Sensitive Detection of Oral Leukoplakia: Analyzing P90 Biomarkers in Saliva and Tissue
Abstract
1. Introduction
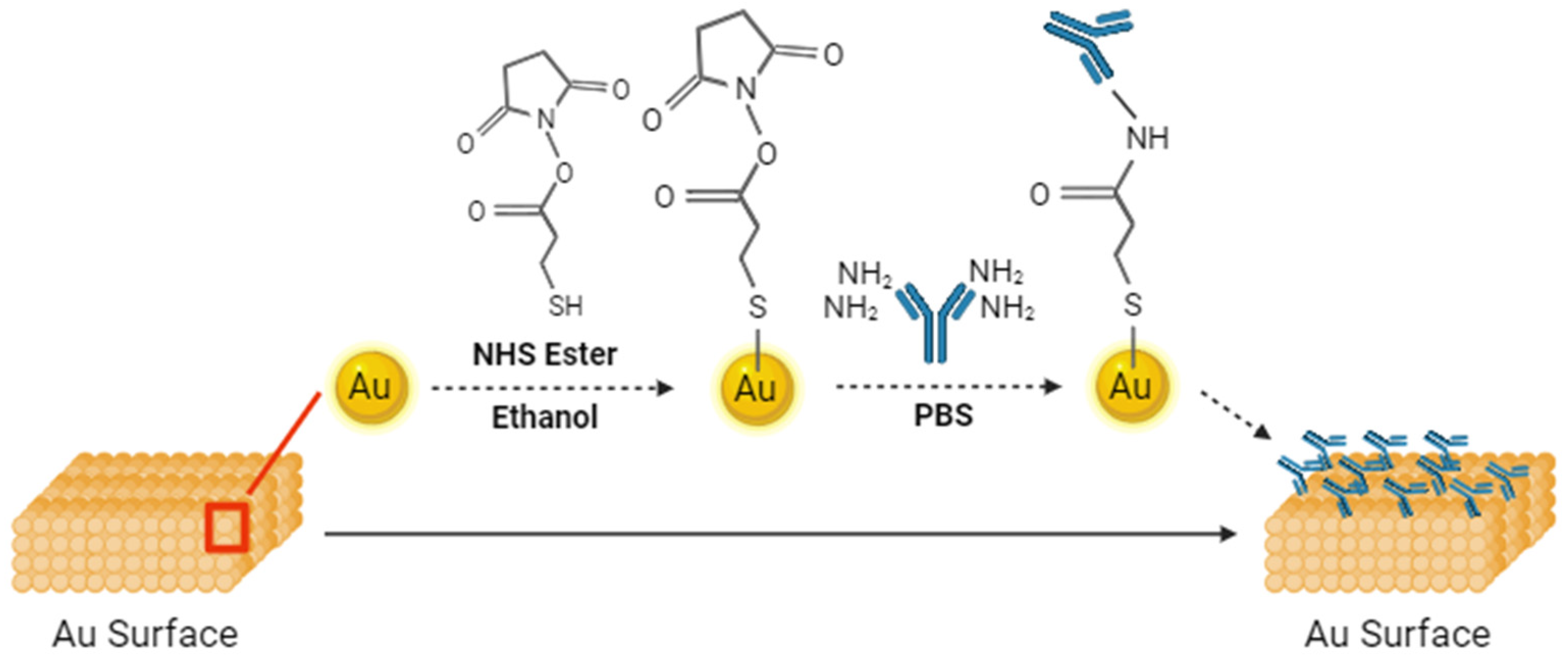
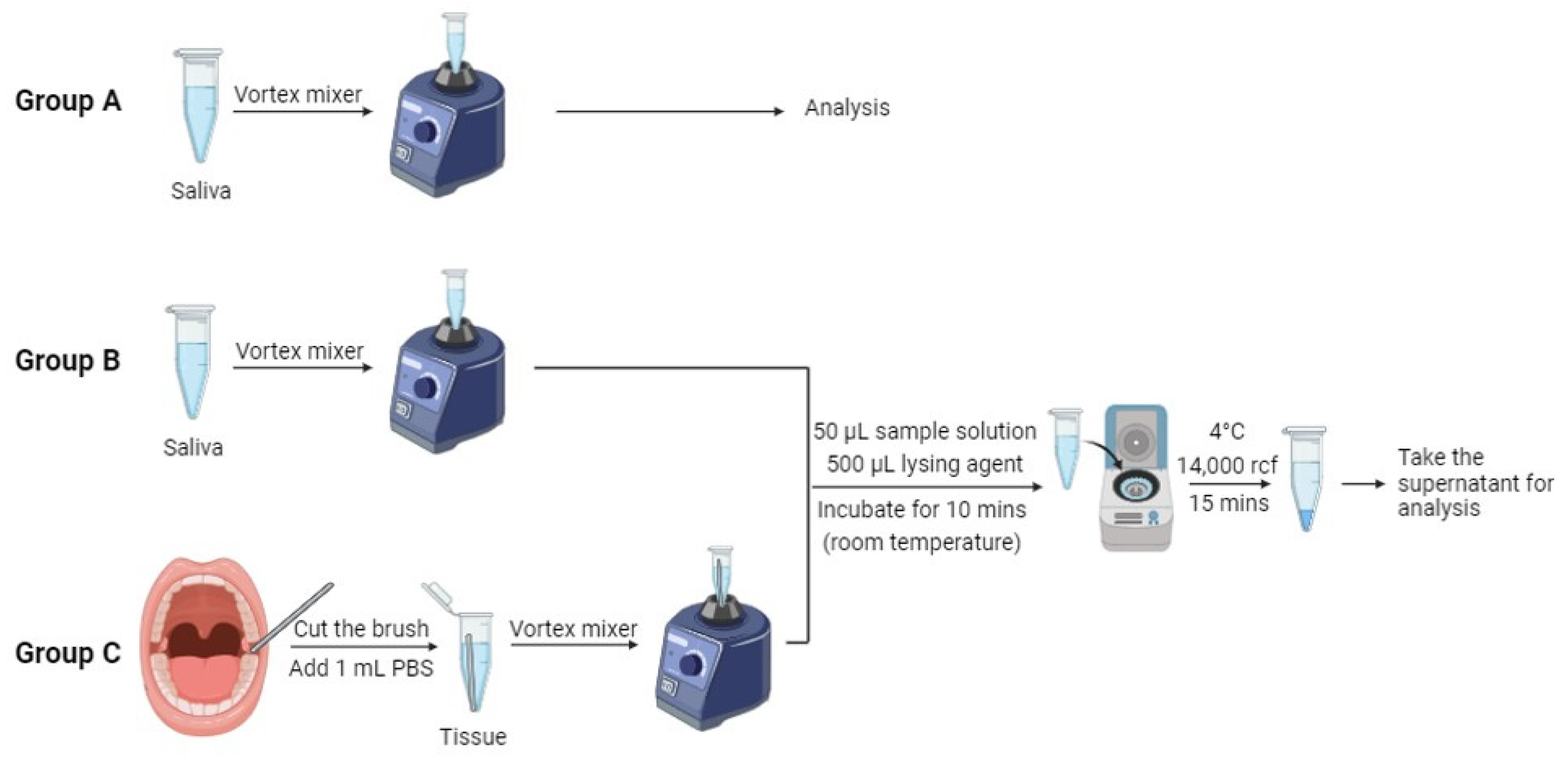
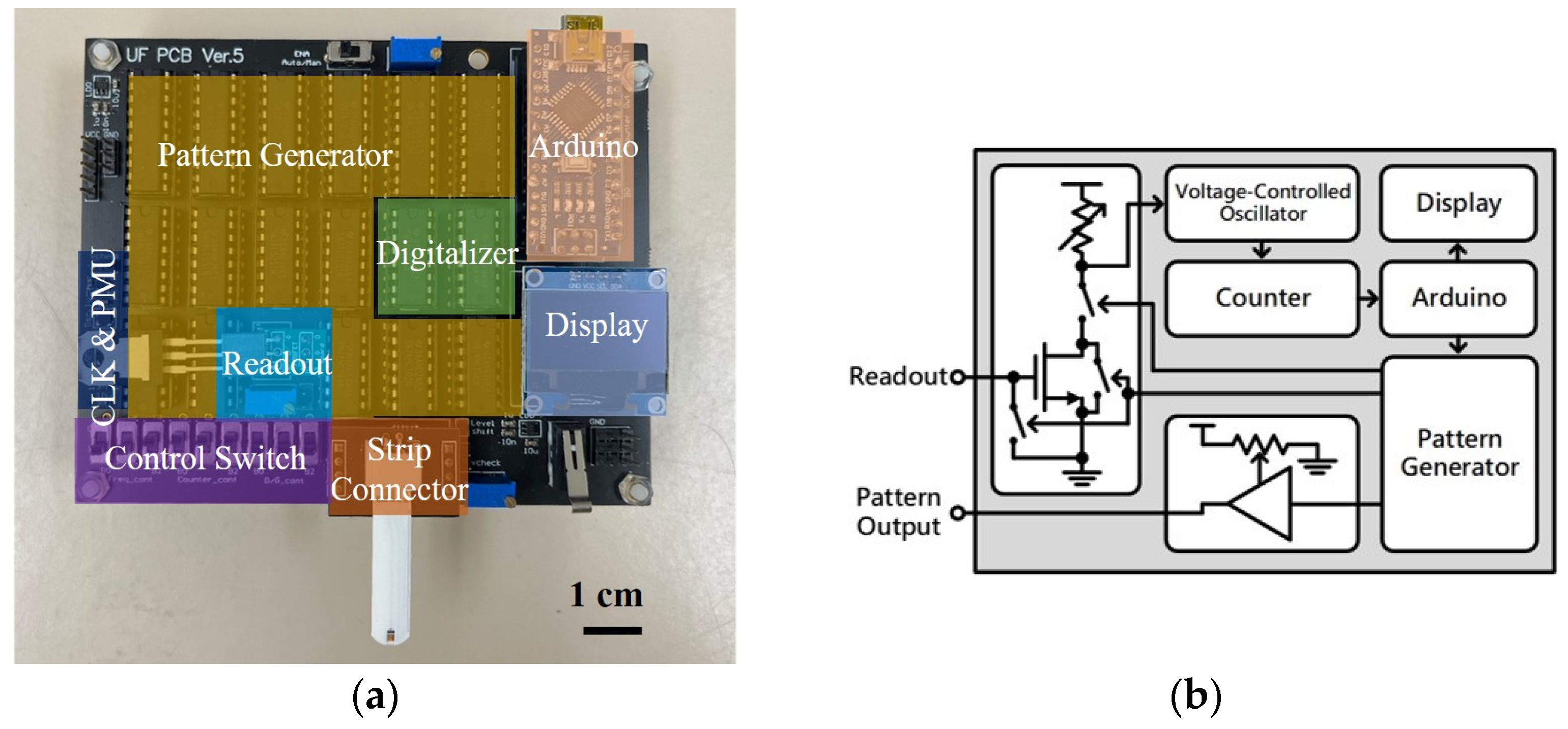
2. Materials and Methods
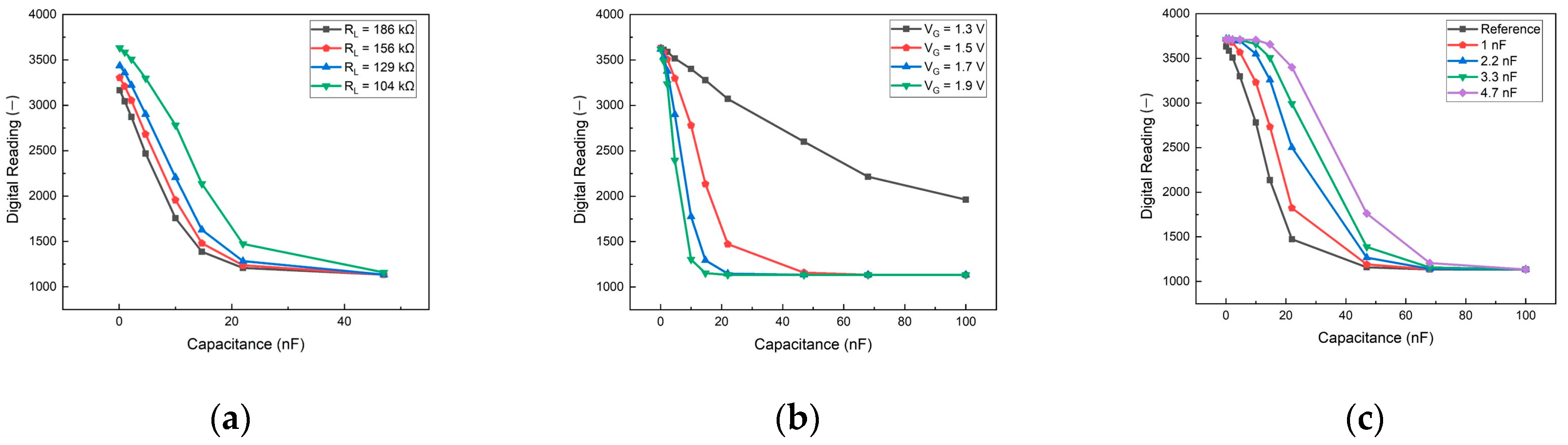
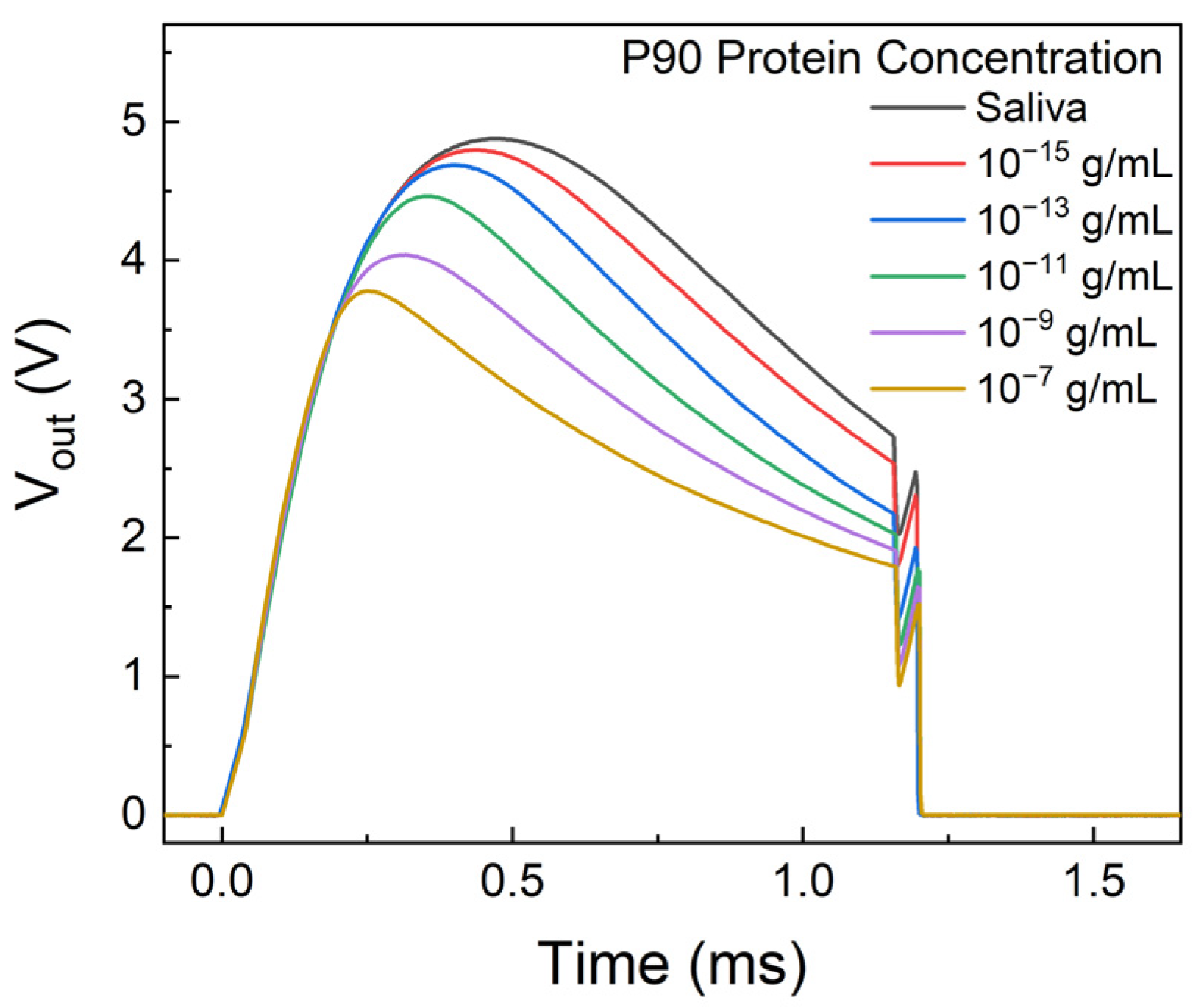
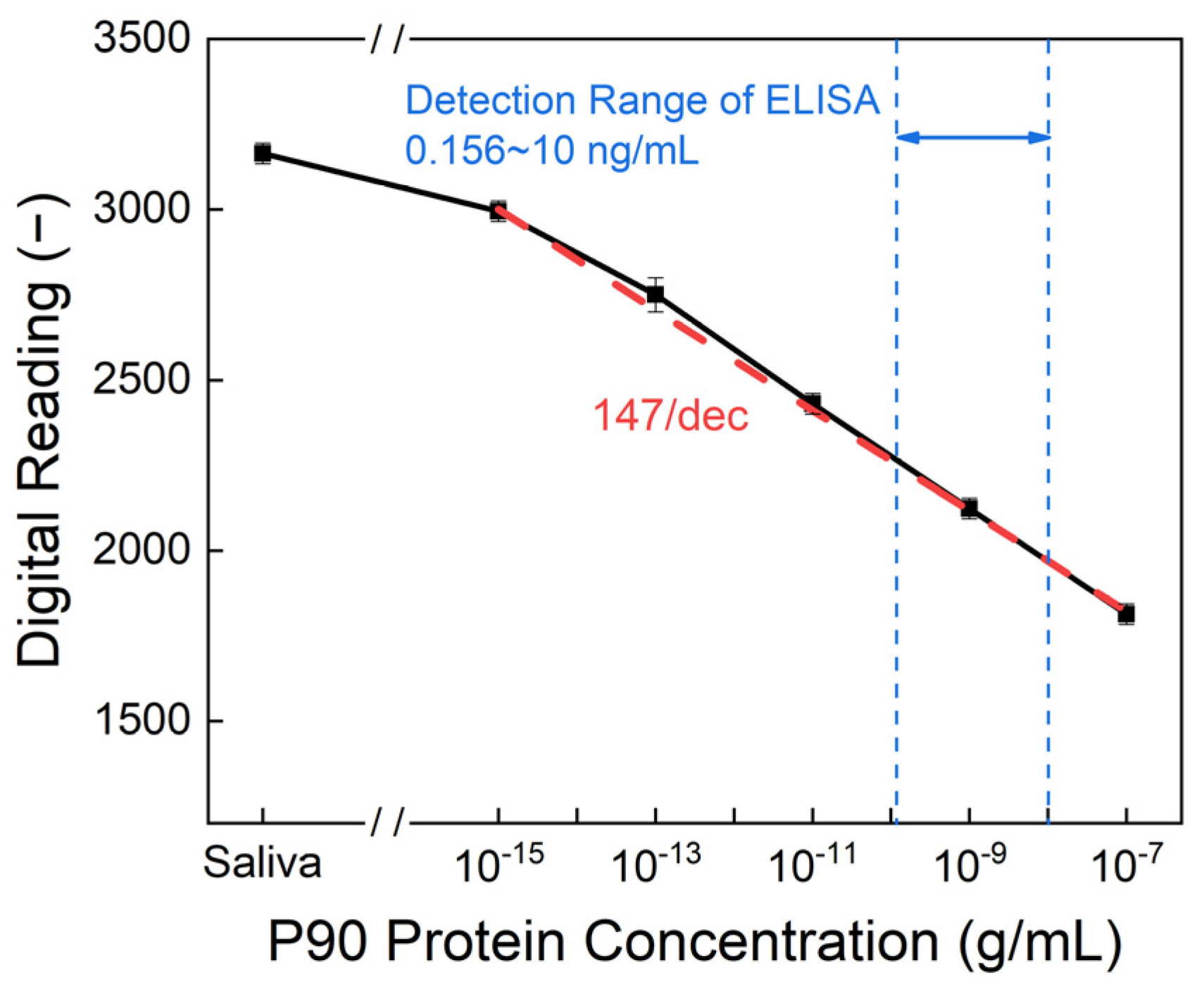
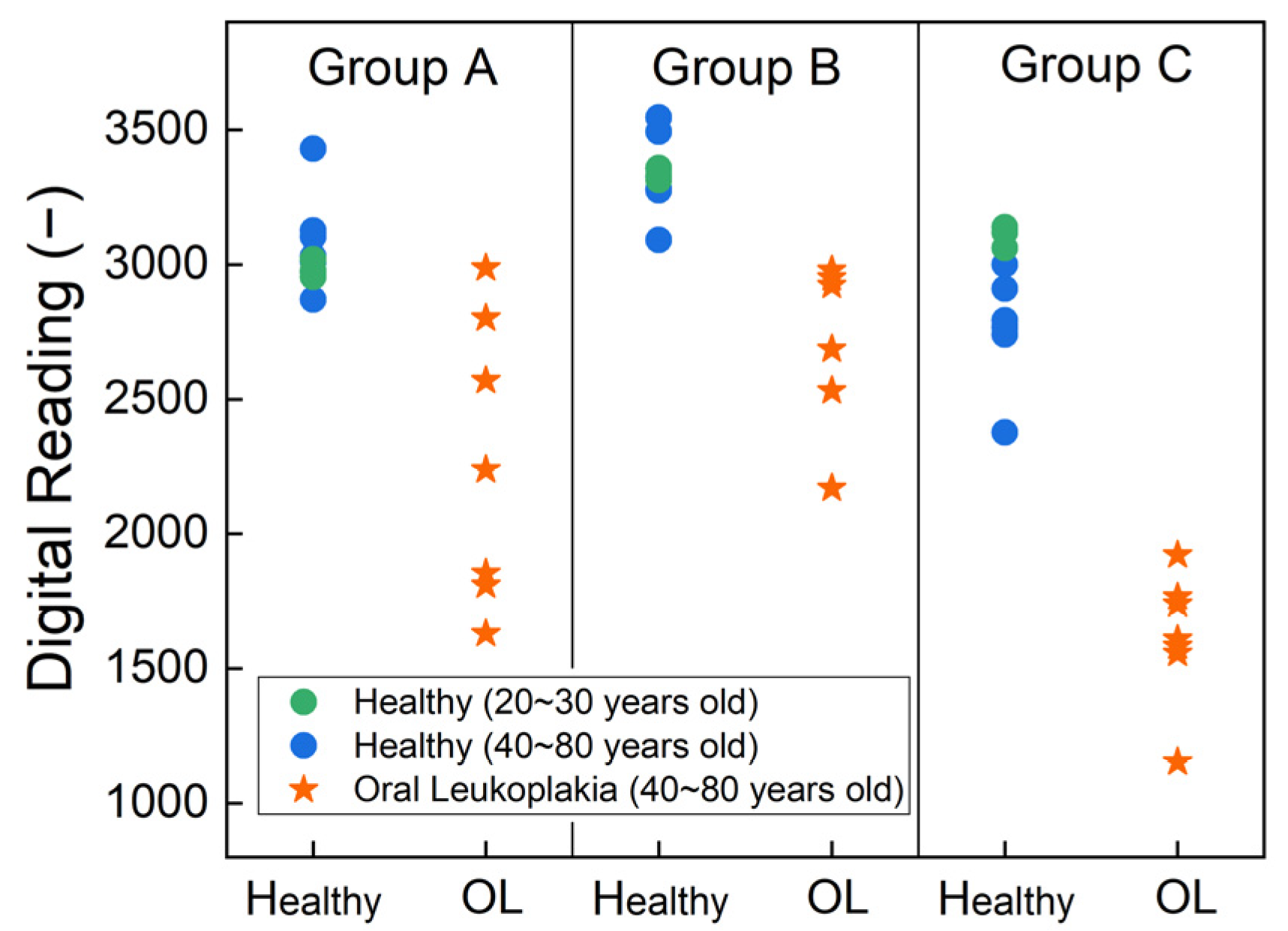
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Y.; Zhang, Q.; Guo, Z.; Deng, G.; Chen, R.; Zheng, Y. Potential noninvasive biomarkers for the malignant transformation of oral leukoplakia: A systematic review and meta-analysis. Cancer Med. 2023, 12, 14718–14730. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Key Statistics for Oral Cavity and Oropharyngeal Cancers. Available online: https://www.cancer.org/cancer/types/oral-cavity-and-oropharyngeal-cancer/about/key-statistics.html (accessed on 19 January 2024).
- Blot, W.J.; McLaughlin, J.K.; Winn, D.M.; Austin, D.F.; Greenberg, R.S.; Preston-Martin, S.; Bernstein, L.; Schoenberg, J.B.; Stemhagen, A.; Fraumeni, J.F., Jr. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988, 48, 3282–3287. [Google Scholar] [PubMed]
- Chaturvedi, P.; Singh, A.; Chien, C.-Y.; Warnakulasuriya, S. Tobacco related oral cancer. BMJ Br. Med. J. 2019, 365, I2142. [Google Scholar] [CrossRef] [PubMed]
- Heller, M.A.; Nyirjesy, S.C.; Balsiger, R.; Talbot, N.; VanKoevering, K.K.; Haring, C.T.; Old, M.O.; Kang, S.Y.; Seim, N.B. Modifiable risk factors for oral cavity cancer in non-smokers: A systematic review and meta-analysis. Oral Oncol. 2023, 137, 106300. [Google Scholar] [CrossRef]
- Tavakoli Pirzaman, A.; Ebrahimi, P.; Niknezhad, S.; Vahidi, T.; Hosseinzadeh, D.; Akrami, S.; Ashrafi, A.M.; Moeen Velayatimehr, M.; Hosseinzadeh, R.; Kazemi, S. Toxic mechanisms of cadmium and exposure as a risk factor for oral and gastrointestinal carcinomas. Hum. Exp. Toxicol. 2023, 42, 09603271231210262. [Google Scholar] [CrossRef]
- Kumar, M.; Nanavati, R.; Modi, T.G.; Dobariya, C. Oral cancer: Etiology and risk factors: A review. J. Cancer Res. Ther. 2016, 12, 458–463. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Engels, E.; Anderson, W.; Gillison, M. Incidence trends for human papillomavirus-related and-unrelated oral squamous cell carcinomas in the United States. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 612–619. [Google Scholar] [CrossRef]
- Goldoni, R.; Scolaro, A.; Boccalari, E.; Dolci, C.; Scarano, A.; Inchingolo, F.; Ravazzani, P.; Muti, P.; Tartaglia, G. Malignancies and biosensors: A focus on oral cancer detection through salivary biomarkers. Biosensors 2021, 11, 396. [Google Scholar] [CrossRef]
- Pearce, A.; Sharp, L.; Hanly, P.; Barchuk, A.; Bray, F.; de Camargo Cancela, M.; Gupta, P.; Meheus, F.; Qiao, Y.-L.; Sitas, F. Productivity losses due to premature mortality from cancer in Brazil, Russia, India, China, and South Africa (BRICS): A population-based comparison. Cancer Epidemiol. 2018, 53, 27–34. [Google Scholar] [CrossRef]
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral cancer and precancer: A narrative review on the relevance of early diagnosis. Int. J. Environ. Res. Public Health 2020, 17, 9160. [Google Scholar] [CrossRef]
- Silverman, S., Jr. Early diagnosis of oral cancer. Cancer 1988, 62, 1796–1799. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-T.; Darvishi, S.; Preet, A.; Huang, T.-Y.; Lin, S.-H.; Girault, H.H.; Wang, L.; Lin, T.-E. A review: Electrochemical biosensors for oral cancer. Chemosensors 2020, 8, 54. [Google Scholar] [CrossRef]
- Megha, D.; Kiruthiga, S.; Lokesh, M.; Pavithra, P.; Indumathy, K.; Nirmala, E. Exploring the comprehensive review of diagnostic methods of oral cancer: A global scenario. World J. Biol. Pharm. Health Sci. 2023, 15, 043–048. [Google Scholar] [CrossRef]
- Bhosle, P.; Pathan, H.; Tapadiya, G.; Alam, M.I. Dusruptive Developments in Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Walsh, T.; Macey, R.; Kerr, A.R.; Lingen, M.W.; Ogden, G.R.; Warnakulasuriya, S. Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions. Cochrane Database Syst. Rev. 2021, 2021, CD010276. [Google Scholar]
- Naito, Y.; Honda, K. Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. J. Pers. Med. 2023, 13, 303. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.G.S.; Bagán, J.V.; Jiménez, Y.; Margaix, M.; Marzal, C. Utility of imaging techniques in the diagnosis of oral cancer. J. Cranio-Maxillofac. Surg. 2015, 43, 1880–1894. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, D.; Natarajan, C.; Mukherjee, A. Advances in oral cancer detection. Adv. Clin. Chem. 2019, 91, 181–200. [Google Scholar] [PubMed]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of salivary biomarkers in oral cancer detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar]
- Huda, W. CT radiation exposure: An overview. Curr. Radiol. Rep. 2015, 3, 80. [Google Scholar] [CrossRef]
- Katz, J.; Jakymiw, A.; Ducksworth, M.K.; Stewart, C.M.; Bhattacharyya, I.; Cha, S.; Chan, E.K. CIP2A expression and localization in oral carcinoma and dysplasia. Cancer Biol. Ther. 2010, 10, 694–699. [Google Scholar] [CrossRef]
- Velmurugan, B.; Wang, H.; Chung, C.; Lee, C.; Huang, L.; Yeh, K.; Lin, S. CIP2A overexpression in Taiwanese oral cancer patients. Cancer Manag. Res. 2019, 11, 2589–2594. [Google Scholar] [CrossRef] [PubMed]
- Xian, M.; Stephany, J.L.; Chiu, C.-W.; Chiang, C.-C.; Ren, F.; Tsai, C.-T.; Shan, S.-S.; Liao, Y.-T.; Esquivel-Upshaw, J.F.; Pearton, S.J. High sensitivity CIP2A detection for oral cancer using a rapid transistor-based biosensor module. J. Vac. Sci. Technol. B 2023, 41, 013201. [Google Scholar] [CrossRef] [PubMed]
- Soofiyani, S.R.; Hejazi, M.S.; Baradaran, B. The role of CIP2A in cancer: A review and update. Biomed. Pharmacother. 2017, 96, 626–633. [Google Scholar] [CrossRef]
- Sangodkar, J.; Farrington, C.C.; McClinch, K.; Galsky, M.D.; Kastrinsky, D.B.; Narla, G. All roads lead to PP 2A: Exploiting the therapeutic potential of this phosphatase. FEBS J. 2016, 283, 1004–1024. [Google Scholar] [CrossRef]
- Nader, C.P.; Cidem, A.; Verrills, N.M.; Ammit, A.J. Protein phosphatase 2A (PP2A): A key phosphatase in the progression of chronic obstructive pulmonary disease (COPD) to lung cancer. Respir. Res. 2019, 20, 222. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Das, S.R.; Brownlee, B.J.; Parate, K.; Davis, T.M.; Stromberg, L.R.; Chan, E.K.; Katz, J.; Iverson, B.D.; Claussen, J.C. CIP2A immunosensor comprised of vertically-aligned carbon nanotube interdigitated electrodes towards point-of-care oral cancer screening. Biosens. Bioelectron. 2018, 117, 68–74. [Google Scholar] [CrossRef]
- Xian, M.; Chiu, C.-W.; Carey, P.H.; Fares, C.; Chen, L.; Wu, R.; Ren, F.; Tsai, C.-T.; Shan, S.-S.; Liao, Y.-T. Digital biosensor for human cerebrospinal fluid detection with single-use sensing strips. J. Vac. Sci. Technol. B 2022, 40, 023202. [Google Scholar] [CrossRef]
- Xian, M.; Luo, H.; Xia, X.; Fares, C.; Carey, P.H.; Chiu, C.-W.; Ren, F.; Shan, S.-S.; Liao, Y.-T.; Hsu, S.-M. Fast SARS-CoV-2 virus detection using disposable cartridge strips and a semiconductor-based biosensor platform. J. Vac. Sci. Technol. B 2021, 39, 033202. [Google Scholar] [CrossRef]
- Wan, H.-H.; Zhu, H.; Chiang, C.-C.; Li, J.-S.; Ren, F.; Tsai, C.-T.; Liao, Y.-T.; Neal, D.; Esquivel-Upshaw, J.F.; Pearton, S.J. High sensitivity saliva-based biosensor in detection of breast cancer biomarkers: HER2 and CA15-3. J. Vac. Sci. Technol. B 2024, 42, 023202. [Google Scholar] [CrossRef]
- Yang, J.; Carey, P.; Ren, F.; Wang, Y.-L.; Good, M.L.; Jang, S.; Mastro, M.A.; Pearton, S. Rapid detection of cardiac troponin I using antibody-immobilized gate-pulsed AlGaN/GaN high electron mobility transistor structures. Appl. Phys. Lett. 2017, 111, 202104. [Google Scholar] [CrossRef]
- Yang, J.; Carey, P.; Ren, F.; Mastro, M.A.; Beers, K.; Pearton, S.; Kravchenko, I.I. Zika virus detection using antibody-immobilized disposable cover glass and AlGaN/GaN high electron mobility transistors. Appl. Phys. Lett. 2018, 113, 032101. [Google Scholar] [CrossRef]
- Böckelman, C.; Hagström, J.; Mäkinen, L.K.; Keski-Säntti, H.; Häyry, V.; Lundin, J.; Atula, T.; Ristimäki, A.; Haglund, C. High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. Br. J. Cancer 2011, 104, 1890–1895. [Google Scholar] [CrossRef] [PubMed]
- Generation, S. Oral Cancer Screenings. Available online: https://www.smilegeneration.com/dental-services/general-dentistry/oral-cancer-screenings/ (accessed on 1 April 2024).


| Healthy (N = 10, 59%) | Oral Leukoplakia (N = 7, 41%) | p-Value | ||
|---|---|---|---|---|
| Gender | Male | 6 (60%) | 3 (43%) | 0.637 |
| Female | 4 (40%) | 4 (57%) | ||
| Age | 20–30 | 3 (30%) | 0 (0%) | 0.045 |
| 40–60 | 5 (50%) | 2 (29%) | ||
| 60–80 | 2 (20%) | 5 (71%) | ||
| Race | Asian | 4 (40%) | 1 (14%) | 0.338 |
| White | 6 (60%) | 6 (86%) |
| Healthy (N = 10, 59%) | Oral Leukoplakia (N = 7, 41%) | p-Value | |
|---|---|---|---|
| Group A | 3049 (152); 3014 [2972, 3084]; (2870, 3430) | 2270 (529); 2238 (1832, 2685]; (1630,2987) | 0.001 |
| Group B | 3336 (123); 3326 [3288, 3358]; (3091, 3545) | 2745 (306); 2922 [2608, 2964]; (2170,2979) | 0.0001 |
| Group C | 2867 (230); 2850 [2766, 3046]; (2376, 3138) | 1618 (241); 1609 [1570, 1752]; (1154,1921) | 0.0008 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, H.-H.; Zhu, H.; Chiang, C.-C.; Li, J.-S.; Ren, F.; Tsai, C.-T.; Liao, Y.-T.; Neal, D.; Katz, J.; Esquivel-Upshaw, J.F. Sensitive Detection of Oral Leukoplakia: Analyzing P90 Biomarkers in Saliva and Tissue. Biosensors 2024, 14, 281. https://doi.org/10.3390/bios14060281
Wan H-H, Zhu H, Chiang C-C, Li J-S, Ren F, Tsai C-T, Liao Y-T, Neal D, Katz J, Esquivel-Upshaw JF. Sensitive Detection of Oral Leukoplakia: Analyzing P90 Biomarkers in Saliva and Tissue. Biosensors. 2024; 14(6):281. https://doi.org/10.3390/bios14060281
Chicago/Turabian StyleWan, Hsiao-Hsuan, Haochen Zhu, Chao-Ching Chiang, Jian-Sian Li, Fan Ren, Cheng-Tse Tsai, Yu-Te Liao, Dan Neal, Joseph Katz, and Josephine F. Esquivel-Upshaw. 2024. "Sensitive Detection of Oral Leukoplakia: Analyzing P90 Biomarkers in Saliva and Tissue" Biosensors 14, no. 6: 281. https://doi.org/10.3390/bios14060281
APA StyleWan, H.-H., Zhu, H., Chiang, C.-C., Li, J.-S., Ren, F., Tsai, C.-T., Liao, Y.-T., Neal, D., Katz, J., & Esquivel-Upshaw, J. F. (2024). Sensitive Detection of Oral Leukoplakia: Analyzing P90 Biomarkers in Saliva and Tissue. Biosensors, 14(6), 281. https://doi.org/10.3390/bios14060281







