Tailoring Carbon Quantum Dots via Precursor Engineering for Fluorescence-Based Biosensing of E. coli
Abstract
1. Introduction
2. Fluorescence E. coli Sensing Assay Using CQDs
3. Hybrid CQDs Material by Integrating Other Nanomaterials
4. Precursor-Driven Functionalization CQDs for Selective E. coli Recognition
4.1. Heteroatom Atom Doping (N, B, S) in CQD Structures
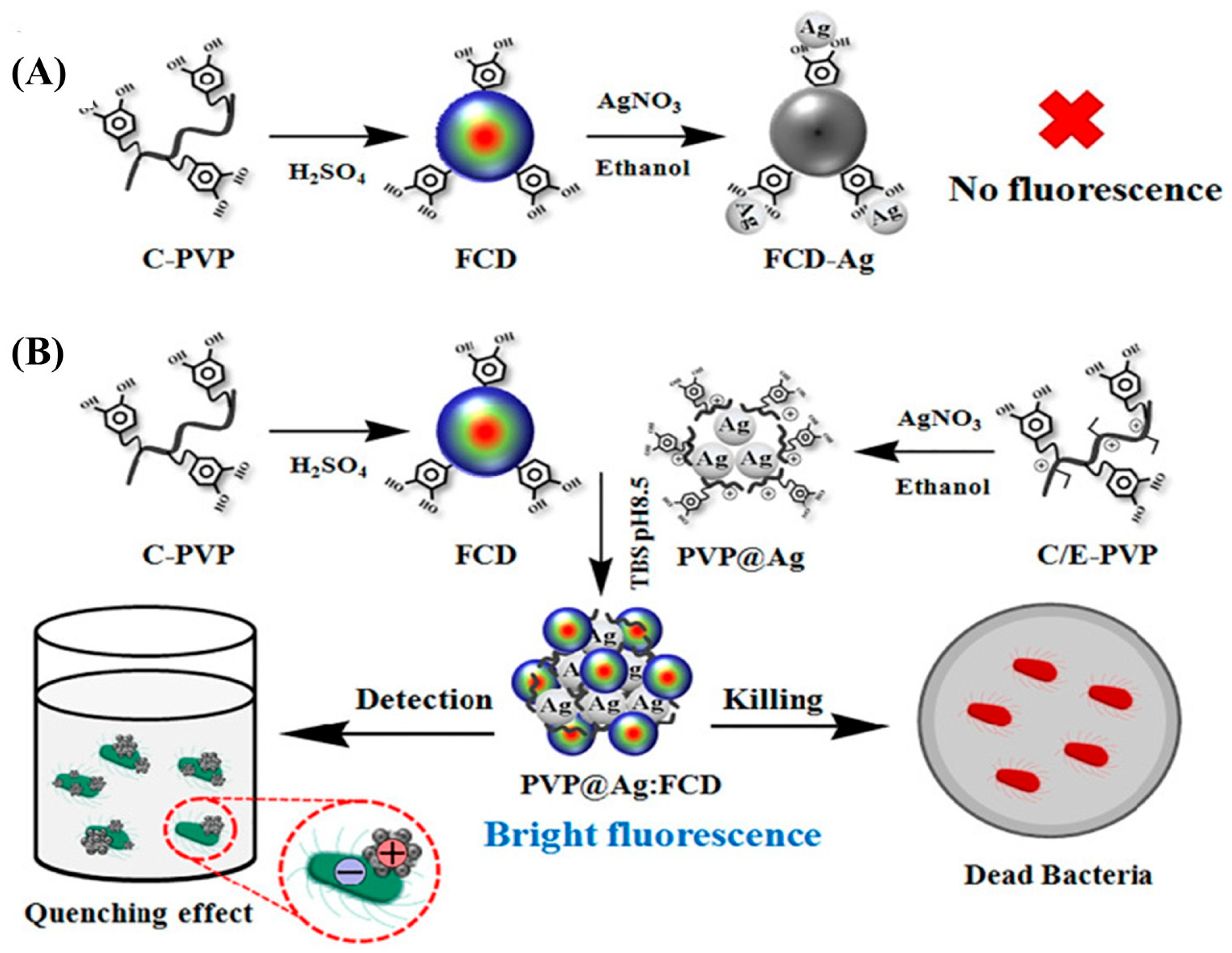
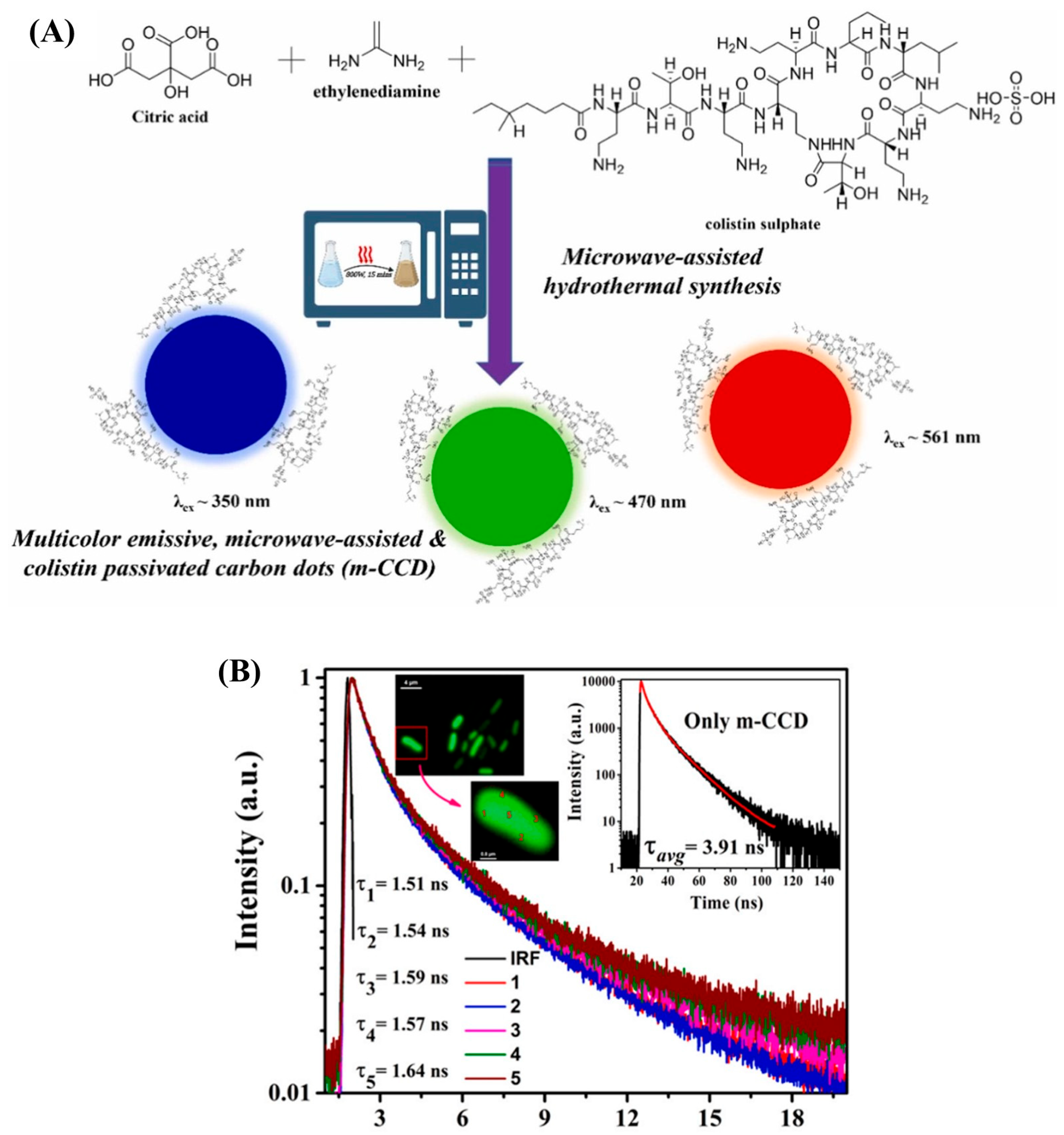
4.2. Using Antibiotics for CQDs Fabrication/Functionalization
4.3. Using Sugars for CQD Fabrication/Functionalization
5. Post-Synthetic Biofunctionalization of CQDs for Target Specificity
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ab | Antibody |
| AgNPs | Silver nanoparticles |
| Arg | L-arginine |
| BONs | Encapsulated breakable organosilica nanocapsules |
| cfu/mL | Colony-forming units per milliliter |
| CQDs | Carbon quantum dots |
| CQDs-Ab | E. coli-specific antibodies |
| CS | Cefminox sodium |
| CSNs | CQD-hybridized silica nanospheres |
| E. coli | Escherichia coli |
| EDX | energy-dispersive X-ray |
| FRET | Förster resonance energy transfer |
| FTIR | Fourier Transform Infrared |
| GO | graphene oxide |
| IFE | Inner filter effect |
| LOD | Limit of detection |
| LPS | Lipopolysaccharides |
| mAb@R-CDs@BONs-NH2 | R-CDs@BONs labeled with anti-E. coli O157:H7 monoclonal antibody |
| Mag | Magnetic |
| Man | Mannose |
| m-CCQDs | multi-emissive colistin-passivated CQDs |
| MNPs@B–N/APBA | Wulff-type boronic acid-functionalized magnetic nanoparticles |
| N-CQDs | N-doping CQDs |
| N,B-CQDs | N-/B-co-doped CQDs |
| Omp | Outer-membrane protein |
| oPD | o-Phenylenediamine |
| PET | Photo-induced electron transfer |
| PVP | Polyvinylpyrrolidone |
| SPA | Spectra Staphylococcal Protein A |
| ss-DNA | single-stranded DNA |
| TA | L-tartaric acid |
| W-CDs | Water soluble carbon dots |
| WHO | World Health Organization |
| XPS | X-ray Photoelectron Spectroscopy |
References
- Pebdeni, A.B.; Roshani, A.; Mirsadoughi, E.; Behzadifar, S.; Hosseini, M. Recent Advances in Optical Biosensors for Specific Detection of E. coli Bacteria in Food and Water. Food Control 2022, 135, 108822. [Google Scholar] [CrossRef]
- Jayan, H.; Zhou, R.; Zheng, Y.; Xue, S.; Yin, L.; El-Seedi, H.R.; Zou, X.; Guo, Z. Microfluidic-SERS Platform with In-Situ Nanoparticle Synthesis for Rapid E. Coli Detection in Food. Food Chem. 2025, 471, 142800. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Gao, Z.; Xie, Y.; Shu, S.; Ke, Y.; Wang, Y.; Deng, B.; Yu, R.; Geng, H. Facile Synthesis of N,B-Co-Doped Carbon Dots with the Gram-Scale Yield for Detection of Iron (III) and E. coli. Nanotechnology 2020, 31, 395702. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Hayat, A.; Mishra, R.K.; Catanante, G.; Shahid, S.A.; Bhand, S.; Marty, J.L. Design of a Fluorescence Aptaswitch Based on the Aptamer Modulated Nano-Surface Impact on the Fluorescence Particles. RSC Adv. 2016, 6, 65579–65587. [Google Scholar] [CrossRef]
- Maruthapandi, M.; Natan, M.; Jacobi, G.; Banin, E.; Luong, J.H.T.; Gedanken, A. Antibacterial Activity against Methicillin-Resistant Staphylococcus aureus of Colloidal Polydopamine Prepared by Carbon Dot Stimulated Polymerization of Dopamine. Nanomaterials 2019, 9, 1731. [Google Scholar] [CrossRef] [PubMed]
- Chaghazardi, M.; Kashanian, S.; Nazari, M.; Omidfar, K.; Joseph, Y.; Rahimi, P. Fluorometric Mercury (II) Detection Using Heteroatom-Doped Carbon and Graphene Quantum Dots. Photonics 2024, 11, 841. [Google Scholar] [CrossRef]
- Zare, I.; Zahed Nasab, S.; Rahi, A.; Ghaee, A.; Koohkhezri, M.; Ramezani Farani, M.; Madadi Gholipour, H.; Atabaki, A.H.; Hamblin, M.R.; Mostafavi, E.; et al. Antimicrobial Carbon Materials-Based Quantum Dots: From Synthesis Strategies to Antibacterial Properties for Diagnostic and Therapeutic Applications in Wound Healing. Coord. Chem. Rev. 2025, 522, 216211. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Li, N.; Yang, G.; Cheng, Z.; Du, X.; Sun, L.; Wang, W.; Li, B. Advances in the Application of Carbon Dots-Based Fluorescent Probes in Disease Biomarker Detection. Colloids Surf. B Biointerfaces 2025, 245, 114360. [Google Scholar] [CrossRef]
- Ma, G.; Li, X.; Cai, J.; Wang, X. Carbon Dots-Based Fluorescent Probe for Detection of Foodborne Pathogens and Its Potential with Microfluidics. Food Chem. 2024, 451, 139385. [Google Scholar] [CrossRef]
- Xu, Y.; Hassan, M.M.; Sharma, A.S.; Li, H.; Chen, Q. Recent Advancement in Nano-Optical Strategies for Detection of Pathogenic Bacteria and Their Metabolites in Food Safety. Crit. Rev. Food Sci. Nutr. 2023, 63, 486–504. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, M.; Li, X.; Fan, Y.; Li, J.; Lu, K.; Wen, H.; Ren, J. Recent Advances of Fluorescent Sensors for Bacteria Detection-A Review. Talanta 2023, 254, 124133. [Google Scholar] [CrossRef]
- Tian, J.; An, M.; Zhao, X.; Wang, Y.; Hasan, M. Advances in Fluorescent Sensing Carbon Dots: An Account of Food Analysis. ACS Omega 2023, 8, 9031–9039. [Google Scholar] [CrossRef]
- Anand, A.; Huang, C.C.; Lai, J.Y.; Bano, D.; Pardede, H.I.; Hussain, A.; Saleem, S.; Unnikrishnan, B. Fluorescent Carbon Dots for Labeling of Bacteria: Mechanism and Prospects—A Review. Anal. Bioanal. Chem. 2024, 416, 3907–3921. [Google Scholar] [CrossRef]
- Yahav, G.; Pawar, S.; Lipovsky, A.; Gupta, A.; Gedanken, A.; Duadi, H.; Fixler, D. Probing Polarity and PH Sensitivity of Carbon Dots in Escherichia coli through Time-Resolved Fluorescence Analyses. Nanomaterials 2023, 13, 2068. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Chaudhary, S. Modulating the Physicochemical and Biological Properties of Carbon Dots Synthesised from Plastic Waste for Effective Sensing of E. coli. Colloids Surf. B Biointerfaces 2020, 196, 111333. [Google Scholar] [CrossRef] [PubMed]
- Bhaisare, M.L.; Gedda, G.; Khan, M.S.; Wu, H.F. Fluorimetric Detection of Pathogenic Bacteria Using Magnetic Carbon Dots. Anal. Chim. Acta 2016, 920, 63–71. [Google Scholar] [CrossRef]
- Tammina, S.K.; Wan, Y.; Li, Y.; Yang, Y. Synthesis of N, Zn-Doped Carbon Dots for the Detection of Fe3+ Ions and Bactericidal Activity against Escherichia coli and Staphylococcus aureus. J. Photochem. Photobiol. B Biol. 2020, 202, 111734. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian-Fard-Fini, S.; Salavati-Niasari, M.; Ghanbari, D. Hydrothermal Green Synthesis of Magnetic Fe3O4−Carbon Dots by Lemon and Grape Fruit Extracts and as a Photoluminescence Sensor for Detecting of E. coli Bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 203, 481–493. [Google Scholar] [CrossRef]
- Roh, S.G.; Robby, A.I.; Phuong, P.T.M.; In, I.; Park, S.Y. Photoluminescence-Tunable Fluorescent Carbon Dots-Deposited Silver Nanoparticle for Detection and Killing of Bacteria. Mater. Sci. Eng. C 2019, 97, 613–623. [Google Scholar] [CrossRef]
- Baig, M.M.F.; Chen, Y.C. Bright Carbon Dots as Fluorescence Sensing Agents for Bacteria and Curcumin. J. Colloid Interface Sci. 2017, 501, 341–349. [Google Scholar] [CrossRef]
- Pathak, A.; Venugopal, P.; Nair, B.G.; Suneesh, P.V.; Satheesh Babu, T.G. Facile PH-Sensitive Optical Detection of Pathogenic Bacteria and Cell Imaging Using Multi-Emissive Nitrogen-Doped Carbon Dots. Microchem. J. 2020, 159, 105324. [Google Scholar] [CrossRef]
- Choi, C.A.; Mazrad; Lee, G.; In, I.; Lee, K.D.; Park, S.Y. Boronate-Based Fluorescent Carbon Dot for Rapid and Selectively Bacterial Sensing by Luminescence off/on System. J. Pharm. Biomed. Anal. 2018, 159, 1–10. [Google Scholar] [CrossRef]
- Wang, F.; Xiao, M.; Qi, J.; Zhu, L. Paper-Based Fluorescence Sensor Array with Functionalized Carbon Quantum Dots for Bacterial Discrimination Using a Machine Learning Algorithm. Anal. Bioanal. Chem. 2024, 416, 3139–3148. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Calic, D.; Schweizer, F.; Zelenitsky, S.; Adam, H.; Lagac-Wiens, P.R.S.; Rubinstein, E.; Gin, A.S.; Hoban, D.J.; Karlowsky, J.A. New Lipoglycopeptides: A Comparative Review of Dalbavancin, Oritavancin and Telavancin. Drugs 2010, 70, 859–886. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmanna, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Mattioni Marchetti, V.; Hrabak, J.; Bitar, I. Fosfomycin Resistance Mechanisms in Enterobacterales: An Increasing Threat. Front. Cell. Infect. Microbiol. 2023, 13, 1178547. [Google Scholar] [CrossRef]
- Rehabbahy; Abu El-Wafa, W.M.; Abouwarda, A.M. Molecular Mechanisms of Fosfomycin Resistance in MDR Escherichia coli Isolates from Urinary Tract Infections. Egypt. J. Med. Microbiol. 2023, 32, 25–29. [Google Scholar] [CrossRef]
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Shen, Z.; Wang, D.; Tang, B.Z. Phospholipid-Mimetic Aggregation-Induced Emission Luminogens for Specific Elimination of Gram-Positive and Gram-Negative Bacteria. ACS Nano 2023, 17, 4239–4249. [Google Scholar] [CrossRef]
- Bafna, J.A.; Sans-Serramitjana, E.; Acosta-Gutiérrez, S.; Bodrenko, I.V.; Hörömpöli, D.; Berscheid, A.; Brötz-Oesterhelt, H.; Winterhalter, M.; Ceccarelli, M. Kanamycin Uptake into Escherichia coli Is Facilitated by OmpF and OmpC Porin Channels Located in the Outer Membrane. ACS Infect. Dis. 2020, 6, 1855–1865. [Google Scholar] [CrossRef]
- Chandra, S.; Chowdhuri, A.R.; Mahto, T.K.; Samui, A.; Sahu, S.K. One-Step Synthesis of Amikacin Modified Fluorescent Carbon Dots for the Detection of Gram-Negative Bacteria like: Escherichia coli. RSC Adv. 2016, 6, 72471–72478. [Google Scholar] [CrossRef]
- Chandra, S.; Mahto, T.K.; Chowdhuri, A.R.; Das, B.; Sahu, S.K. One Step Synthesis of Functionalized Carbon Dots for the Ultrasensitive Detection of Escherichia coli and Iron (III). Sens. Actuators B Chem. 2017, 245, 835–844. [Google Scholar] [CrossRef]
- Putri, F.A.R.; Mudasir, M.; Morita, K.; Suherman, S. Microwave-Assisted Synthesis of Amikacin Modified n, s Co-Doped Carbon Dots for Escherichia coli Detection. Chemosensors 2019, 7, 61. [Google Scholar] [CrossRef]
- Pathak, A.; Navaneeth, P.; Gupta, M.; Pradeep, A.; Nair, B.G.; Suneesh, P.V.; Elangovan, R.; Sundberg, L.R.; Marjomäki, V.; Babu, T.G.S. Revolutionizing Gram-Negative Bacteria Detection: FLIM and Multicolor Imaging Based Selective Interaction Study Using Colistin Passivated Carbon Dots. Sens. Actuators B Chem. 2023, 395, 134433. [Google Scholar] [CrossRef]
- Kuang, Y.; Song, M.; Zhou, X.; Mi, J.; Zhang, Z.; Liu, G.; Shen, Z.; Liu, Z.; Chen, C.; Wu, M.X.; et al. Cefminox Sodium Carbon Nanodots for Treatment and Bacterial Detection of Bloodstream Infection. Chem. Eng. J. 2023, 470, 143988. [Google Scholar] [CrossRef]
- Audio Haryanto, N.; Tri Wahyuni, E.; Ilmi, M.; Morita, K.; Oki, Y. Carbon Dots Modification for Escherichia coli Detection: Variation of Colistin Sulphate Concentration. Orient. J. Chem. 2019, 35, 49–55. [Google Scholar] [CrossRef]
- Zhong, D.; Zhuo, Y.; Feng, Y.; Yang, X. Employing Carbon Dots Modified with Vancomycin for Assaying Gram-Positive Bacteria like Staphylococcus aureus. Biosens. Bioelectron. 2015, 74, 546–553. [Google Scholar] [CrossRef]
- Zheng, L.; Qi, P.; Zhang, D. Identification of Bacteria by a Fluorescence Sensor Array Based on Three Kinds of Receptors Functionalized Carbon Dots. Sens. Actuators B Chem. 2019, 286, 206–213. [Google Scholar] [CrossRef]
- Whitfield, C. Biosynthesis and Assembly of Capsular Polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Krammer, E.M.; De Ruyck, J.; Roos, G.; Bouckaert, J.; Lensink, M.F. Targeting Dynamical Binding Processes in the Design of Non-Antibiotic Anti-Adhesives by Molecular Simulation—The Example of FimH. Molecules 2018, 23, 1641. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.I.; Chang, H.T.; Lin, C.H.; Shen, Y.W.; Unnikrishnan, B.; Li, Y.J.; Huang, C.C. One-Step Synthesis of Biofunctional Carbon Quantum Dots for Bacterial Labeling. Biosens. Bioelectron. 2015, 68, 1–6. [Google Scholar] [CrossRef]
- Lai, I.P.J.; Harroun, S.G.; Chen, S.Y.; Unnikrishnan, B.; Li, Y.J.; Huang, C.C. Solid-State Synthesis of Self-Functional Carbon Quantum Dots for Detection of Bacteria and Tumor Cells. Sens. Actuators B Chem. 2016, 228, 465–470. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Y.; Guo, T.; Yang, T.; Chen, M.; Wang, J. Green Preparation of Carbon Dots with Papaya as Carbon Source for Effective Fluorescent Sensing of Iron (III) and Escherichia coli. Biosens. Bioelectron. 2016, 85, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Farooq, U.; Chen, W.; Ullah, M.W.; Wang, S. Fluorimetric Detection of Single Pathogenic Bacterium in Milk and Sewage Water Using Ph-Sensitive Fluorescent Carbon Dots and MALDI-TOF MS. Microorganisms 2020, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Gao, X.; Tao, Z.; Wang, X.; Lin, X.; Wang, S.; Liu, Y. Sugar-Metabolism-Triggered Pathogenic Bacteria Identification Based on PH-Sensitive Fluorescent Carbon Dots. Sens. Actuators B Chem. 2020, 316, 128063. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Zhang, P.; Yan, Z.; Zhou, Y.; Du, Y.; Qu, C.; Song, Y.; Zhou, D.; Qu, S.; et al. Cell-Based Fluorescent Microsphere Incorporated with Carbon Dots as a Sensitive Immunosensor for the Rapid Detection of Escherichia coli O157 in Milk. Biosens. Bioelectron. 2021, 179, 113057. [Google Scholar] [CrossRef]
- Song, Y.; Ostermeyer, G.P.; Du, D.; Lin, Y. Carbon Nanodot-Hybridized Silica Nanospheres Assisted Immunoassay for Sensitive Detection of Escherichia coli. Sens. Actuators B Chem. 2021, 349, 130730. [Google Scholar] [CrossRef]
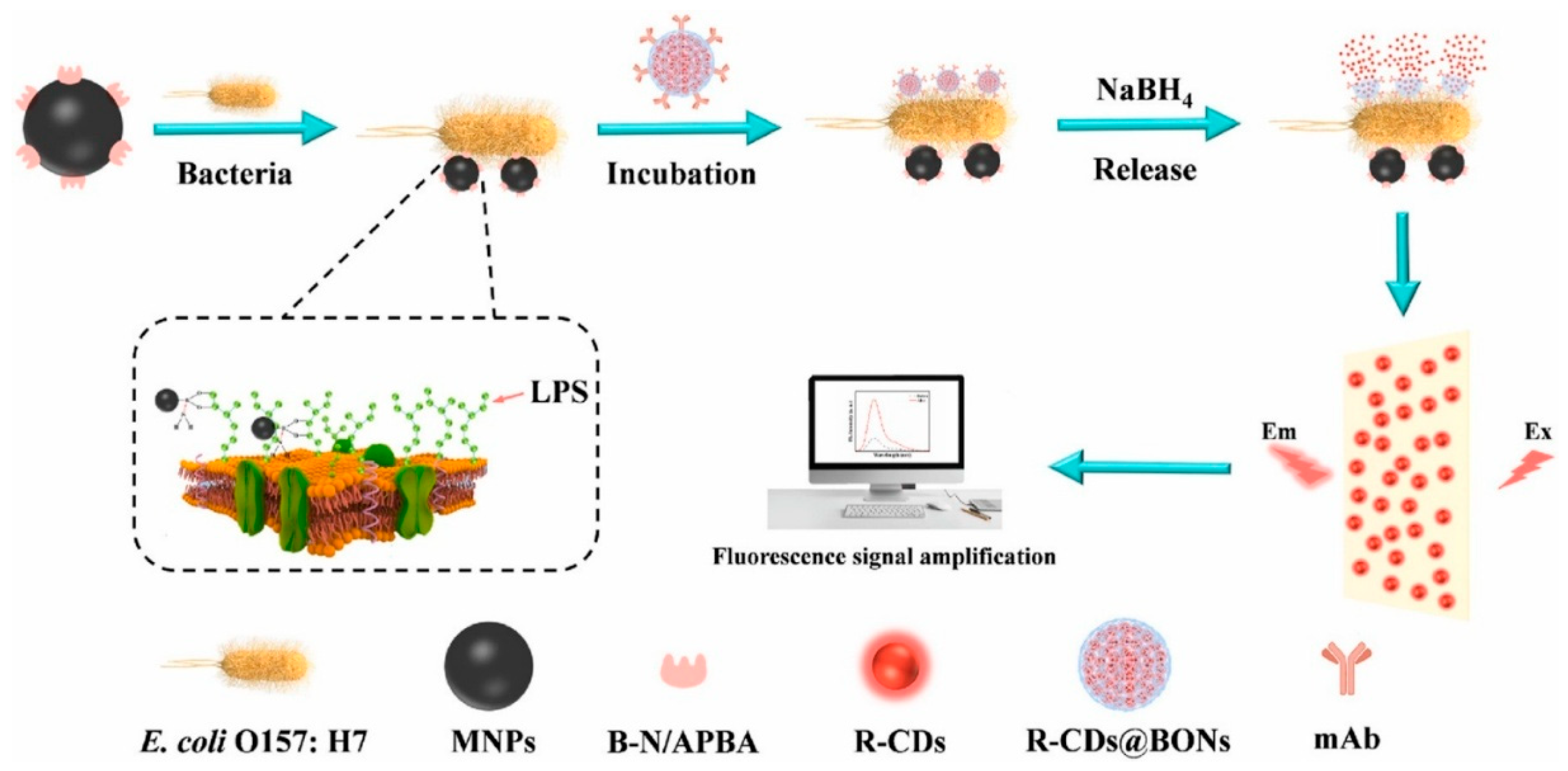
- Ling, Z.; Xu, Q.; Song, Y.; Zhang, W.; Xu, H. Fluorescent Biosensor Based on Magnetic Separation Platform and Spore-like Breakable Organosilica Nanocapsules Controlled-Release Carbon Dots for the Detection of Escherichia coli O157:H7. Talanta 2024, 276, 126273. [Google Scholar] [CrossRef]
- Wang, S.; Liang, N.; Hu, X.; Li, W.; Guo, Z.; Zhang, X.; Huang, X.; Li, Z.; Zou, X.; Shi, J. Carbon Dots and Covalent Organic Frameworks Based FRET Immunosensor for Sensitive Detection of Escherichia coli O157:H7. Food Chem. 2024, 447, 138663. [Google Scholar] [CrossRef]
- Majdinasab, M.; Daneshi, M.; Louis Marty, J. Recent Developments in Non-Enzymatic (Bio)Sensors for Detection of Pesticide Residues: Focusing on Antibody, Aptamer and Molecularly Imprinted Polymer. Talanta 2021, 232, 122397. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.; Xu, Y.; Gan, Z.; Zou, X.; Shi, J.; Huang, X.; Li, Z.; Li, Y. Green One-Step Synthesis of Carbon Quantum Dots from Orange Peel for Fluorescent Detection of Escherichia coli in Milk. Food Chem. 2021, 339, 127775. [Google Scholar] [CrossRef]
- Pan, T.; Shan, X.; Jiang, D.; Qi, L.; Wang, W.; Chen, Z. Fluorometric Aptasensor for Determination of Escherichia coli O157:H7 by FRET Effect between Aminated Carbon Quantum Dots and Graphene Oxide. Anal. Sci. 2021, 37, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Hou, X.; Ga, L.; Ai, J. Aptamer-carbon quantum dots and silver nanoparticles construct a FRET sensor for sensitive detection of E. coli. Biomed. Anal. 2024, 1, 162–173. [Google Scholar] [CrossRef]
- Bai, X.; Ga, L.; Du, Y.; Ai, J. Carbon Quantum Dot-Based Label-Free Fluorescent Biosensor to Detect E. coli. IEEE Sens. J. 2024, 24, 25284–25290. [Google Scholar] [CrossRef]
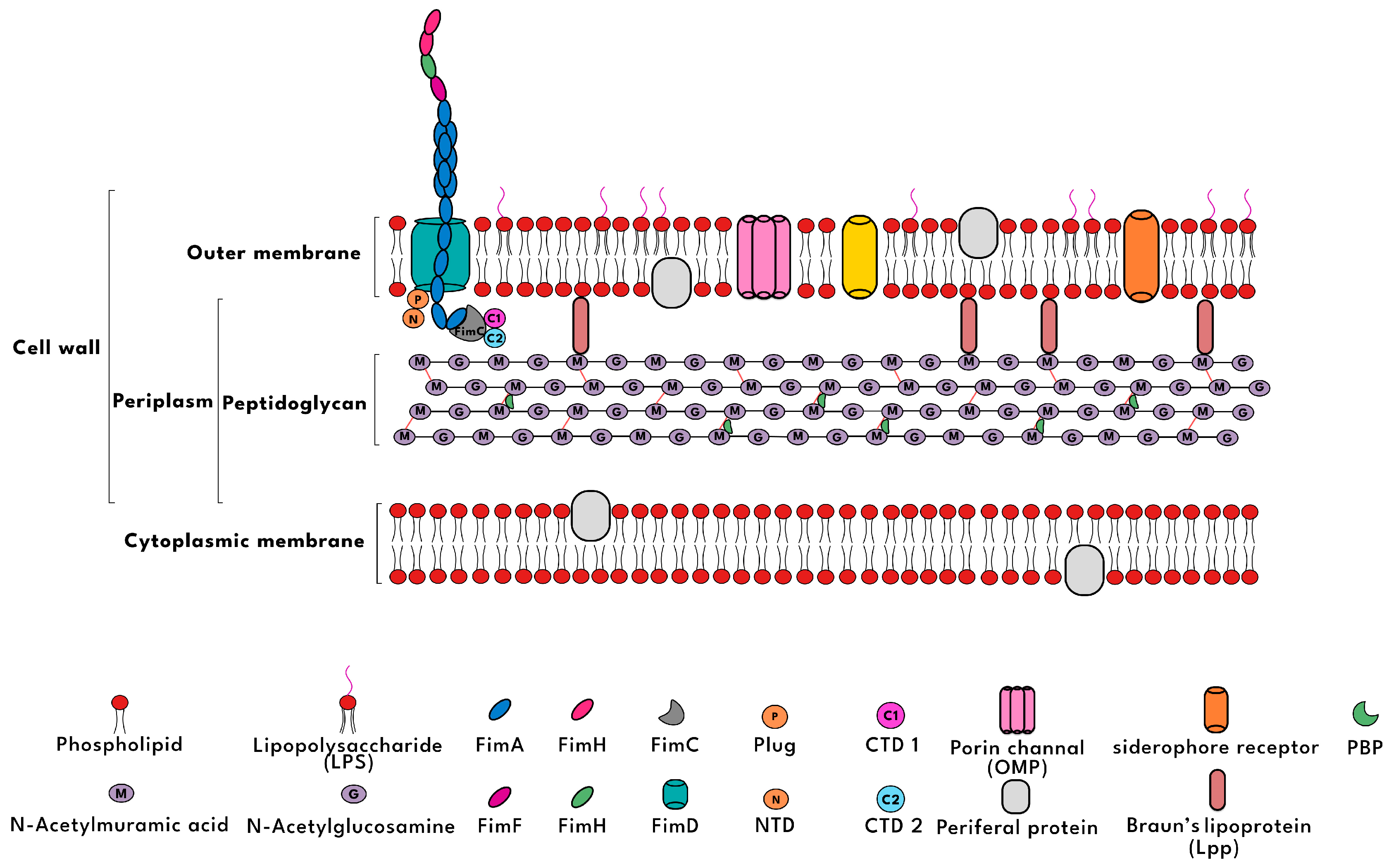

| Antibiotic Class | Examples | Mechanism of Entry | Target and Mode of Action | Effectiveness Against Gram-Negative Bacteria |
|---|---|---|---|---|
| β-lactams and Monobactams | Penicillin, Cephalosporins, Aztreonam | Enter through porins (OmpF, OmpC) or passively diffuse | Target peptidoglycan transpeptidase (PBP) to inhibit cell wall synthesis | Effective, depends on porin access |
| Polymyxins | Colistin, Polymyxin B | Bind to LPS, alter membrane permeability, penetrate | Disrupt inner membrane integrity, causing cell lysis | Highly effective |
| Glycopeptides | Vancomycin, Teicoplanin | Cannot penetrate outer membrane due to size | Bind to D-alanine–D-alanine in peptidoglycan, blocking cell wall extension | Ineffective |
| Lipopeptides | Daptomycin | Unable to pass through outer membrane due to LPS | Targets bacterial membrane but cannot bind to Gram-negative membranes | Ineffective |
| Fosfomycin | Fosfomycin | Enters via GlpT/UhpT transporters | Inhibits MurA enzyme, first step in peptidoglycan synthesis | Variable, resistance rising in some Gram-negative |
| Cycloserine | D-cycloserine | Diffuses passively or via porin channels | Inhibits alanine racemase and D-Ala-D-Ala ligase in cell wall synthesis | Limited; used mainly for MDR-TB, weak Gram-neg coverage |
| Chloramphenicol (Amphenicols) | Chloramphenicol | Passive diffusion (due to small size) | Inhibits protein synthesis by binding to 50S ribosomal subunit | Moderate (some Gram-neg coverage, but resistance concerns) |
| Tetracyclines | Doxycycline, Tetracycline | Enters via passive diffusion or OmpF/OmpC porins | Inhibits protein synthesis by binding 30S ribosomal subunit | Effective against many Gram-negatives |
| Aminoglycoside | Amikacin, Gentamicin | Requires O2-dependent active transport (porins aid entry) | Binds irreversibly to 30S ribosomal subunit, causing misreading of mRNA | Highly effective against many aerobic Gram-negatives |
| CQD Type | Precursor | Synthesis Method | QY % | Linear Range (cfu/mL) | LOD (cfu/mL) | Refs |
|---|---|---|---|---|---|---|
| CQD | Urea, Citric acid, NaOH | Hydrothermal (160 °C, 6 h) | - | - | - | [15] |
| CQD | Plastic polybags, cups and bottles | Thermal calcination (300 °C, 2 h) followed by hydrothermal (200 °C, 5 h) | 60, 65, 69 | 0–40 × 108 | down to 108 | [16] |
| Mag-CQDs | Acetic acid (4%), chitosan, Fe3O4 NPs | Hydrothermal (180 °C, 12 h) | - | 4.0 × 102–3.4 × 103 | 3.5 × 102 | [17] |
| N,Zn-CQD | Glucosamine, Zinc acetate, water | Hydrothermal (130 °C, 1 h) | 74 | 0.25–125 µM | 0.15 µM | [18] |
| Fe3O4-CQDs | Magnetite, H2O, lemon turmeric | Hydrothermal (180 °C, 6 h) | - | - | - | [19] |
| CQD | Pvp, CCDP, ethanol, H2SO4, N2 | Hydrothermal (70 °C, 10 h) | - | - | - | [20] |
| CQD | Egg White | Thermal (200 °C, 4 h) | 43 | 0.997–0.999 | 40 nm (40 ng/mL) | [21] |
| N-CQDs | Citric acid, glycine | Incubation at 70 °C for 12 h, then Hydrothermal (230 °C, 6 h) | 27.2 | - | - | [22] |
| N,B-CQDs | TA, Arg, and H3BO3 | Hydrothermal (180 °C, 10 h) | 14.5 | 102–107 | 165 | [4] |
| CQDs@amikacin | Di ammonium hydrogen citrate and amikacin | Hydrothermal (180 °C, 4 h) | 12.35 | 7.625 × 102–3.904 × 105 | 552 | [32] |
| CQDs@ colistin | Di ammonium hydrogen citrate and colistin sulfate | Pyrolysis (180 °C, 1 h) | 7.56 | 3.81 × 102–2.44 × 104 | 460 | [33] |
| N,S-CQDs@amikacin | Citric acid, thiourea, and amikacin | Microwave-assisted treatment (180 °C, 30 min) | - | 0–1.20 × 104 | 3.04 | [34] |
| CQDs@ colistin | Citric acid, ethylenediamine and colistin sulfate | Microwave-assisted (15 min) | 25.1 | 3.40 × 105–9.80 × 105 and 6.90 × 107–4.14 × 108 | 3.68–4.89 × 104 | [35] |
| cefminox sodium-CQDs | cefminox sodium | microwave | 0.5 × 106–1 × 109 | 3.7 × 105 | [36] | |
| CQDs@ colistin | Ammonium citric and colistin sulfate | Pyrolysis (180 °C, 1 h) | - | 102–104 | - | [37] |
| CQD@van | Citric acid, Urea, Water, phosphate buffer, EDC, NHS, Vancomycin | Hydrothermal (Microwave, 750 W) | - | 3.18 × 105–1.59 ×108 | 9.4 | [38] |
| (PM-BA-Van)CQD’s | Ammonium citrate/3-Aminophenylboronic acid, vancomycin hydrochloride/Polymaxin B sulfate | Thermal (4 h, 180) | - | - | - | [39] |
| Man-CQDs | Ammonium citrate and Man | Pyrolysis (180 °C, 2 h) | 9.8 | 102–108 | 450 | [42] |
| CQD Man-FCQD FA-CQD | Ammonium citrate for CQD synthesizing; Man and FA for functionalizing | Pyrolysis (180 °C for 2 h) for CQD fabrication; Pyrolysis (180 °C for 2 h) for CQD functionalization with Man and FA | 9 | 0–108 0–108 0–108 | - 100 - | [43] |
| W-CQDs | Papaya peel | Hydrothermal (200 °C, 5 h) | 18.98 | 105–108 | 9.5 × 104 | [44] |
| p-CQD | Sucrose, H2SO4, NaOH | Hydrothermal | - | - | 3.5 × 102 | [45] |
| CQD | Dopamine and oPD | Hydrothermal (200 °C, 8 h) | - | 103–107 | 21 | [46] |
| CQD Type | Precursor | Synthesis Method | QY % | Linear Range (cfu/mL) | LOD (cfu/mL) | Refs |
|---|---|---|---|---|---|---|
| Ab-CQDs-microsphere | Citric acid, urea, and CaCl2 | Hydrothermal (250 °C) | - | 2.4 × 102–2.4 × 107 | 2.4 × 102 | [47] |
| Ab–CSN | Aminosalicylic acid | Solvothermal (200 °C, 18 h) | 16.4 | 0–104 | 2.4 | [48] |
| mAb@R-CDs@BONs-NH2 | reduced glutathione, formamide | Hydrothermal (160 °C, 1 h) | 101–106 | 25 | [49] | |
| CQD-Ab-COF | Ascorbic acid | Hydrothermal (180 °C, 8 h) | 50.8 | 0–106 | 7 | [24] |
| NH2-CQD-apt + GO | Polyethyleneimine and citric acid monohydrate | Hydrothermal (180 °C for 2 h) | - | 102–107 | 89 | [53] |
| Apt-CQDs + AgNPs | White grapefruit peels and ethylene glycol | Microwave oven (3 min) | - | 2 × 103–2 × 108 | 77 | [54] |
| Apt + CQDs + AgNPs | Celery leaves | Hydrothermal (200 °C, 12 h) | - | 2 × 102–2 × 107 | 185 | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazari, M.; Zinatizadeh, A.; Mohammadi, P.; Kashanian, S.; Amiri, M.; Valipour, N.; Joseph, Y.; Rahimi, P. Tailoring Carbon Quantum Dots via Precursor Engineering for Fluorescence-Based Biosensing of E. coli. Biosensors 2025, 15, 635. https://doi.org/10.3390/bios15100635
Nazari M, Zinatizadeh A, Mohammadi P, Kashanian S, Amiri M, Valipour N, Joseph Y, Rahimi P. Tailoring Carbon Quantum Dots via Precursor Engineering for Fluorescence-Based Biosensing of E. coli. Biosensors. 2025; 15(10):635. https://doi.org/10.3390/bios15100635
Chicago/Turabian StyleNazari, Maryam, Alireza Zinatizadeh, Parviz Mohammadi, Soheila Kashanian, Mandana Amiri, Nona Valipour, Yvonne Joseph, and Parvaneh Rahimi. 2025. "Tailoring Carbon Quantum Dots via Precursor Engineering for Fluorescence-Based Biosensing of E. coli" Biosensors 15, no. 10: 635. https://doi.org/10.3390/bios15100635
APA StyleNazari, M., Zinatizadeh, A., Mohammadi, P., Kashanian, S., Amiri, M., Valipour, N., Joseph, Y., & Rahimi, P. (2025). Tailoring Carbon Quantum Dots via Precursor Engineering for Fluorescence-Based Biosensing of E. coli. Biosensors, 15(10), 635. https://doi.org/10.3390/bios15100635






