Development of a DNA-Based Lateral Flow Strip Membrane Assay for Rapid Screening and Genotyping of Six High-Incidence STD Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Clinical Samples
2.3. Nucleic Acid Extraction from the Clinical Specimens and Polymerase Chain Reaction (PCR) Amplification
2.4. Composition of Various Solutions Used
2.5. Preparation of 9G DNAChips for Selection of Six STD Genotyping Probes
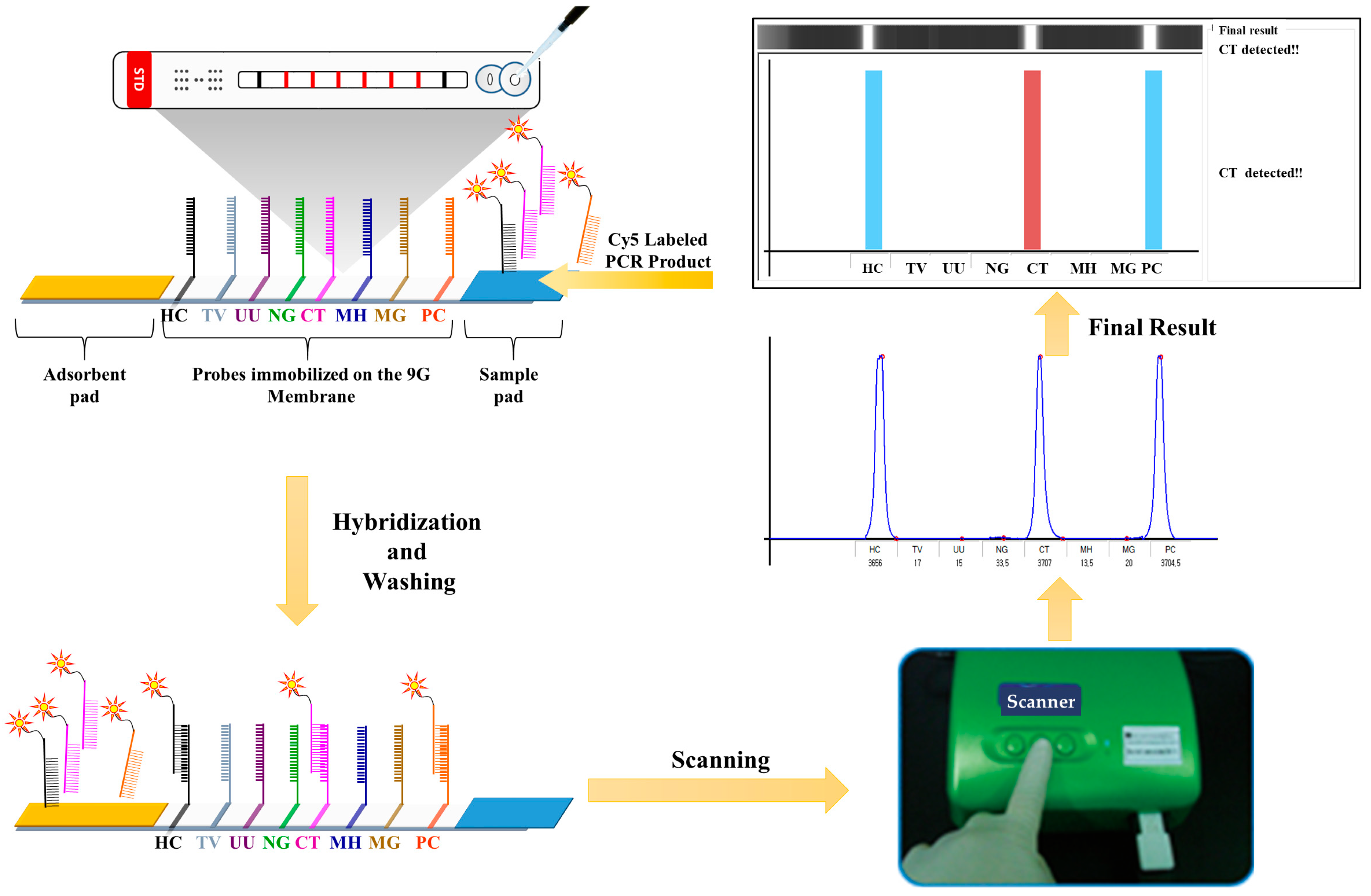
2.6. Typical Method for Preparation of the 6STD Genotyping 9G Membranes
2.7. General Procedure of Hybridization, Washing, and Scanning on 6STD Genotyping 9G Membranes
3. Results
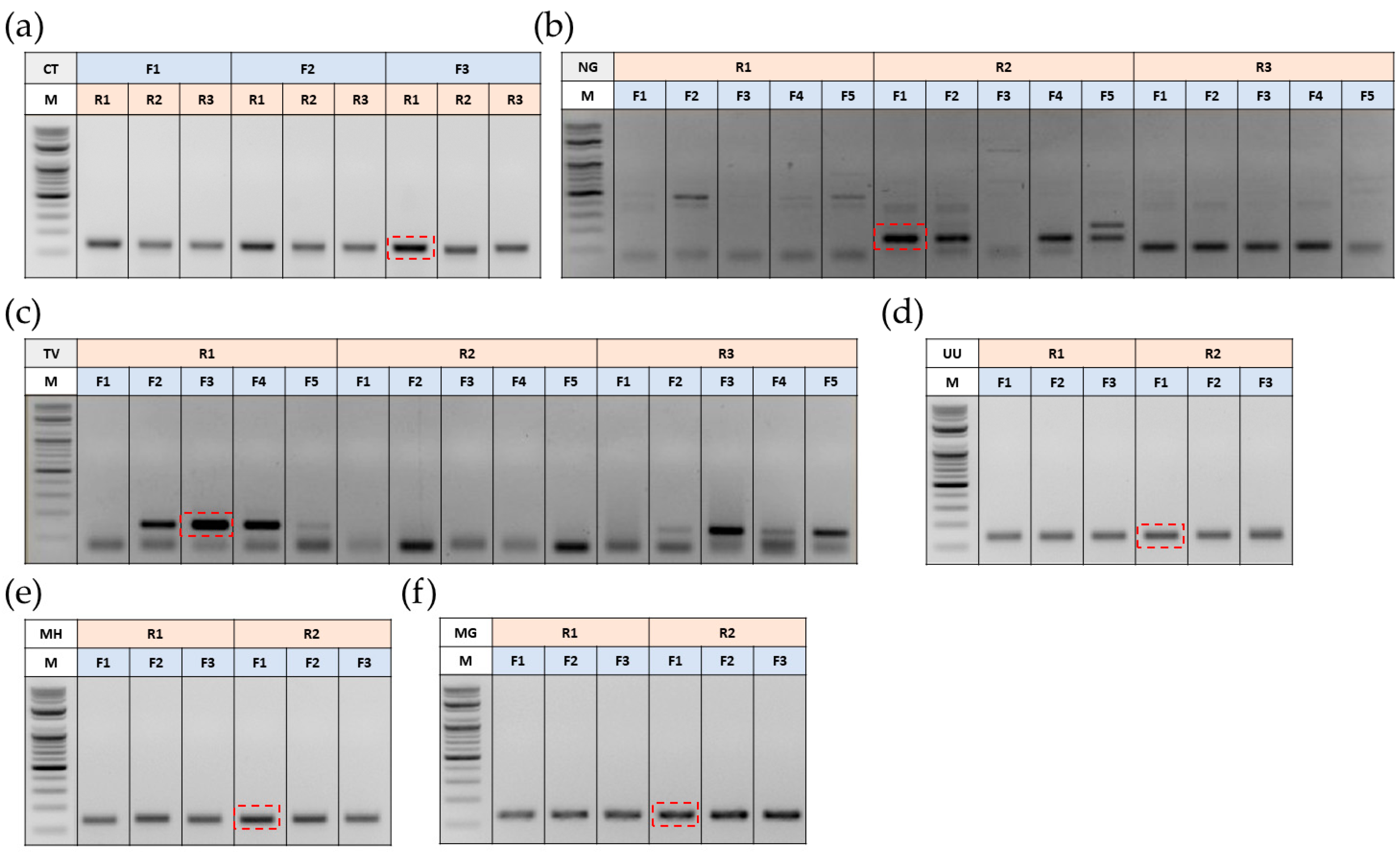
3.1. Optimization of Primers for Polymerase Chain Reaction (PCR)
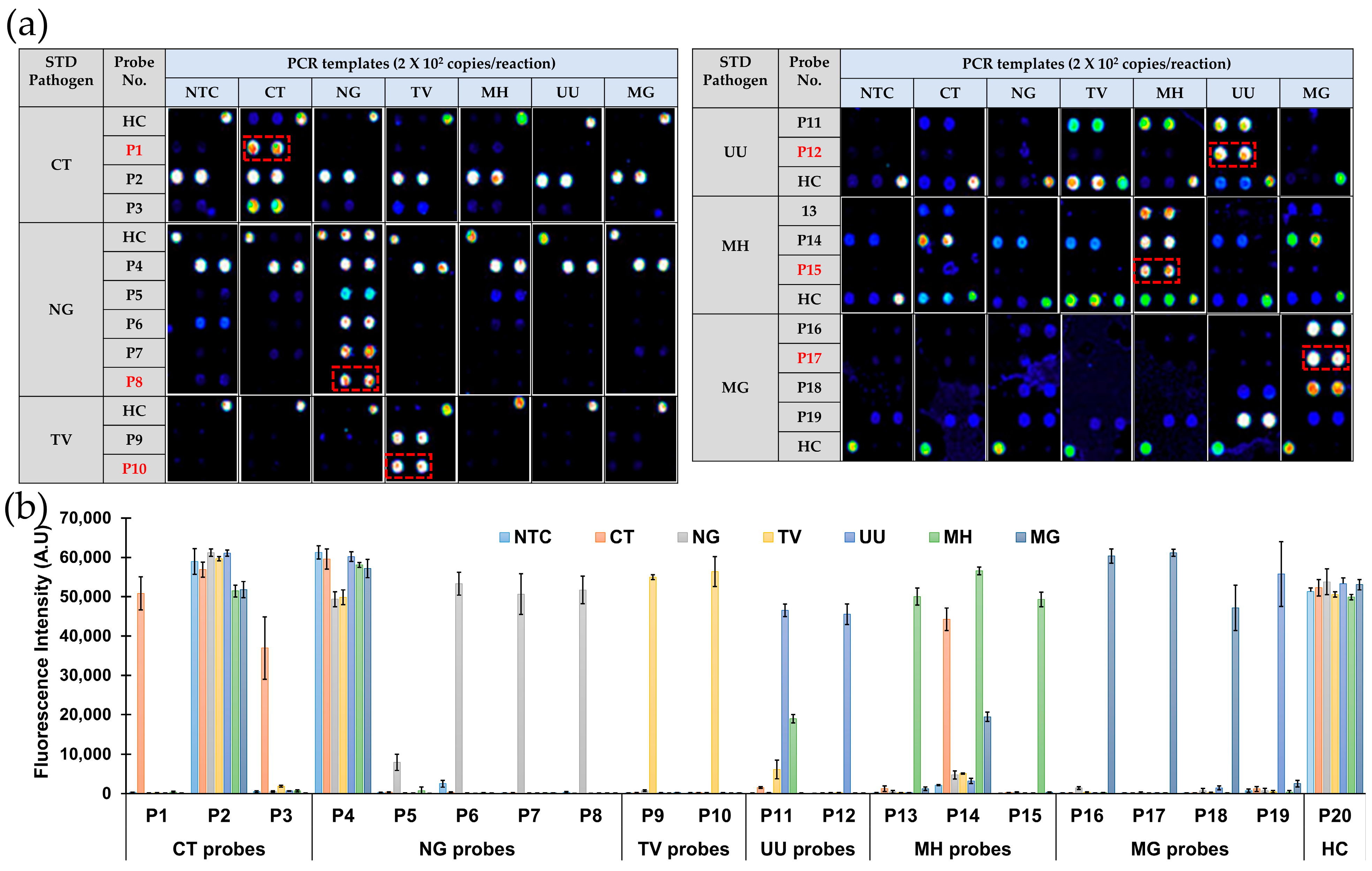
3.2. Selection of Oligonucleotide Probes for the Development of 6STD Genotyping 9G Membranes
3.3. Development of 6STD Genotyping 9G Membranes
3.4. Evaluation of 6STD Genotyping 9G Membranes
3.4.1. Specificity
3.4.2. Sensitivity (Limit of Detection)
3.5. Detection and Genotyping of STD Pathogens in Clinical Samples (Urine) Using 6STD Genotyping 9G Membrane Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021. Accountability for the Global Health Sector Strategies 2016–2021: Actions for Impact. Geneva: World Health Organization. 2021. Available online: https://apps.who.int/iris/handle/10665/341412 (accessed on 4 March 2024).
- Gökengin, D.; Noori, T.; Alemany, A.; Bienkowski, C.; Liegon, G.; İnkaya, A.Ç.; Carrillo, J.; Stary, G.; Knapp, K.; Mitja, O.; et al. Prevention strategies for sexually transmitted infections, HIV, and viral hepatitis in Europe. Lancet Reg. Health–Eur. 2023, 34, 100736. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Sun, Y.; Han, M.; Wang, B.; Xiao, F.; Zhou, Y.; Gao, Y.; Fitzpatrick, T.; Yuan, T.; Li, P.; et al. Incidence trends of five common sexually transmitted infections excluding HIV from 1990 to 2019 at the global, regional, and national levels: Results from the global burden of disease study 2019. Front. Med. 2022, 9, 851635. [Google Scholar] [CrossRef]
- Tsevat, D.G.; Wiesenfeld, H.C.; Parks, C.; Peipert, J.F. Sexually transmitted diseases and infertility. Am. J. Obstet. Gynecol. 2017, 216, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jamison, C.D.; Coleman, J.S.; Mmeje, O. Improving women’s health and combatting sexually transmitted infections through expedited partner therapy. Obstet. Gynecol. 2019, 133, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Hart, R.J. Physiological aspects of female fertility: Role of the environment, modern lifestyle, and genetics. Physiol. Rev. 2016, 96, 873–909. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.; Schmiedel, S. Sexually transmitted diseases, STD. Dtsch. Med. Wochenschr. 2022, 147, 1407–1422. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, S.; Hamill, M.M.; Ghanem, K.G. Diagnosis and treatment of sexually transmitted infections: A review. JAMA 2022, 327, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Giammanco, A.; Virruso, R.; Fasciana, T. Current and future trends in the laboratory diagnosis of sexually transmitted infections. Int. J. Environ. Res. Public Health 2021, 18, 1038. [Google Scholar] [CrossRef]
- Meyer, T. Diagnostic Procedures to Detect Chlamydia trachomatis Infections. Microorganisms 2016, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Macaluso, M.; Vermund, S.H.; Hook III, E.W. Relative accuracy of nucleic acid amplification tests and culture in detecting chlamydia in asymptomatic men. J. Clin. Microbiol. 2001, 39, 3927–3937. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Frickmann, H.; Loderstädt, U. Testing as Prevention of Resistance in Bacteria Causing Sexually Transmitted Infections—A Population-Based Model for Germany. Antibiotics 2021, 10, 929. [Google Scholar] [CrossRef]
- Adamson, P.C.; Loeffelholz, M.J.; Klausner, J.D. Point-of-care testing for sexually transmitted infections: A review of recent developments. Arch. Pathol. Lab. Med. 2020, 144, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Janssen, K.J.; Hoebe, C.J.; Dukers-Muijrers, N.H.; Eppings, L.; Lucchesi, M.; Wolffs, P.F. Viability-PCR shows that NAAT detects a high proportion of DNA from non-viable Chlamydia trachomatis. PLoS ONE 2016, 11, e0165920. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiang, T.T.; Li, J.; Yin, Y.P.; Chen, X.S. Performance of point-of-care tests for the detection of chlamydia trachomatis infections: A systematic review and meta-analysis. EClinicalMedicine 2021, 37, 100961. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Munson, E. Molecular Diagnostics Update for the Emerging (If Not Already Widespread) Sexually Transmitted Infection Agent Mycoplasma genitalium: Just About Ready for Prime Time. J. Clin. Microbiol. 2017, 55, 2894–2902. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhan, L.; Qin, Z.; Sackrison, J.; Bischof, J.C. Ultrasensitive and highly specific lateral flow assays for point-of-care diagnosis. ACS Nano 2021, 15, 3593–3611. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Song, K.-S.; Warkad, S.D.; Kim, T. A Novel Method That Allows SNP Discrimination with 160:1 Ratio for Biosensors Based on DNA-DNA Hybridization. Biosensors 2021, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Unemo, M.; Bradshaw, C.S.; Hocking, J.S.; de Vries, H.J.; Francis, S.C.; Mabey, D.; Marrazzo, J.M.; Sonder, G.J.; Schwebke, J.R.; Hoornenborg, E.; et al. Sexually transmitted infections: Challenges ahead. Lancet Infect. Dis. 2017, 17, e235–e279. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Report on Global Sexually Transmitted Infection Surveillance, 2018. 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/277258/9789241565691-eng.pdf (accessed on 4 March 2024).
- Coorevits, L.; Traen, A.; Bingé, L.; Van Dorpe, J.; Praet, M.; Boelens, J.; Padalko, E. Identifying a consensus sample type to test for Chlamydia trachomatis, Neisseria gonorrhoeae, Mycoplasma genitalium, Trichomonas vaginalis and human papillomavirus. Clin. Microbiol. Infect. 2018, 24, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Luo, J.; Chen, Y.; Chen, L.; Hu, H.; Qiu, T.; Liu, X.; Xu, X.; Chen, Y.; Zhang, Z.; et al. Prevalence of syphilis and Chlamydia trachomatis infection among female sex workers in Jiangsu, China: Results from a multicenter cross-sectional and venue-based study. Front. Public Health 2022, 10, 1018724. [Google Scholar] [CrossRef] [PubMed]
- Butcher, R.; Jarju, S.; Obayemi, D.; Bashorun, A.O.; Vasileva, H.; Bransbury-Hare, H.; Agboghoroma, O.; Drammeh, L.; Holland, M.; Harding-Esch, E.; et al. Prevalence of five treatable sexually transmitted infections among women in Lower River region of The Gambia. BMC Infect. Dis. 2023, 23, 471. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Jang, T.S.; Jeon, J.S.; Kim, J.K. Coinfections with multiple sexually transmitted pathogens in Republic of Korea, 2018–2020. J. Clin. Lab. Anal. 2022, 36, e24682. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, J.; Lee, K.A. Prevalence of sexually transmitted infections among healthy Korean women: Implications of multiplex PCR pathogen detection on antibiotic therapy. J. Infect. Chemother. 2014, 20, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Osterman, A.; Forster, J.; Jakob, L.; Suerbaum, S.; Flaig, M.J.; Schubert, S.; Wagener, J. A retrospective evaluation of the Euroarray STI-11 multiplex system for the detection of eight STI causing agents. Sci. Rep. 2023, 13, 11382. [Google Scholar] [CrossRef]
- Nimse, S.B.; Song, K.S.; Kim, J.; Ta, V.T.; Nguyen, V.T.; Kim, T. A generalized probe selection method for DNA chips. Chem. Commun. 2011, 47, 12444–12446. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; An, H.; Kim, J.; Nguyen, V.T.; Ta, V.T.; Kim, T. HPV 9G DNAChip: Based on the 9G DNAChip technology. J. Virol. Methods 2012, 183, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; Kim, J.; Kim, J.; Nguyen, V.T.; Ta, V.T.; Kim, T. 9G DNAChip: Microarray based on the multiple interactions of 9 consecutive guanines. Chem. Commun. 2011, 47, 7101–7103. [Google Scholar] [CrossRef] [PubMed]
- Sayyed, D.R.; Nimse, S.B.; Song, K.; Kim, T. Detection of multiple mutations in a single codon of genomic DNA. Chem. Commun. 2014, 50, 12344–12347. [Google Scholar] [CrossRef] [PubMed]
- Gerbase, A.C.; Rowley, J.T.; Heymann, D.H.; Berkley, S.F.; Piot, P. Global prevalence and incidence estimates of selected curable STDs. Sex. Transm. Infect. 1998, 74, S12–S16. [Google Scholar] [PubMed]
- Newman, L.; Rowley, J.; Vander Hoorn, S.; Wijesooriya, N.S.; Unemo, M.; Low, N.; Stevens, G.; Gottlieb, S.; Kiarie, J.; Temmerman, M. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 2015, 10, e0143304. [Google Scholar] [CrossRef]
- Hu, H.; Chen, Y.; Shi, L.; Liu, X.; Xu, Z.; Sun, L.; Zhao, X.; Zhou, Y.; Lu, J.; Zhang, Z.; et al. Prevalence of syphilis and chlamydia trachomatis infection among men who have sex with men in Jiangsu province, China: A cross-sectional survey. Front. Public Health 2022, 10, 1006254. [Google Scholar] [CrossRef] [PubMed]
- Warr, A.J.; Pintye, J.; Kinuthia, J.; Drake, A.L.; Unger, J.A.; Mcclelland, R.S.; Matemo, D.; Osborn, L.; John-Stewart, G. Sexually transmitted infections during pregnancy and subsequent risk of stillbirth and infant mortality in Kenya: A prospective study. Sex. Transm. Infect. 2019, 95, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, S.M.; Reno, H.E.L. Management of Patients with Sexually Transmitted Infections in the Emergency Department. Emerg. Med. Clin. N. Am. 2018, 36, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Panchanadeswaran, S.; Johnson, S.C.; Mayer, K.H.; Srikrishnan, A.K.; Sivaran, S.; Zelaya, C.E.; Go, V.F.; Solomon, S.; Bentley, M.E.; Celentano, D.D. Gender differences in the prevalence of sexually transmitted infections and genital symptoms in an urban setting in southern India. Sex. Trans. Infect. 2006, 82, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.K.; Park, S.C.; Kim, Y.G.; Choi, H.; Ko, J.I.; Kim, M.K.; Jeong, Y.B.; Shin, Y.S. The prevalence of causative organisms of community-acquired urethritis in an age group at high risk for sexually transmitted infections in Korean Soldiers. BMJ Mil. Health 2017, 163, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Waites, K.B.; Crabb, D.M.; Ratliff, A.E.; Geisler, W.M.; Atkinson, T.P.; Xiao, L. Latest Advances in Laboratory Detection of Mycoplasma genitalium. J. Clin. Microbiol. 2023, 61, e0079021. [Google Scholar] [CrossRef] [PubMed]
- Muralidhar, S. Molecular methods in the laboratory diagnosis of sexually transmitted infections. Indian J. Sex. Transm. Dis. AIDS 2015, 36, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Rumyantseva, T.; Golparian, D.; Nilsson, C.S.; Johansson, E.; Falk, M.; Fredlund, H.; Van Dam, A.; Guschin, A.; Unemo, M. Evaluation of the new AmpliSens multiplex real-time PCR assay for simultaneous detection of Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma genitalium, and Trichomonas vaginalis. APMIS 2015, 123, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Park, D.C.; Lee, D.S.; Choe, H.S.; Cho, Y.H. Evaluation of Seeplex®® STD6 ACE Detection kit for the diagnosis of six bacterial sexually transmitted infections. J. Infect. Chemother. 2012, 18, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.M.; Xu, J.X.; Jiang, L.X.; Deng, L.R.; Gu, Z.M.; Xie, X.Y.; Ji, H.C.; Wang, W.H.; Li, L.M.; Tian, C.N.; et al. Design and evaluation of a novel multiplex real-time PCR melting curve assay for the simultaneous detection of nine sexually transmitted disease pathogens in genitourinary secretions. Front. Cell. Infect. Microbiol. 2019, 9, 382. [Google Scholar] [CrossRef] [PubMed]
- Ashshi, A.M.; Batwa, S.A.; Kutbi, S.Y.; Malibary, F.A.; Batwa, M.; Refaat, B. Prevalence of 7 sexually transmitted organisms by multiplex real-time PCR in Fallopian tube specimens collected from Saudi women with and without ectopic pregnancy. BMC Infect. Dis. 2015, 15, 569. [Google Scholar] [CrossRef] [PubMed]
- Trecker, M.A.; Dillon, J.A.R.; Lloyd, K.; Hennink, M.; Waldner, C.L. Demographic and behavioural characteristics predict bacterial STI reinfection and coinfection among a cross-sectional sample of laboratory-confirmed gonorrhea cases in a local health region from Saskatchewan, Canada. Can. J. Public Health 2015, 106, e17–e21. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.S.; Lee, D.S.; Lee, S.J.; Hong, S.H.; Park, D.C.; Lee, M.K.; Kim, T.H.; Cho, Y.H. Performance of Anyplex™ II multiplex real-time PCR for the diagnosis of seven sexually transmitted infections: Comparison with currently available methods. Int. J. Infect. Dis. 2013, 17, e1134–e1140. [Google Scholar] [CrossRef] [PubMed]
- Samra, Z.; Rosenberg, S.; Madar-Shapiro, L. Direct simultaneous detection of 6 sexually transmitted pathogens from clinical specimens by multiplex polymerase chain reaction and auto-capillary electrophoresis. Diagn. Microbiol. Infect. Dis. 2011, 70, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Omidfar, K.; Riahi, F.; Kashanian, S. Lateral flow assay: A summary of recent progress for improving assay performance. Biosensors 2023, 13, 837. [Google Scholar] [CrossRef] [PubMed]
- Kweon, O.J.; Choi, J.H.; Song, U.H.; Park, A.J. Performance evaluation of a DNA chip assay in the identification of major genitourinary pathogens. J. Microbiol. Methods 2015, 109, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Healey, S.; Regan, P.; Laksanalamai, P.; Hu, Z. PCR-based methodologies for detection and characterization of Listeria monocytogenes and Listeria ivanovii in foods and environmental sources. Food Sci. Hum. Wellness 2017, 6, 39–59. [Google Scholar] [CrossRef]


| STD Pathogen | Probe Name | Probe Sequence (5′ to 3′) | Tm (°C) | GC % |
|---|---|---|---|---|
| CT | P1 | GGGGGGGGG AAATACAAA TGAGGCTGATGACTAGGATG | 61.0 | 50.0 |
| P2 | GGGGGGGGG AAATACAAA CATGGGAGTTGGTTTTACCT | 60.8 | 45.0 | |
| P3 | GGGGGGGGG AAATACAAA CCAAGGTGAGGCTGATGACT | 64.7 | 55.0 | |
| NG | P4 | GGGGGGGGG AAATACAAA AGTGGGGGATACCAGAAGTA | 60.4 | 50.0 |
| P5 | GGGGGGGGG AAATACAAA ACCGCAAGGAGTCCGCTTAC | 67.8 | 60.0 | |
| P6 | GGGGGGGGG AAATACAAA ACCACGGTATGCTTCATGAC | 62.9 | 50.0 | |
| P7 | GGGGGGGGG AAATACAAA TCCGCTGACCACGGTATGCT | 70.4 | 60.0 | |
| P8 | GGGGGGGGG AAATACAAA CGCTGACCACGGTATGCTTC | 68.2 | 60.0 | |
| TV | P9 | GGGGGGGGG AAATACAAA TGGATGACTCGGTGAAATCA | 64.7 | 45.0 |
| P10 | GGGGGGGGG AAATACAAA AATCACGTTATCTAGAGGAAGG | 58.4 | 40.9 | |
| UU | P11 | GGGGGGGGG AAATACAAA GCTAACCTTTTGGAGGCATG | 63.8 | 50.0 |
| P12 | GGGGGGGGG AAATACAAA ATGCGTCTAGGGTAGGATCG | 63.3 | 55.0 | |
| MH | P13 | GGGGGGGGG AAATACAAA TTGCTAACCTCGGAGGCGAC | 68.9 | 60.0 |
| P14 | GGGGGGGGG AAATACAAA CTAAGGTAGGACTGGTGACT | 55.4 | 50.0 | |
| P15 | GGGGGGGGG AAATACAAA GACCGCCTAAGTTAGGACTG | 60.8 | 55 | |
| MG | P16 | GGGGGGGGG AAATACAAA GAAGTGCATGTCAAGGATAGC | 61.7 | 47.6 |
| P17 | GGGGGGGGG AAATACAAA CCTTTATTGGAAGTGCATGTC | 61.4 | 42.9 | |
| P18 | GGGGGGGGG AAATACAAA CCTTTATTGGAAGTGCTTGTC | 60.6 | 42.9 | |
| P19 | GGGGGGGGG AAATACAAA CTATCCTTTATTGGAAGTGC | 56.0 | 40.0 | |
| HC | P20 * | GGGGGGGGG AAATACAAA AAGGATAAGGAAGAAGCCTG | 59.4 | 45.0 |
| GGGGGGGGG AAATACAAA GTTCTAGTTTTAATAACTAACAC | 48.8 | 26.1 | ||
| GGGGGGGGG AAATACAAA CTAGAGAAAGAAGGGGCTTT | 58.7 | 45.0 | ||
| GGGGGGGGG AAATACAAA CCTACGAGAACGTGGGGATG | 67.0 | 60.0 |
| STD Pathogens | Positive Samples | Negative Samples |
|---|---|---|
| TV | 10 | 60 |
| UU | 10 | |
| NG | 10 | |
| CT | 10 | |
| MH | 10 | |
| MG | 10 |
| STD Pathogens | Total Samples | Results of 6STD Genotyping 9G Membrane | Results of Sequencing Analysis |
|---|---|---|---|
| TV | 10 | 10 | 10 |
| UU | 10 | 10 | 10 |
| NG | 10 | 10 | 10 |
| CT | 10 | 10 | 10 |
| MH | 10 | 10 | 10 |
| MG | 10 | 10 | 10 |
| Negative | 60 | 60 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, G.; Song, K.-S.; Nimse, S.B.; Kim, T. Development of a DNA-Based Lateral Flow Strip Membrane Assay for Rapid Screening and Genotyping of Six High-Incidence STD Pathogens. Biosensors 2024, 14, 260. https://doi.org/10.3390/bios14050260
Choi G, Song K-S, Nimse SB, Kim T. Development of a DNA-Based Lateral Flow Strip Membrane Assay for Rapid Screening and Genotyping of Six High-Incidence STD Pathogens. Biosensors. 2024; 14(5):260. https://doi.org/10.3390/bios14050260
Chicago/Turabian StyleChoi, Gunho, Keum-Soo Song, Satish Balasaheb Nimse, and Taisun Kim. 2024. "Development of a DNA-Based Lateral Flow Strip Membrane Assay for Rapid Screening and Genotyping of Six High-Incidence STD Pathogens" Biosensors 14, no. 5: 260. https://doi.org/10.3390/bios14050260
APA StyleChoi, G., Song, K.-S., Nimse, S. B., & Kim, T. (2024). Development of a DNA-Based Lateral Flow Strip Membrane Assay for Rapid Screening and Genotyping of Six High-Incidence STD Pathogens. Biosensors, 14(5), 260. https://doi.org/10.3390/bios14050260







