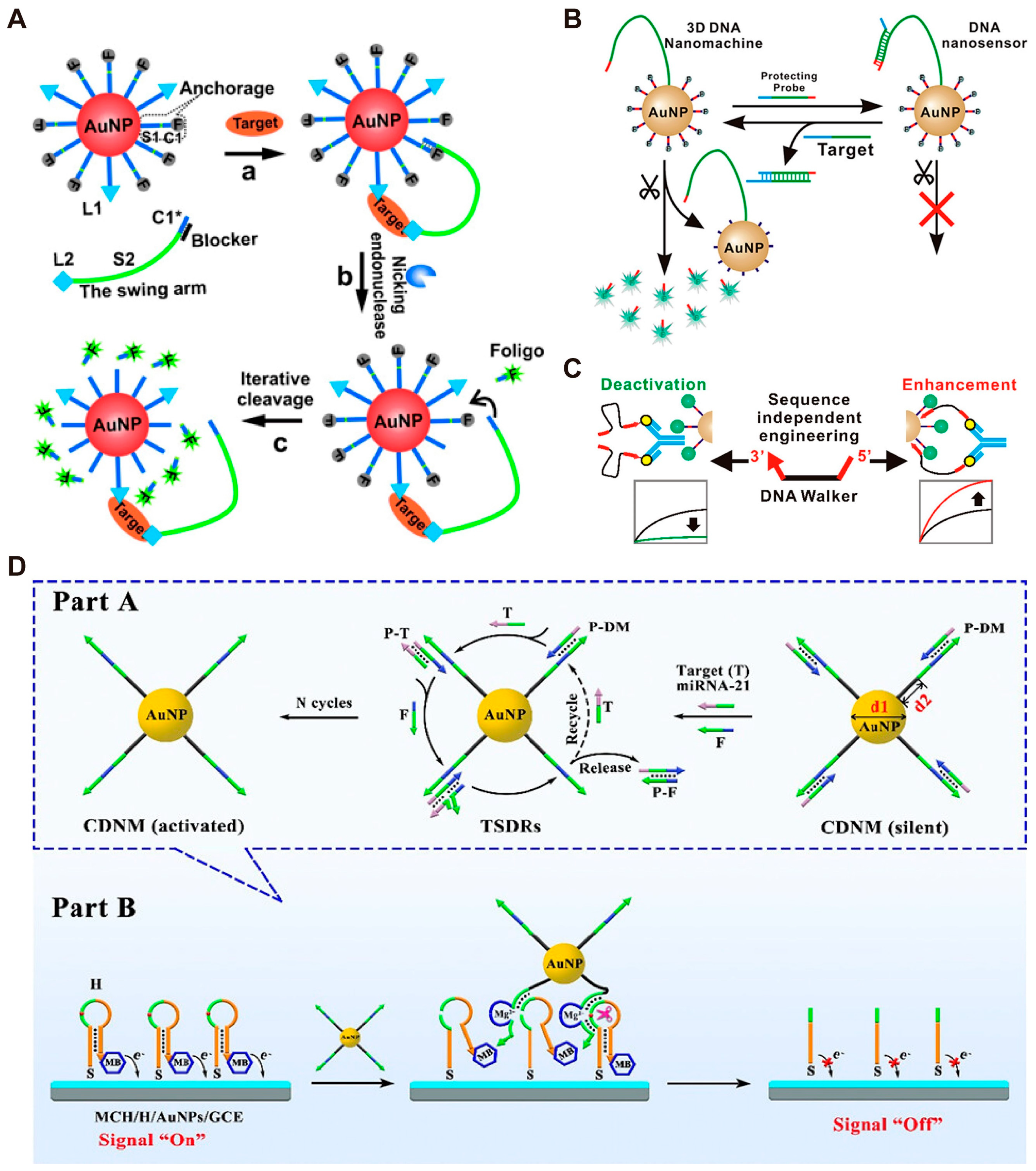
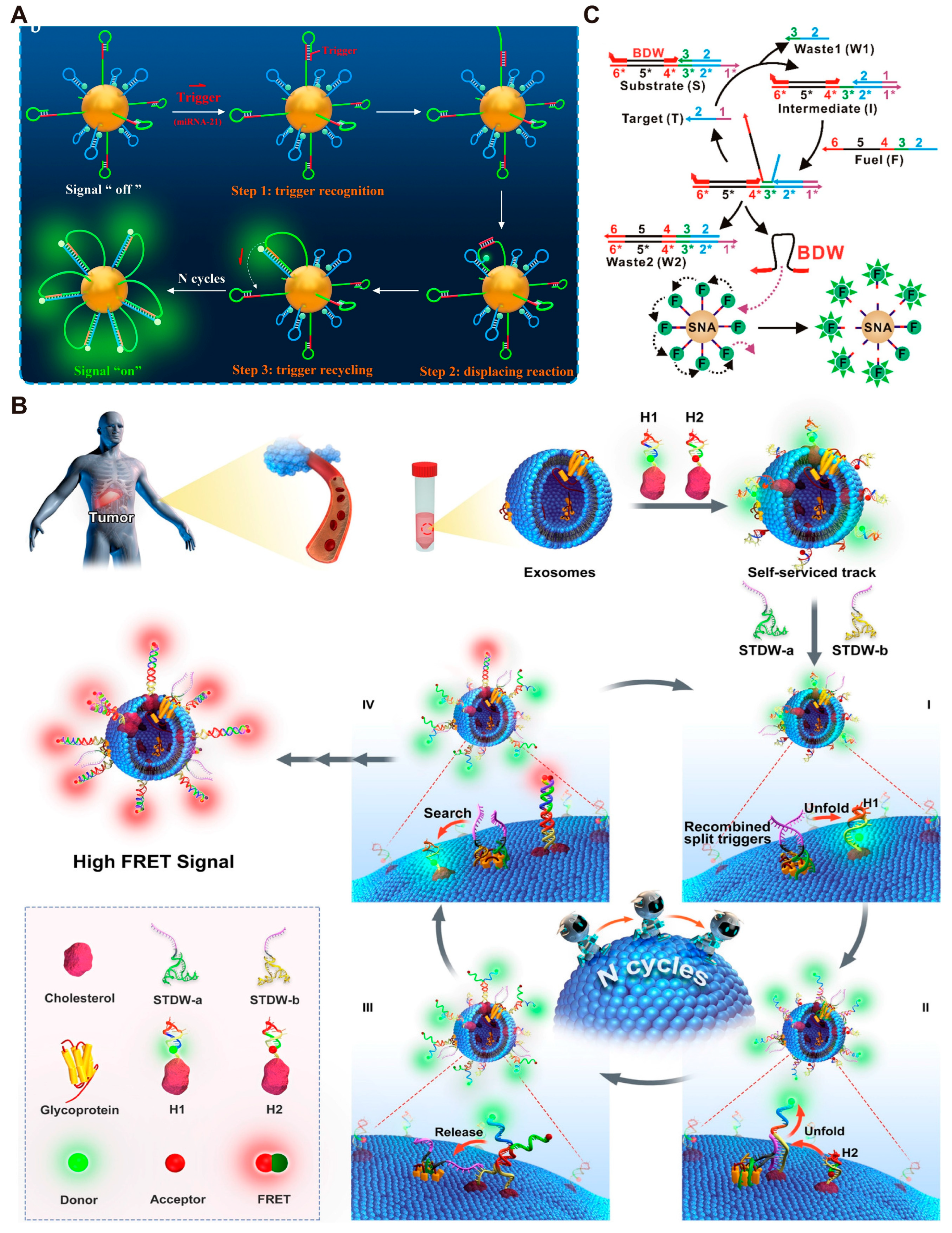
3.1. DMMs for Amplified Detection of Biomarkers
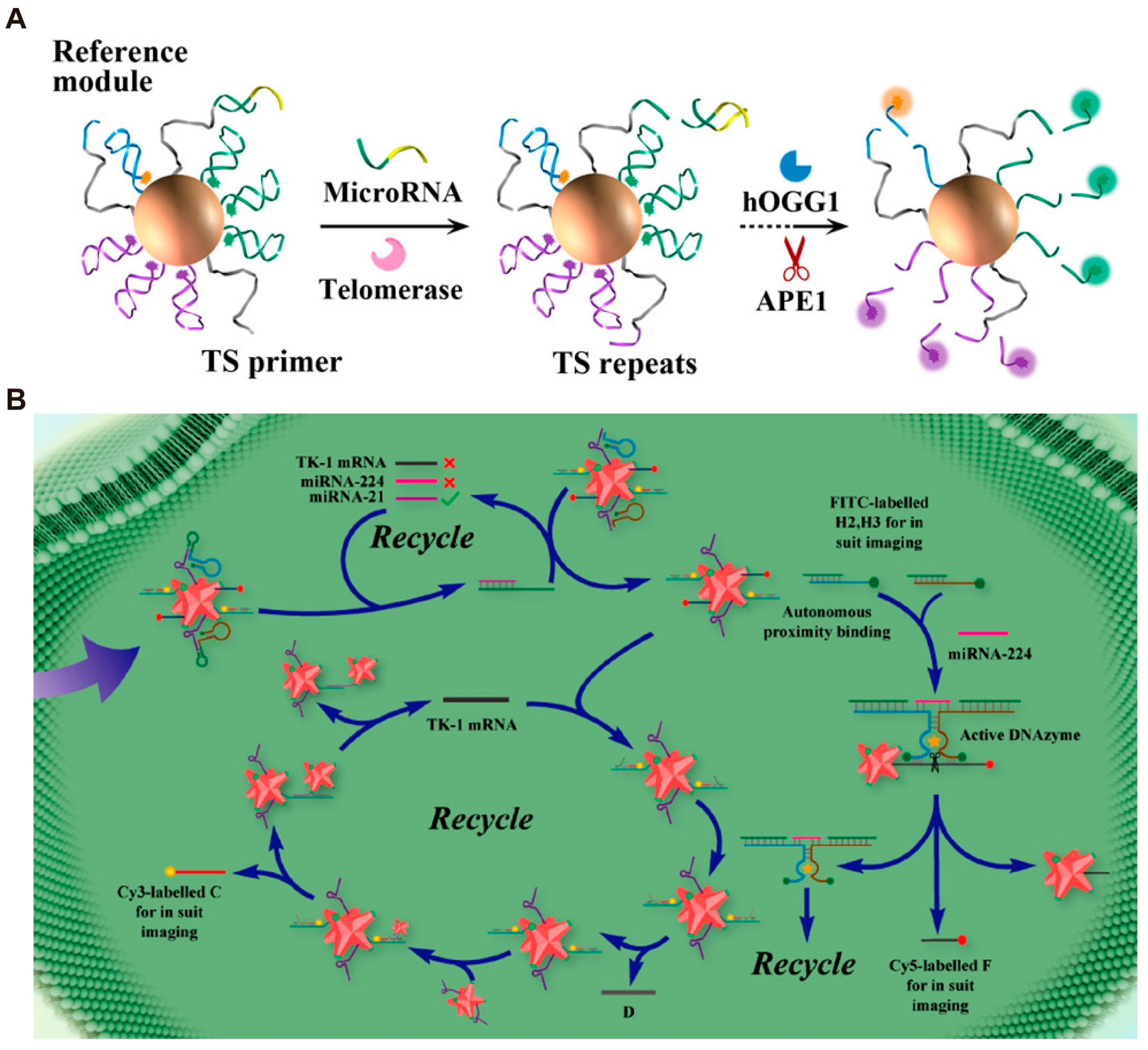
3.2. DMMs for Multiplex Detection of Biomarkers
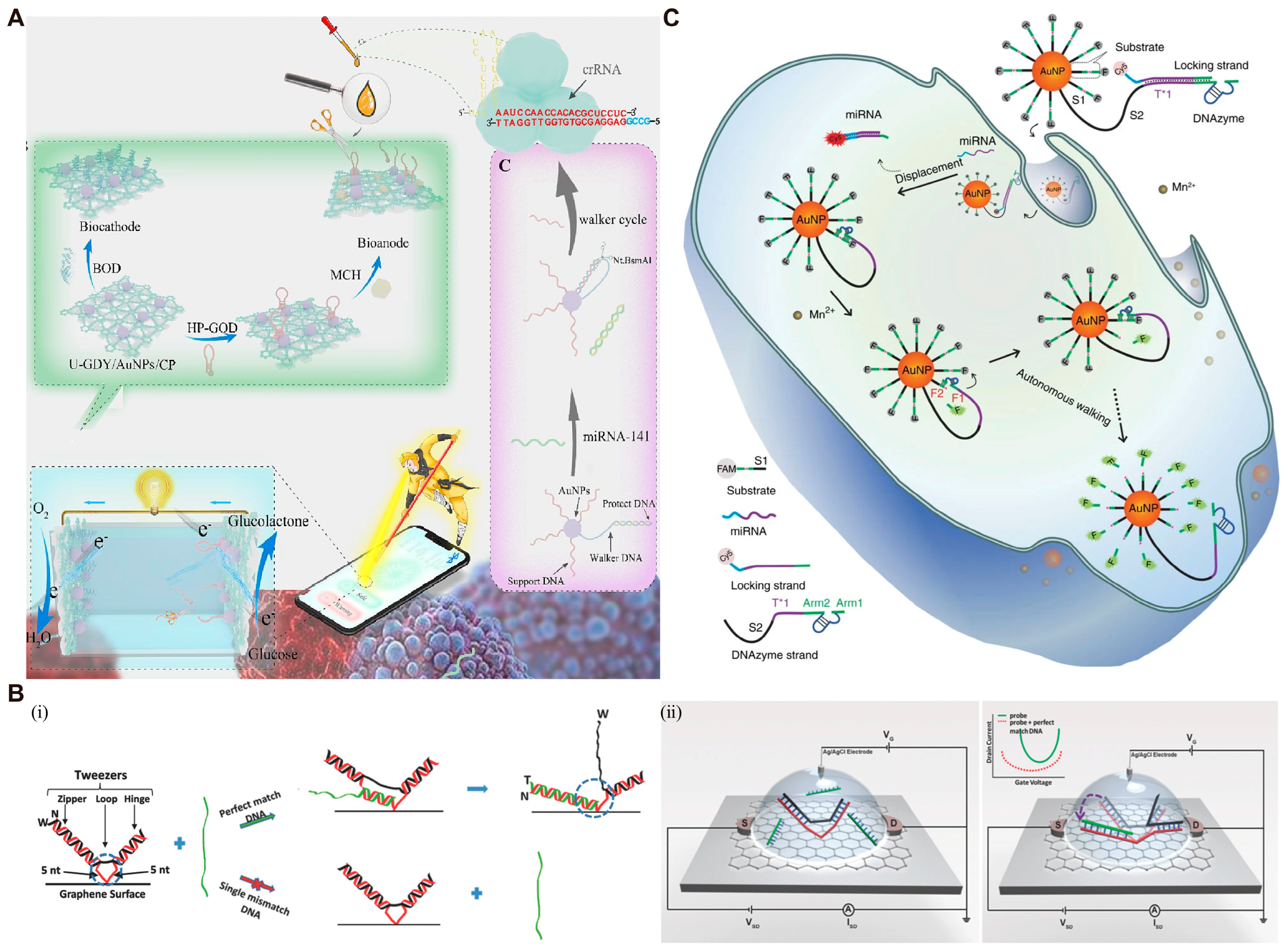
3.3. DMMs for Real-Time Monitoring of Biomarkers
3.4. DMMs for Spatial Recognition Detection of Biomarkers
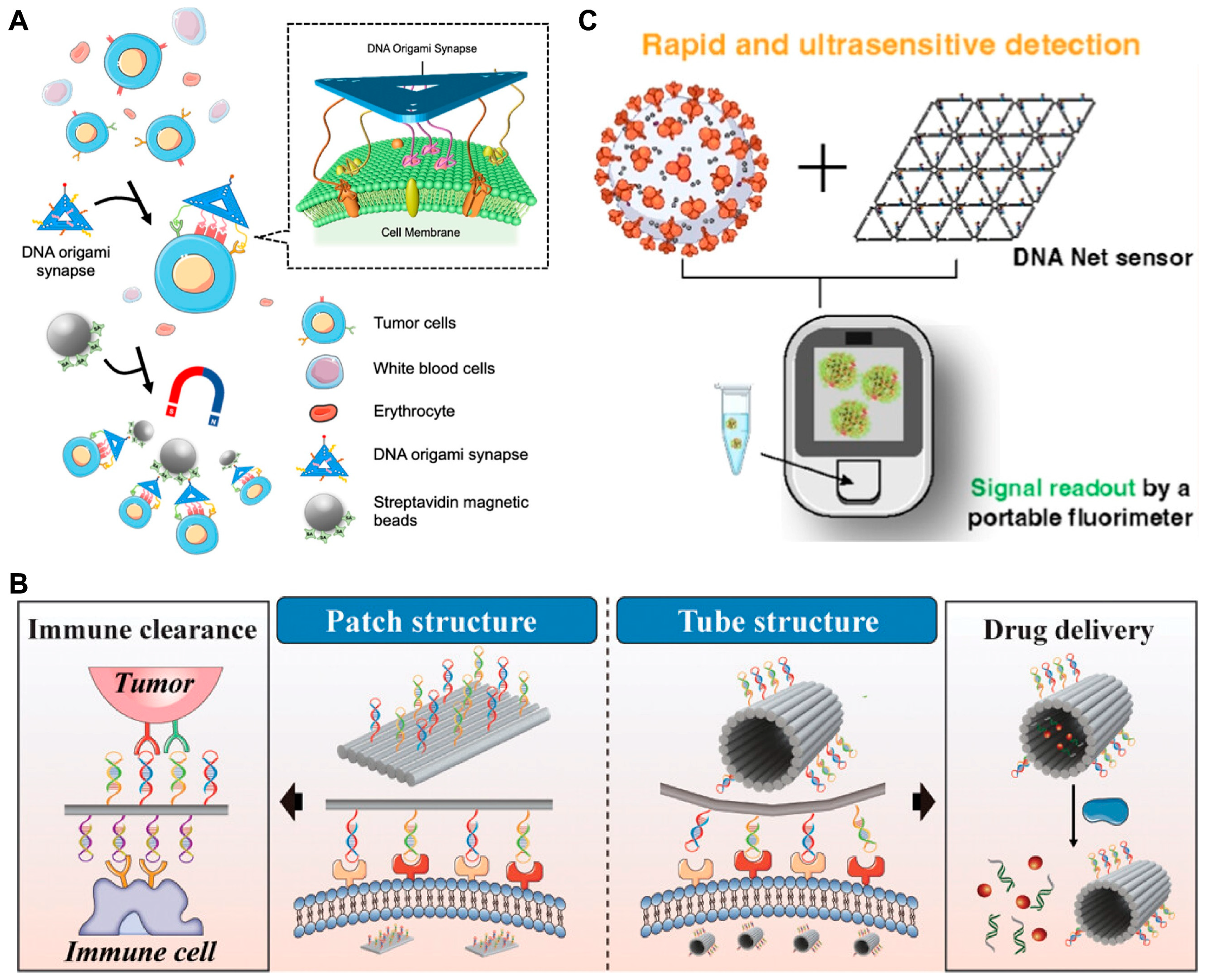
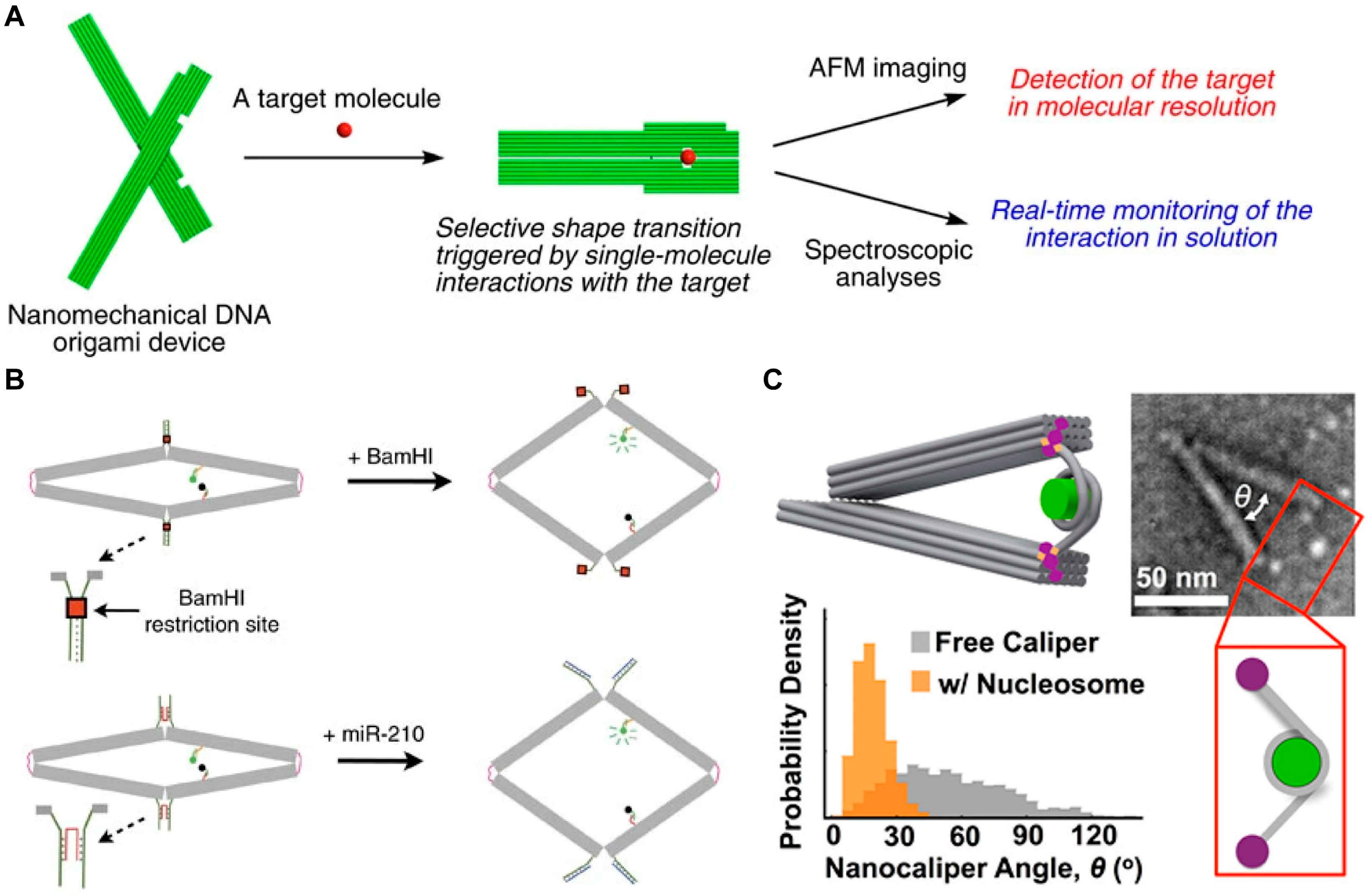
3.5. DMMs for Single-Molecule Detection of Biomarkers
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tasbas, M.N.; Sahin, E.; Erbas-Cakmak, S. Bio-inspired molecular machines and their biological applications. Coord. Chem. Rev. 2021, 443, 214039. [Google Scholar] [CrossRef]
- Song, C.; Wang, Z.G.; Ding, B.Q. Smart Nanomachines Based on DNA Self-Assembly. Small 2013, 9, 2382–2392. [Google Scholar] [CrossRef] [PubMed]
- Feynman, R.P. There’s plenty of room at the bottom. J. Microelectromech. Syst. 1992, 1, 60–66. [Google Scholar] [CrossRef]
- Livoreil, A.; Dietrichbuchecker, C.O.; Sauvage, J.P. Electrochemically Triggered Swinging of a 2-Catenate. J. Am. Chem. Soc. 1994, 116, 9399–9400. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, M.C.; Dietrich-Buchecker, C.; Sauvage, J.P. Towards synthetic molecular muscles:: Contraction and stretching of a linear rotaxane dimer. Angew. Chem. Int. Ed. 2000, 39, 3284–3287. [Google Scholar] [CrossRef]
- Liu, Y.; Flood, A.H.; Bonvallett, P.A.; Vignon, S.A.; Northrop, B.H.; Tseng, H.R.; Jeppesen, J.O.; Huang, T.J.; Brough, B.; Baller, M.; et al. Linear artificial molecular muscles. J. Am. Chem. Soc. 2005, 127, 9745–9759. [Google Scholar] [CrossRef]
- Chang, J.C.; Tseng, S.H.; Lai, C.C.; Liu, Y.H.; Peng, S.M.; Chiu, S.H. Mechanically interlocked daisy-chain-like structures as multidimensional molecular muscles. Nat. Chem. 2017, 9, 128–134. [Google Scholar] [CrossRef]
- Badjic, J.D.; Balzani, V.; Credi, A.; Silvi, S.; Stoddart, J.F. A molecular elevator. Science 2004, 303, 1845–1849. [Google Scholar] [CrossRef] [PubMed]
- Badjic, J.D.; Ronconi, C.M.; Stoddart, J.F.; Balzani, V.; Silvi, S.; Credi, A. Operating molecular elevators. J. Am. Chem. Soc. 2006, 128, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Boursalian, G.B.; Nijboer, E.R.; Dorel, R.; Pfeifer, L.; Markovitch, O.; Blokhuis, A.; Feringa, B. All-Photochemical Rotation of Molecular Motors with a Phosphorus Stereoelement. J. Am. Chem. Soc. 2020, 142, 16868–16876. [Google Scholar] [CrossRef] [PubMed]
- Bach, N.N.; Josef, V.; Maid, H.; Dube, H. Active Mechanical Threading by a Molecular Motor. Angew. Chem. Int. Ed. 2022, 61, e202201882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, Y.Y.; Liu, W.G.; Chen, H.L.; Shen, D.K.; Song, B.; Cai, K.; Wu, H.; Jiao, Y.; Feng, Y.N.; et al. An electric molecular motor. Nature 2023, 613, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Galbadage, T.; Liu, D.D.; Alemany, L.B.; Pal, R.; Tour, J.M.; Gunasekera, R.S.; Cirillo, J.D. Molecular Nanomachines Disrupt Bacterial Cell Wall, Increasing Sensitivity of Extensively Drug-Resistant Klebsiella pneumoniae to Meropenem. ACS Nano 2019, 13, 14377–14387. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.D.; García-López, V.; Gunasekera, R.S.; Nilewski, L.G.; Alemany, L.B.; Aliyan, A.; Jin, T.; Wang, G.F.; Tour, J.M.; Pal, R. Near-Infrared Light Activates Molecular Nanomachines to Drill into and Kill Cells. Acs Nano 2019, 13, 6813–6823. [Google Scholar] [CrossRef] [PubMed]
- García-López, V.; Chen, F.; Nilewski, L.G.; Duret, G.; Aliyan, A.; Kolomeisky, A.B.; Robinson, J.T.; Wang, G.F.; Pal, R.; Tour, J.M. Molecular machines open cell membranes. Nature 2017, 548, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.H.; Chen, J.W.; Luan, Y.F.; Vainikka, P.A.; Thallmair, S.; Marrink, S.J.; Feringa, B.; van Rijn, P. Unidirectional rotating molecular motors dynamically interact with adsorbed proteins to direct the fate of mesenchymal stem cells. Sci. Adv. 2020, 6, eaay2756. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, S.; Hu, H.; Cheng, Y.; Zou, K.; Song, J.; Deng, J.; Li, L.; Zhang, X.-B.; Ke, G.; et al. Selective In Situ Analysis of Mature microRNAs in Extracellular Vesicles Using a DNA Cage-Based Thermophoretic Assay. Angew. Chem. Int. Ed. 2023, 62, e202303121. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, T.; Shi, S.; Gao, Y.; Cai, X.; Lin, Y. A dynamic DNA tetrahedron framework for active targeting. Nat. Protoc. 2023, 18, 1028–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, P.; Li, W.; Ye, L.; Li, L.; Li, Z.; Li, M. Near-Infrared Light-Activatable Spherical Nucleic Acids for Conditional Control of Protein Activity. Angew. Chem. Int. Ed. 2022, 61, e202117562. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chen, Y.; Zhao, S.; Tang, R.; Nie, Z.; Xing, H. A Biomimetic Approach for Spatially Controlled Cell Membrane Engineering Using Fusogenic Spherical Nucleic Acid. Angew. Chem. Int. Ed. 2022, 61, e202111647. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jiang, S.; Loza, O.; Fahmi, N.E.; Sulc, P.; Stephanopoulos, N. Rapid Photoactuation of a DNA Nanostructure using an Internal Photocaged Trigger Strand. Angew. Chem. Int. Ed. 2018, 57, 9341–9345. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, J.; Moshe, M.; Willner, I. Coherent Activation of DNA Tweezers: A “SET-RESET” Logic System. Angew. Chem. Int. Ed. 2009, 48, 3834–3837. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zeng, J.; Huang, J.; Cheng, H.; Chen, B.; Hu, X.; He, X.; Zhou, Y.; Wang, K. A Self-Serviced-Track 3D DNA Walker for Ultrasensitive Detection of Tumor Exosomes by Glycoprotein Profiling. Angew. Chem. Int. Ed. 2022, 61, e202116932. [Google Scholar] [CrossRef] [PubMed]
- Valero, J.; Famulok, M. Regeneration of Burnt Bridges on a DNA Catenane Walker. Angew. Chem. Int. Ed. 2020, 59, 16366–16370. [Google Scholar] [CrossRef] [PubMed]
- Ketterer, P.; Willner, E.M.; Dietz, H. Nanoscale rotary apparatus formed from tight-fitting 3D DNA components. Sci. Adv. 2016, 2, e1501209. [Google Scholar] [CrossRef] [PubMed]
- Lund, K.; Manzo, A.J.; Dabby, N.; Michelotti, N.; Johnson-Buck, A.; Nangreave, J.; Taylor, S.; Pei, R.; Stojanovic, M.N.; Walter, N.G.; et al. Molecular robots guided by prescriptive landscapes. Nature 2010, 465, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, X.P.; Shen, Z.Y.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, A.T.; Bazrafshan, A.S.; Yi, J.; Eisman, J.T.; Yehl, K.M.; Bian, T.; Mugler, A.; Salaita, K. Highly Polyvalent DNA Motors Generate 100+pN of Force via Autochemophoresis. Nano Lett. 2019, 19, 6977–6986. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Chen, Y.; Zhang, X.; Liu, H.; Wang, R.; Wang, K.; Williams, K.R.; Tan, W. An autonomous and controllable light-driven DNA walking device. Angew. Chem. Int. Ed. 2012, 51, 2457–2460. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Li, F.; Yang, Y.; Wang, W.; Song, Y.; Liu, D. Light-driven conformational switch of i-motif DNA. Angew. Chem. Int. Ed. 2007, 46, 2515–2517. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Song, W.; Zhao, H.Y.; Ma, X.; Yang, S.Y.; Qiao, X.J.; Sheng, Q.L.; Yue, T.L. DNA walker-assisted aptasensor for highly sensitive determination of Ochratoxin A. Biosens. Bioelectron. 2021, 182, 113171. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Song, H.J.; Zhao, X.; Liu, R.; Lv, Y. Multiplex DNA Walking Machines for Lung Cancer-Associated miRNAs. Anal. Chem. 2022, 94, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, X.Y.; Xia, J.; Li, X.; Yang, T.; Wang, J.H. Ratiometric 3D DNA Machine Combined with Machine Learning Algorithm for Ultrasensitive and High-Precision Screening of Early Urinary Diseases. ACS Nano 2021, 15, 19522–19534. [Google Scholar] [CrossRef]
- Xiao, M.S.; Zou, K.; Li, L.; Wang, L.H.; Tian, Y.; Fan, C.H.; Pei, H. Stochastic DNA Walkers in Droplets for Super-Multiplexed Bacterial Phenotype Detection. Angew. Chem. Int. Ed. 2019, 58, 15448–15454. [Google Scholar] [CrossRef]
- Mills, A.; Aissaoui, N.; Maurel, D.; Elezgaray, J.; Morvan, F.; Vasseur, J.J.; Margeat, E.; Quast, R.B.; Kee-Him, J.L.; Saint, N.; et al. A modular spring-loaded actuator for mechanical activation of membrane proteins. Nat. Commun. 2022, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.C.; Chen, T.S.; Li, W.X.; Mao, D.S.; Liu, C.B.; Wu, Q.; Huang, N.; Hu, S.; Sun, F.Y.; Pan, Q.H.; et al. Throwing and manipulating and cheating with a DNA nano-dice. Nat. Commun. 2023, 14, 2440. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.F.; Marras, A.E.; Huang, C.M.; Castro, C.E.; Su, H.J. Paper Origami-Inspired Design and Actuation of DNA Nanomachines with Complex Motions. Small 2018, 14, e1802580. [Google Scholar] [CrossRef] [PubMed]
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.B.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A cargo-sorting DNA robot. Science 2017, 357, eaan6558. [Google Scholar] [CrossRef] [PubMed]
- Liber, M.; Tomov, T.E.; Tsukanov, R.; Berger, Y.; Nir, E. A Bipedal DNA Motor that Travels Back and Forth between Two DNA Origami Tiles. Small 2015, 11, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.J.; Both, S.; Zhou, C.; Kuzyk, A.; Lindfors, K.; Weiss, T.; Liu, N. Gold nanocrystal-mediated sliding of doublet DNA origami filaments. Nat. Commun. 2018, 9, 1454. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Jiang, Y.J.; Zhang, L.C.; Liu, H.; Li, Y.F.; Li, C.M.; Huang, C.Z. Rational fabrication of a DNA walking nanomachine on graphene oxide surface for fluorescent bioassay. Biosens. Bioelectron. 2022, 211, 114349. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Centola, M.; Valero, J.; Matthies, M.; Sulc, P.; Famulok, M. A Self-Regulating DNA Rotaxane Linear Actuator Driven by Chemical Energy. J. Am. Chem. Soc. 2021, 143, 13292–13298. [Google Scholar] [CrossRef] [PubMed]
- Valero, J.; Pal, N.; Dhakal, S.; Walter, N.G.; Famulok, M. A bio-hybrid DNA rotor-stator nanoengine that moves along predefined tracks. Nat. Nanotechnol. 2018, 13, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Centola, M.; Poppleton, E.; Ray, S.; Centola, M.; Welty, R.; Valero, J.; Walter, N.G.; Sulc, P.; Famulok, M. A rhythmically pulsing leaf-spring DNA-origami nanoengine that drives a passive follower. Nat. Nanotechnol. 2023, 19, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, M.S.; Mao, C.D. An autonomous DNA nanomotor powered by a DNA enzyme. Angew. Chem. Int. Ed. 2004, 43, 3554–3557. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Yan, H.; Daniell, X.G.; Turberfield, A.J.; Reif, J.H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004, 43, 4906–4911. [Google Scholar] [CrossRef] [PubMed]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A self-assembled nanoscale robotic arm controlled by electric fields. Science 2018, 359, 296–300. [Google Scholar] [CrossRef]
- Pumm, A.K.; Engelen, W.; Kopperger, E.; Isensee, J.; Vogt, M.; Kozina, V.; Kube, M.; Honemann, M.N.; Bertosin, E.; Langecker, M.; et al. A DNA origami rotary ratchet motor. Nature 2022, 607, 492–498. [Google Scholar] [CrossRef]
- Shi, X.; Pumm, A.K.; Maffeo, C.; Kohler, F.; Feigl, E.; Zhao, W.X.; Verschueren, D.; Golestanian, R.; Aksimentiev, A.; Dietz, H.; et al. A DNA turbine powered by a transmembrane potential across a nanopore. Nat. Nanotechnol. 2023, 19, 338–344. [Google Scholar] [CrossRef]
- Vogt, M.; Langecker, M.; Gouder, M.; Kopperger, E.; Rothfischer, F.; Simmel, F.C.; List, J. Storage of mechanical energy in DNA nanorobotics using molecular torsion springs. Nat. Phys. 2023, 19, 741–751. [Google Scholar] [CrossRef]
- Akter, M.; Keya, J.J.; Kayano, K.; Kabir, A.M.R.; Inoue, D.; Hess, H.; Sada, K.; Kuzuya, A.; Asanuma, H.; Kakugo, A. Cooperative cargo transportation by a swarm of molecular machines. Sci. Robot. 2022, 7, eabm0677. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xie, G.H.; Liu, P.; Kong, X.Y.; Song, Y.L.; Wen, L.P.; Jiang, L. Light-Driven ATP Transmembrane Transport Controlled by DNA Nanomachines. J. Am. Chem. Soc. 2018, 140, 16048–16052. [Google Scholar] [CrossRef]
- Qi, X.J.; Lu, C.H.; Liu, X.Q.; Shimron, S.; Yang, H.H.; Willner, I. Autonomous Control of Interfacial Electron Transfer and the Activation of DNA Machines by an Oscillatory pH System. Nano Lett. 2013, 13, 4920–4924. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.B.; Jing, X.X.; Liu, M.M.; Li, F.; Li, M.; Li, Q.; Shi, J.Y.; Li, J.; Wang, L.H.; Mao, X.H.; et al. Mechano-fluorescence actuation in single synaptic vesicles with a DNA framework nanomachine. Sci. Robot. 2022, 7, eabq5151. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Cecconello, A.; Elbaz, J.; Credi, A.; Willner, I. A Three-Station DNA Catenane Rotary Motor with Controlled Directionality. Nano Lett. 2013, 13, 2303–2308. [Google Scholar] [CrossRef] [PubMed]
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347, 1446–1452. [Google Scholar] [CrossRef] [PubMed]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- Liu, M.H.; Fu, J.L.; Hejesen, C.; Yang, Y.H.; Woodbury, N.W.; Gothelf, K.; Liu, Y.; Yan, H. A DNA tweezer-actuated enzyme nanoreactor. Nat. Commun. 2013, 4, 2127. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.B.; Seeman, N.C. A precisely controlled DNA biped walking device. Nano Lett. 2004, 4, 1203–1207. [Google Scholar] [CrossRef]
- Yao, D.; Bhadra, S.; Erhu, X.; Liang, H.; Ellington, A.D.; Jung, C. Dynamic Programming of a DNA Walker Controlled by Protons. Acs Nano 2020, 14, 4007–4013. [Google Scholar] [CrossRef] [PubMed]
- Oishi, M.; Saito, K. Simple Single-Legged DNA Walkers at Diffusion-Limited Nanointerfaces of Gold Nanoparticles Driven by a DNA Circuit Mechanism. Acs Nano 2020, 14, 3477–3489. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Omabegho, T.; Sha, R.; Seeman, N.C. A Bipedal DNA Brownian Motor with Coordinated Legs. Science 2009, 324, 67–71. [Google Scholar] [CrossRef]
- Marras, A.E.; Zhou, L.F.; Su, H.J.; Castro, C.E. Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. USA 2015, 112, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.F.; Both, S.; Weiss, T.; Liu, N. DNA-Assembled Multilayer Sliding Nanosystems. Nano Lett. 2019, 19, 6385–6390. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.F.; Urban, M.J.; Both, S.; Duan, X.Y.; Kuzyk, A.; Weiss, T.; Liu, N. DNA-assembled nanoarchitectures with multiple components in regulated and coordinated motion. Sci. Adv. 2019, 5, eaax6023. [Google Scholar] [CrossRef]
- Peil, A.; Xin, L.; Both, S.; Shen, L.Y.; Ke, Y.G.; Weiss, T.; Zhan, P.F.; Liu, N. DNA Assembly of Modular Components into a Rotary Nanodevice. Acs Nano 2022, 16, 5284–5291. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Zhu, D.; Yao, G.; Su, S.; Chao, J.; Liu, H.; Zuo, X.; Wang, L.; Shi, J.; Wang, L.; et al. An Exonuclease III-Powered, On-Particle Stochastic DNA Walker. Angew. Chem. Int. Ed. 2017, 56, 1855–1858. [Google Scholar] [CrossRef]
- Yehl, K.; Mugler, A.; Vivek, S.; Liu, Y.; Zhang, Y.; Fan, M.Z.; Weeks, E.R.; Salaita, K. High-speed DNA-based rolling motors powered by RNase H. Nat. Nanotechnol. 2016, 11, 184–190. [Google Scholar] [CrossRef]
- Xin, L.; Zhou, C.; Duan, X.Y.; Liu, N. A rotary plasmonic nanoclock. Nat. Commun. 2019, 10, 5394. [Google Scholar] [CrossRef] [PubMed]
- You, M.X.; Zhu, Z.; Liu, H.P.; Gulbakan, B.; Han, D.; Wang, R.W.; Williams, K.R.; Tan, W.H. Pyrene-Assisted Efficient Photolysis of Disulfide Bonds in DNA-Based Molecular Engineering. ACS Appl. Mater. Interfaces 2010, 2, 3601–3605. [Google Scholar] [CrossRef] [PubMed]
- You, M.X.; Huang, F.J.; Chen, Z.; Wang, R.W.; Tan, W.H. Building a Nanostructure with Reversible Motions Using Photonic Energy. Acs Nano 2012, 6, 7935–7941. [Google Scholar] [CrossRef] [PubMed]
- Skugor, M.; Valero, J.; Murayama, K.; Centola, M.; Asanuma, H.; Famulok, M. Orthogonally Photocontrolled Non-Autonomous DNA Walker. Angew. Chem. Int. Ed. 2019, 58, 6948–6951. [Google Scholar] [CrossRef] [PubMed]
- Han, X.G.; Zhou, Z.H.; Yang, F.; Deng, Z.X. Catch and Release: DNA Tweezers that Can Capture, Hold, and Release an Object under Control. J. Am. Chem. Soc. 2008, 130, 14414–14415. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Lamm, G.; Pack, G.R. The triplex-hairpin transition in cytosine-rich DNA. Biophys. J. 2004, 87, 3954–3973. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.A.P.; Zheng, L.W.; Eisenstein, M.; Soh, H.T. Rational design of aptamer switches with programmable pH response. Nat. Commun. 2020, 11, 2946. [Google Scholar] [CrossRef]
- Flynn, C.D.; Chang, D.; Mahmud, A.; Yousefi, H.; Das, J.; Riordan, K.T.; Sargent, E.H.; Kelley, S.O. Biomolecular sensors for advanced physiological monitoring. Nat. Rev. Bioeng. 2023, 1, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Wang, Y.D.; Xu, X.; Liu, Y.L.; Lin, B.Q.; Zhang, M.X.; Zhang, J.L.; Wan, S.; Yang, C.Y.; Tan, W.H. Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.M.; Jiang, J.; Peng, K.F.; Chai, Y.Q.; Yuan, R. Two kinds of DNA enzyme-powered bidirectional one-dimensional DNA walking nanomachine for payload release and biosensing. Biosens. Bioelectron. 2021, 175, 112848. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hu, Y.L.; Pan, Q.S.; Yi, J.T.; Zhang, J.; He, M.M.; He, M.Y.; Chen, T.T.; Chu, X. A microRNA-triggered self-powered DNAzyme walker operating in living cells. Biosens. Bioelectron. 2019, 136, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lin, Y.W.; Lau, A.; Tang, Y.N.; Chen, J.B.; Le, X.C. Binding-Induced Molecular Amplifier as a Universal Detection Platform for Biomolecules and Biomolecular Interaction. Anal. Chem. 2018, 90, 8651–8657. [Google Scholar] [CrossRef]
- Li, W.; Rong, Y.C.; Wang, J.Y.; Li, T.Z.; Wang, Z.G. MnO2 switch-bridged DNA walker for ultrasensitive sensing of cholinesterase activity and organophosphorus pesticides. Biosens. Bioelectron. 2020, 169, 112605. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Li, Y.Y.; Mason, S.D.; Chen, F.F.; Chen, J.B.; Zhou, R.X.; Liu, J.W.; Hou, X.D.; Li, F. Concentric DNA Amplifier That Streamlines In-Solution Biorecognition and On-Particle Biocatalysis. Anal. Chem. 2020, 92, 3220–3227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Q.; Lai, M.D.; Zuehlke, A.; Peng, H.Y.; Li, X.F.; Le, X.C. Binding-Induced DNA Nanomachines Triggered by Proteins and Nucleic Acids. Angew. Chem. Int. Ed. 2015, 54, 14326–14330. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Tang, Y.A.; Mason, S.D.; Chen, J.B.; Li, F. Enzyme-Powered Three-Dimensional DNA Nanomachine for DNA Walking, Payload Release, and Biosensing. Acs Nano 2016, 10, 2324–2330. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Xu, X.W.; Jiang, W. A track-regenerated DNA walker: Construction and its derived sensing application. Sens. Actuators B Chem. 2020, 314, 128053. [Google Scholar] [CrossRef]
- Yang, P.; Zhou, R.X.; Kong, C.P.; Fan, L.; Dong, C.; Chen, J.B.; Hou, X.D.; Li, F. Stimuli-Responsive Three-Dimensional DNA Nanomachines Engineered by Controlling Dynamic Interactions at Biomolecule-Nanoparticle Interfaces. ACS Nano 2021, 15, 16870–16877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gao, X.; Ho, Y.P.; Liu, M.; Han, Y.; Li, D.L.; Yuan, H.M.; Zhang, C.Y. Controllable Assembly of a Quantum Dot-Based Aptasensor Guided by CRISPR/Cas12a for Direct Measurement of Circulating Tumor Cells in Human Blood. Nano Lett. 2024, 24, 2360–2368. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, X.L.; Chai, Y.Q.; Yuan, R. Controllable Three-Dimensional DNA Nanomachine-Mediated Electrochemical Biosensing Platform for Rapid and Ultrasensitive Detection of MicroRNA. Anal. Chem. 2023, 95, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Williamson, P.; Piskunen, P.; Ijäs, H.; Butterworth, A.; Linko, V.; Corrigan, D.K. Signal Amplification in Electrochemical DNA Biosensors Using Target-Capturing DNA Origami Tiles. ACS Sens. 2023, 8, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lei, S.; Zou, L.N.; Li, G.P.; Xu, L.L.; Ye, B.X. Highly ordered 3D electrochemical DNA biosensor based on dual orientation controlled rolling motor and graftable tetrahedron DNA. Biosens. Bioelectron. 2020, 147, 111759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xia, Y.; Liu, G.Q.; Ouyang, X.P. A miniature integrated nuclear reactor design with gravity independent autonomous circulation. Nucl. Eng. Des. 2018, 340, 9–16. [Google Scholar] [CrossRef]
- Zhu, L.P.; Zhang, M.Q.; Ye, J.; Yan, M.X.; Zhu, Q.J.; Huang, J.S.; Yang, X.R. Ratiometric Electrochemiluminescent/Electrochemical Strategy for Sensitive Detection of MicroRNA Based on Duplex-Specific Nuclease and Multilayer Circuit of Catalytic Hairpin Assembly. Anal. Chem. 2020, 92, 8614–8622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Lv, S.Z.; Lin, Z.Z.; Tang, D.P. CdS:Mn quantum dot-functionalized g-C3N4 nanohybrids as signal-generation tags for photoelectrochemical immunoassay of prostate specific antigen coupling DNAzyme concatamer with enzymatic biocatalytic precipitation. Biosens. Bioelectron. 2017, 95, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.D.; Gao, Z.Q.; Wei, Q.H.; Chen, G.N.; Tang, D.P. Hemin/G-quadruplex-based DNAzyme concatamers for in situ amplified impedimetric sensing of copper(II) ion coupling with DNAzyme-catalyzed precipitation strategy. Biosens. Bioelectron. 2015, 74, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Lin, Y.X.; Zhang, K.Y.; Li, M.J.; Tang, D.P. Reduced graphene oxide/BiFeO3 nanohybrids-based signal-on photoelectrochemical sensing system for prostate-specific antigen detection coupling with magnetic microfluidic device. Biosens. Bioelectron. 2018, 101, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, C.; Deng, S.; Jiang, Y.; Zhang, P.; Yang, H.; Xiang, L.; Lyu, Y.; Cai, R.; Tan, W. Dual-responsive 3D DNA nanomachines cascaded hybridization chain reactions for novel self-powered flexible microRNA-detecting platform. Biosens. Bioelectron. 2024, 252, 116149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Yang, G.M.; Zhao, J.W.; He, Y.; Yuan, R.; Chen, S.H. Dynamic 3D DNA Rolling Walkers via Directional Movement on a Lipid Bilayer Supported by Au@Fe3O4 Nanoparticles for Sensitive Detection of MiRNA-16. Anal. Chem. 2022, 94, 8346–8353. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.D.; Tang, D.P. Recent advances in DNA walker machines and their applications coupled with signal amplification strategies: A critical review. Anal. Chim. Acta 2021, 1171, 338523. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Li, Y.X.; Zhang, P.; He, L.; Feng, Y.T.; Feng, Y.Q.; Qian, C.; Tian, Y.H.; Duan, Y.X. Catalytic hairpin assembly as cascade nucleic acid circuits for fluorescent biosensor: Design, evolution and application. TrAC Trends Anal. Chem. 2022, 151, 116582. [Google Scholar] [CrossRef]
- Liu, J.M.; Zhang, Y.; Xie, H.B.; Zhao, L.; Zheng, L.; Ye, H.M. Applications of Catalytic Hairpin Assembly Reaction in Biosensing. Small 2019, 15, e1902989. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Zhang, X.L.; Liu, J.L.; Zhou, Y.; Zhang, P.; Chai, Y.Q.; Yuan, R. Dual-layer 3D DNA nanostructure: The next generation of ultrafast DNA nanomachine for microRNA sensing and intracellular imaging. Biosens. Bioelectron. 2023, 237, 115517. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Luo, Z.W.; Zhang, C.Y.; Sun, R.; Zhou, C.; Sun, C.J. Entropy driven circuit as an emerging molecular tool for biological sensing: A review. TrAC Trends Anal. Chem. 2021, 134, 116142. [Google Scholar] [CrossRef]
- Mason, S.D.; Wang, G.A.; Yang, P.; Li, Y.Y.; Li, F. Probing and Controlling Dynamic Interactions 27 at Biomolecule-Nanoparticle Interfaces Using Stochastic DNA Walkers. Acs Nano 2019, 13, 8106–8113. [Google Scholar] [CrossRef] [PubMed]
- He, M.Q.; Wang, K.; Wang, W.J.; Yu, Y.L.; Wang, J.H. Smart DNA Machine for Carcinoembryonic Antigen Detection by Exonuclease III-Assisted Target Recycling and DNA Walker Cascade Amplification. Anal. Chem. 2017, 89, 9292–9298. [Google Scholar] [CrossRef]
- Schweitzer, B.; Kingsmore, S. Combining nucleic acid amplification and detection. Curr. Opin. Biotechnol. 2001, 12, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ji, X.H.; Du, M.Y.; Tian, S.B.; He, Z.K. Rational construction of a DNA nanomachine for HIV nucleic acid ultrasensitive sensing. Nanoscale 2018, 10, 17206–17211. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.F.; Zhang, S.X.; Chen, L.; Li, M.L.; Zhang, Y.T.; Zhou, N.D. Self-Assembled DNA Nanoflowers Triggered by a DNA Walker for Highly Sensitive Electrochemical Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2021, 13, 4905–4914. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.Y.; Xiong, X.W.; Zhu, Y.; Xiao, M.S.; Li, L.; Pei, H. DNA computational device-based smart biosensors. TrAC Trends Anal. Chem. 2023, 159, 116911. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, K.R.; Liu, Z.J.; Zhang, Y.B.; Liu, P.F.; Ye, S.Y.; Zhang, Y.W.; Liang, G.X. An “on-off” signal-switchable electrochemiluminescence biosensor for ultrasensitive detection of dual microRNAs based on DNAzyme-powered DNA walker. Sens. Actuators B Chem. 2021, 348, 130660. [Google Scholar] [CrossRef]
- Freeman, R.; Liu, X.Q.; Willner, I. Amplified Multiplexed Analysis of DNA by the Exonuclease III-Catalyzed Regeneration of the Target DNA in the Presence of Functionalized Semiconductor Quantum Dots. Nano Lett. 2011, 11, 4456–4461. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Sun, H.; Yu, C.; Lu, P.; Feng, F.; Zhang, J.; Li, W.; Yao, L. Force-Encoding DNA Nanomachines for Simultaneous and Direct Detection of Multiple Pathogenic Bacteria in Blood. Anal. Chem. 2024, 96, 4314–4321. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Nizak, C.; Surana, S.; Halder, S.; Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013, 8, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Jiang, Q.; Wang, Y.N.; Ding, B.Q. Biomedical Applications of DNA-Based Molecular Devices. Adv. Healthc. Mater. 2019, 8, e1801658. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.S.; Xiang, K.K.; Wang, C.Y.; Zhang, Y.T.; Fan, G.C.; Wang, W.J.; Han, H.Y. DNA Nanotweezers for Biosensing Applications: Recent Advances and Future Prospects. ACS Sens. 2022, 7, 3–20. [Google Scholar] [CrossRef]
- Yang, W.T.; Shen, Y.; Zhang, D.Y.; Li, C.; Yuan, R.; Xu, W.J. Programmed Dual-Functional DNA Tweezer for Simultaneous and Recognizable Fluorescence Detection of microRNA and Protein. Anal. Chem. 2019, 91, 7782–7789. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Y.; Wu, Z.Y.; Sun, Q.X.; Zhuo, Y.; Chai, Y.Q.; Yuan, R. Simply Constructed and Highly Efficient Classified Cargo-Discharge DNA Robot: A DNA Walking Nanomachine Platform for Ultrasensitive Multiplexed Sensing. Anal. Chem. 2019, 91, 8123–8128. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Chen, F.; Bai, M.; Can, X.W.; Huang, P.; Zhao, Y.X. All-in-One Synchronized DNA Nanodevices Facilitating Multiplexed Cell Imaging. Anal. Chem. 2019, 91, 4696–4701. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.H.; Fu, Y.Q.; Qiu, Z.L.; Wang, Y.; Wang, W.D.; Gu, W.X.; Li, Z.; Wu, S.Y.; Gao, F.L. Multitarget Reaction Programmable Automatic Diagnosis and Treatment Logic Device. ACS Nano 2021, 15, 19150–19164. [Google Scholar] [CrossRef] [PubMed]
- Shahhosseini, M.; Beshay, P.E.; Akbari, E.; Roki, N.; Lucas, C.R.; Avendano, A.; Song, J.W.; Castro, C.E. Multiplexed Detection of Molecular Interactions with DNA Origami Engineered Cells in 3D Collagen Matrices. ACS Appl. Mater. Interfaces 2022, 14, 55307–55319. [Google Scholar] [CrossRef] [PubMed]
- Chieng, A.; Wan, Z.J.; Wang, S.P. Recent Advances in Real-Time Label-Free Detection of Small Molecules. Biosensors 2024, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Paek, S.H.; Choi, D.; Lee, M.K.; Park, J.N.; Cho, H.M.; Paek, S.H. Real-time Monitoring of Biomarkers in Serum for Early Diagnosis of Target Disease. BioChip J. 2020, 14, 2–17. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.B.; Huang, K.J.; Wang, R.J.; Li, J.Q. Cascade amplification strategy based on ultra-thin graphdiyne and CRISPR/Cas for real-time detection of tumor biomarker. Chem. Eng. J. 2023, 466, 143230. [Google Scholar] [CrossRef]
- Hwang, M.T.; Wang, Z.J.; Ping, J.L.; Ban, D.K.; Shiah, Z.C.; Antonschmidt, L.; Lee, J.; Liu, Y.S.; Karkisaval, A.G.; Johnson, A.T.C.; et al. DNA Nanotweezers and Graphene Transistor Enable Label-Free Genotyping. Adv. Mater. 2018, 30, e1802440. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.Y.; Li, X.F.; Zhang, H.Q.; Le, X.C. A microRNA-initiated DNAzyme motor operating in living cells. Nat. Commun. 2017, 8, 14378. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Swetha, M.G.; Goswami, D.; Gupta, G.D.; Mayor, S.; Krishnan, Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009, 4, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Surana, S.; Chakraborty, S.; Koushika, S.P.; Krishnan, Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat. Commun. 2011, 2, 339. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, A.; Devany, J.; Veetil, A.T.; Suresh, B.; Pillai, K.S.; Schwake, M.; Krishnan, Y. A DNA-based voltmeter for organelles. Nat. Nanotechnol. 2021, 16, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Dan, K.; Veetil, A.T.; Chakraborty, K.; Krishnan, Y. DNA nanodevices map enzymatic activity in organelles. Nat. Nanotechnol. 2019, 14, 252–259. [Google Scholar] [CrossRef]
- Prakash, V.; Tsekouras, K.; Venkatachalapathy, M.; Heinicke, L.; Pressé, S.; Walter, N.G.; Krishnan, Y. Quantitative Mapping of Endosomal DNA Processing by Single Molecule Counting. Angew. Chem. Int. Ed. 2019, 58, 3073–3076. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Prakash, V.; Halder, S.; Chakraborty, K.; Krishnan, Y. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 2015, 10, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.M.; Chen, G.F.; Gong, L.; Ge, K.Z.; Pan, W.Z.; Li, N.; Machuki, J.O.; Yu, Y.Y.; Geng, D.Q.; Dong, H.F.; et al. DNAzyme-Powered Three-Dimensional DNA Walker Nanoprobe for Detection Amyloid β-Peptide Oligomer in Living Cells and in Vivo. Anal. Chem. 2020, 92, 9247–9256. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Lin, Z.; Chen, L.; Zou, Z.Y.; Zhang, J.; Dou, Q.H.; Wu, J.C.; Chen, J.L.; Wu, M.H.; Niu, L.; et al. Modular DNA-Origami-Based Nanoarrays Enhance Cell Binding Affinity through the “Lock-and-Key” Interaction. J. Am. Chem. Soc. 2023, 145, 5447–5455. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.X.; Chi, H.L.; Fu, X.Y.; Chen, J.L.; Dong, L.Y.; Jiang, S.Q.; Li, Y.; Chen, J.Y.; Cheng, M.; Min, Q.H.; et al. Tunable Multivalent Aptamer-Based DNA Nanostructures To Regulate Multiheteroreceptor-Mediated Tumor Recognition. J. Am. Chem. Soc. 2024, 146, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Kwon, P.S.; Ren, S.; Kwon, S.J.; Kizer, M.E.; Kuo, L.; Xie, M.; Zhu, D.; Zhou, F.; Zhang, F.M.; Kim, D.; et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020, 12, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, N.; Xiong, Y.Y.; Ren, S.K.; Dwivedy, A.; Magazine, N.; Zhou, L.F.; Jin, X.H.; Zhang, T.Y.; Cunningham, B.T.; Yao, S.R.; et al. Net-Shaped DNA Nanostructures Designed for Rapid/Sensitive Detection and Potential Inhibition of the SARS-CoV-2 Virus. J. Am. Chem. Soc. 2022, 145, 20214–20228. [Google Scholar] [CrossRef] [PubMed]
- Kuzuya, A.; Sakai, Y.; Yamazaki, T.; Xu, Y.; Komiyama, M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011, 2, 449. [Google Scholar] [CrossRef]
- Ke, Y.G.; Meyer, T.; Shih, W.M.; Bellot, G. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat. Commun. 2016, 7, 10935. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Yue, L.; Li, Z.Y.; Zhang, J.J.; Tian, H.; Willner, I. Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities. Nat. Commun. 2019, 10, 4963. [Google Scholar] [CrossRef] [PubMed]
- Funke, J.J.; Ketterer, P.; Lieleg, C.; Schunter, S.; Korber, P.; Dietz, H. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv. 2016, 2, e1600974. [Google Scholar] [CrossRef]
- Le, J.V.; Luo, Y.; Darcy, M.A.; Lucas, C.R.; Goodwin, M.F.; Poirier, M.G.; Castro, C.E. Probing Nucleosome Stability with a DNA Origami Nanocaliper. Acs Nano 2016, 10, 7073–7084. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, C.H.; Pei, H.; Shi, J.Y.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Li, S.; Zeng, Y.; Lyu, Y. DNA-Based Molecular Machines: Controlling Mechanisms and Biosensing Applications. Biosensors 2024, 14, 236. https://doi.org/10.3390/bios14050236
Ma C, Li S, Zeng Y, Lyu Y. DNA-Based Molecular Machines: Controlling Mechanisms and Biosensing Applications. Biosensors. 2024; 14(5):236. https://doi.org/10.3390/bios14050236
Chicago/Turabian StyleMa, Chunran, Shiquan Li, Yuqi Zeng, and Yifan Lyu. 2024. "DNA-Based Molecular Machines: Controlling Mechanisms and Biosensing Applications" Biosensors 14, no. 5: 236. https://doi.org/10.3390/bios14050236
APA StyleMa, C., Li, S., Zeng, Y., & Lyu, Y. (2024). DNA-Based Molecular Machines: Controlling Mechanisms and Biosensing Applications. Biosensors, 14(5), 236. https://doi.org/10.3390/bios14050236



