Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring
Abstract
1. Introduction
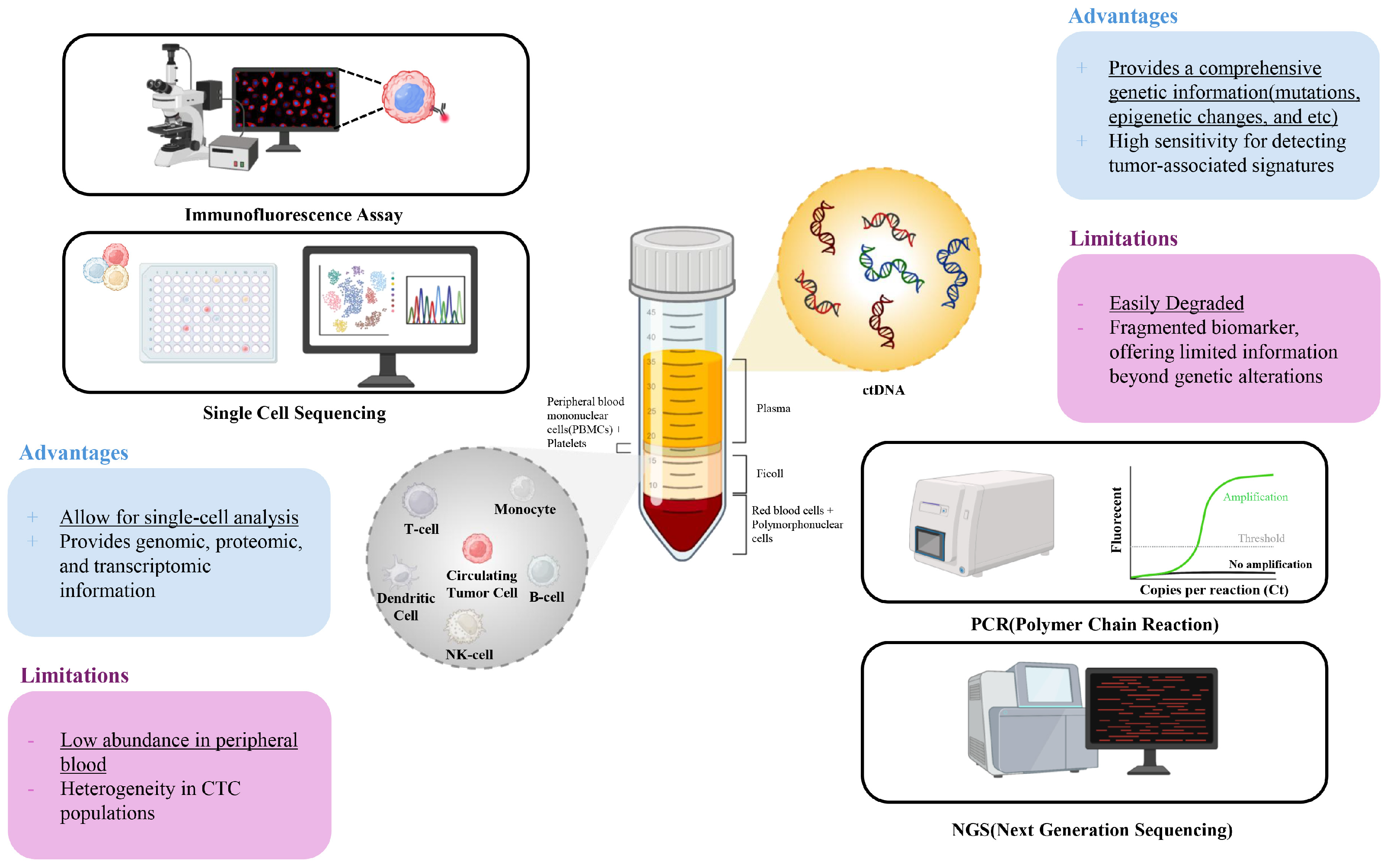
2. Circulating Tumor Cells (CTCs): Clinical Utility and Limitations
3. Circulating Tumor DNA (ctDNA): Introduction to Clinical Utility and Limitations
4. Integrated Analysis of CTCs and ctDNA
4.1. Advantages of Integrated Analysis of CTCs
4.2. Breast Cancer
| Type | Samples | CTC | ctDNA | Summary | Ref. | ||
|---|---|---|---|---|---|---|---|
| Isolation | Analysis | Extraction | Analysis | ||||
| MBC | 193 MBC patients (cfDNA and CTC both analyzed) | CellSearch System (EpCAM-positive selection) | IF staining (EpCAM+, CK+, DAPI+, CD45−) | Qiagen Circulating Nucleic Acids Kit | Quantitatively analyzed using RealTime PCR System (96 bp single copy TaqMan assay) | High cfDNA levels and CTC counts associated with poor OS. Combined analysis of CTCs and cfDNA enhanced prognostic accuracy. | [46] |
| MBC | 18 MBC patients with matched blood samples | AdnaTest EMT-2/StemCell Select System (targeting EpCAM, EGFR, and HER2) | IF staining (CK+, CD45−, DAPI+) Variants analyzed using customized QIAseq Targeted DNA Panel Kit | QIAamp Circulating Nucleic Acid Kit | Variants analyzed using customized QIAseq Targeted DNA Panel Kit (17 cancer-related genes) | 94% of patients had tumor-specific variants when combining cfDNA and CTC gDNA. Unique variants observed in AR and ERBB2 for CTC gDNA and PIK3CA and ESR1 for cfDNA. | [49] |
| MBC | 227 blood samples from 117 MBC patients | CellSearch System (EpCAM-positive selection) | IF staining (CK+, CD45−, DAPI+) | QIAamp Circulating Nucleic Acid Kit | Quantitatively analyzed using Qubit fluorometry and qPCR targeting ALU repeats | High CTC counts and cfDNA levels were independently associated with poor OS. Combined high levels showed >17-fold increased risk of death. | [47] |
| TNBC | 196 TNBC patients (142 ctDNA; 123 CTC analyzed) | Anti-EpCAM magnetic beads with microfluidic device | IF staining (CK+, CD45−, DAPI+) | Qiagen Circulating Nucleic Acids Kit | FoundationACT and Foundation One Liquid assays (detection of point mutations, CNAs, and rearrangements) | Combination of ctDNA and CTCs associated with increased sensitivity for detecting DDFS and DFS. Patients positive for both markers had significantly worse outcomes. | [48] |
| MBC | 57 MBC patients (57 CTC/ctDNA analyzed + 31 samples for targeted CTC sequencing) | DEPArray system (EpCAM-positive selection) | IF staining (EpCAM+, CK+, DAPI+, CD45−) NGS with AmpliSeq panel | QIAamp Circulating Nucleic Acid Kit | NGS with a AmpliSeq panel (50-gene panel) targeting mutations in PIK3CA, TP53, ESR1, KRAS, etc. | Significant mutational heterogeneity observed in CTCs. High cfDNA and CTC levels associated with poor overall survival. | [34] |
| BC | 114 paired CTC and cfDNA samples including 79 BC patients | Anti-EpCAM magnetic bead-based positive selection | Methylation-specific PCR (MSP) targeting SOX17 promoter analyzed from DNA and KRT19 analyzed from mRNA using RTq-PCR | High Pure Viral nucleic acid kit | SOX17 promoter methylation analysis using MSP | SOX17 promoter methylation detected in 86.0% of primary tumors, with a strong correlation between cfDNA and CTC methylation patterns. | [50] |
4.3. Lung Cancer
4.4. Gastrointestinal Cancer
4.5. Other Types of Cancer
5. Limitations and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer Treatment and Survivorship Statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Alinia, S.; Ahmadi, S.; Mohammadi, Z.; Rastkar Shirvandeh, F.; Asghari-Jafarabadi, M.; Mahmoudi, L.; Safari, M.; Roshanaei, G. Exploring the Impact of Stage and Tumor Site on Colorectal Cancer Survival: Bayesian Survival Modeling. Sci. Rep. 2024, 14, 4270. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Jeong, W.J.; Jafari, R.; Kubiatowicz, L.J.; Nair, A.; Poellmann, M.J.; Hong, R.S.; Liu, E.W.; Owen, R.H.; Rawding, P.A.; et al. Bimodal liquid biopsy for cancer immunotherapy based on peptide engineering and nanoscale analysis. Biosens. Bioelectron. 2022, 213, 114445. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, H. Tumor Heterogeneity and the Potential Role of Liquid Biopsy in Bladder Cancer. Cancer Commun. 2021, 41, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.H.; Allison, D.H.R.; Feng, Y.; Jour, G.; Park, K.; Zhou, F.; Moreira, A.L.; Shen, G.; Feng, X.; Sabari, J. Comparison of Solid Tissue Sequencing and Liquid Biopsy Accuracy in Identification of Clinically Relevant Gene Mutations and Rearrangements in Lung Adenocarcinomas. Mod. Pathol. 2021, 34, 2168–2174. [Google Scholar] [CrossRef]
- Bayle, A.; Peyraud, F.; Belcaid, L.; Brunet, M.; Aldea, M.; Clodion, R.; Dubos, P.; Vasseur, D.; Nicotra, C.; Geraud, A. Liquid Versus Tissue Biopsy for Detecting Actionable Alterations According to the ESMO Scale for Clinical Actionability of Molecular Targets in Patients with Advanced Cancer: A Study from the French National Center for Precision Medicine (PRISM). Ann. Oncol. 2022, 33, 1328–1331. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E. Liquid Versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Deng, Z.; Wu, S.; Wang, Y.; Shi, D. Circulating Tumor Cell Isolation for Cancer Diagnosis and Prognosis. eBioMedicine 2022, 83, 104237. [Google Scholar] [CrossRef]
- Zhu, Z.; Hu, E.; Shen, H.; Tan, J.; Zeng, S. The Functional and Clinical Roles of Liquid Biopsy in Patient-derived Models. J. Hematol. Oncol. 2023, 16, 36. [Google Scholar] [CrossRef]
- Ring, A.; Nguyen-Sträuli, B.D.; Wicki, A.; Aceto, N. Biology, Vulnerabilities and Clinical Applications of Circulating Tumour Cells. Nat. Rev. Cancer 2023, 23, 95–111. [Google Scholar] [CrossRef]
- Boukovala, M.; Westphalen, C.B.; Probst, V. Liquid Biopsy into the Clinics: Current Evidence and Future Perspectives. J. Liq. Biopsy 2024, 4, 100146. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shiratsuchi, H.; Palanisamy, N.; Nagrath, S.; Ramnath, N. Expanded Circulating Tumor Cells from a Patient with ALK- Positive Lung Cancer Present with EML4-ALK Rearrangement Along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J. Thorac. Oncol. 2017, 12, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Stejskal, P.; Goodarzi, H.; Srovnal, J.; Hajdúch, M.; Van ’T Veer, L.J.; Magbanua, M.J.M. Circulating Tumor Nucleic Acids: Biology, Release Mechanisms, and Clinical Relevance. Mol. Cancer 2023, 22, 15. [Google Scholar] [CrossRef]
- Seyfoori, A.; Seyyed Ebrahimi, S.A.; Samandari, M.; Samiei, E.; Stefanek, E.; Garnis, C.; Akbari, M. Microfluidic-assisted CTC Isolation and in Situ Monitoring Using Smart Magnetic Microgels. Small 2023, 19, 2205320. [Google Scholar] [CrossRef]
- Franken, A.; Kraemer, A.; Sicking, A.; Watolla, M.; Rivandi, M.; Yang, L.; Warfsmann, J.; Polzer, B.M.; Friedl, T.W.P.; Meier-Stiegen, F. Comparative Analysis of Epcam High-expressing and Low-expressing Circulating Tumour Cells with Regard to Their Clonal Relationship and Clinical Value. Br. J. Cancer 2023, 128, 1742–1752. [Google Scholar] [CrossRef]
- Bu, J.; Nair, A.; Kubiatowicz, L.J.; Poellmann, M.J.; Jeong, W.J.; Reyes-Martinez, M.; Armstrong, A.J.; George, D.J.; Wang, A.Z.; Zhang, T.; et al. Surface engineering for efficient capture of circulating tumor cells in renal cell carcinoma: From nanoscale analysis to clinical application. Biosens. Bioelectron. 2020, 162, 112250. [Google Scholar] [CrossRef]
- Bu, J.; Kang, Y.T.; Lee, Y.S.; Kim, J.; Cho, Y.H.; Moon, B.I. Lab on a fabric: Mass producible and low-cost fabric filters for the high-throughput viable isolation of circulating tumor cells. Biosens. Bioelectron. 2017, 91, 747–755. [Google Scholar] [CrossRef]
- Shim, J.E.; Bu, J.; Lee, M.K.; Cho, Y.H.; Kim, T.H.; Bu, J.U.; Han, S.W. Viable and high-throughput isolation of heterogeneous circulating tumor cells using tapered-slit filters. Sens. Actuators B Chem. 2020, 321, 128369. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but Not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Bu, J.; Cho, Y.H.; Han, S.W. Enhancement of Isolation Sensitivity for the Viable Heterogeneous Circulating Tumor Cells Swelled by Hypo-osmotic Pressure. RSC Adv. 2017, 7, 49684–49693. [Google Scholar] [CrossRef]
- Kim, Y.; Bu, J.; Cho, Y.H.; Son, I.T.; Kang, S.B. A viable circulating tumor cell isolation device with high retrieval efficiency using a reversibly deformable membrane barrier. J. Micromech. Microeng. 2017, 27, 025015. [Google Scholar] [CrossRef]
- Kang, Y.T.; Kim, Y.J.; Bu, J.; Chen, S.; Cho, Y.H.; Lee, H.M.; Ryu, C.J.; Lim, Y.; Han, S.W. Epithelial and mesenchymal circulating tumor cell isolation and discrimination using dual-immunopatterned device with newly-developed anti-63B6 and anti-EpCAM. Sens. Actuators B Chem. 2018, 260, 320–330. [Google Scholar] [CrossRef]
- Green, B.J.; Marazzini, M.; Hershey, B.; Fardin, A.; Li, Q.; Wang, Z.; Giangreco, G.; Pisati, F.; Marchesi, S.; Disanza, A. Pillarx: A Microfluidic Device to Profile Circulating Tumor Cell Clusters Based on Geometry, Deformability, and Epithelial State. Small 2022, 18, 2106097. [Google Scholar] [CrossRef]
- Poellmann, M.J.; Bu, J.; Liu, S.; Wang, A.Z.; Seyedin, S.N.; Chandrasekharan, C.; Hong, H.; Kim, Y.S.; Caster, J.M.; Hong, S. Nanotechnology and machine learning enable circulating tumor cells as a reliable biomarker for radiotherapy responses of gastrointestinal cancer patients. Biosens. Bioelectron. 2023, 226, 115117. [Google Scholar] [CrossRef]
- Negishi, R.; Yamakawa, H.; Kobayashi, T.; Horikawa, M.; Shimoyama, T.; Koizumi, F.; Sawada, T.; Oboki, K.; Omuro, Y.; Funasaka, C. Transcriptomic Profiling of Single Circulating Tumor Cells Provides Insight into Human Metastatic Gastric Cancer. Commun. Biol. 2022, 5, 20. [Google Scholar] [CrossRef]
- Bu, J.; Lee, T.H.; Jeong, W.-J.; Poellmann, M.J.; Mudd, K.; Eun, H.S.; Liu, E.W.; Hong, S.; Hyun, S.H. Enhanced Detection of Cell-free DNA (cfdna) Enables Its Use as a Reliable Biomarker for Diagnosis and Prognosis of Gastric Cancer. PLoS ONE 2020, 15, e0242145. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, L.; Wang, G.; Lv, J.; He, Y.; Shao, P.L.; Hu, R.; Xiao, H.; Tang, J.; Zhang, B. A nano-magnetic size selective cfDNA extraction platform for liquid biopsy with enhanced precision. J. Chromatogr. B 2022, 1199, 123236. [Google Scholar] [CrossRef]
- Janku, F.; Huang, H.J.; Pereira, D.Y.; Kobayashi, M.; Chiu, C.H.; Call, S.G.; Woodbury, K.T.; Chao, F.; Marshak, D.R.; Chiu, R.Y.T. A Novel Method for Liquid-phase Extraction of Cell-free DNA for Detection of Circulating Tumor DNA. Sci. Rep. 2021, 11, 19653. [Google Scholar] [CrossRef]
- Arisi, M.F.; Dotan, E.; Fernandez, S.V. Circulating Tumor DNA in Precision Oncology and Its Applications in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 4441. [Google Scholar] [CrossRef]
- Dang, D.K.; Park, B.H. Circulating Tumor DNA: Current Challenges for Clinical Utility. J. Clin. Investig. 2022, 132, e154941. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid Biopsy in Cancer: Current Status, Challenges and Future Prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Chen, L.; Li, K.; Liu, R.; Sun, L.; Han, T. Circulating Tumor DNA: Current Implementation Issues and Future Challenges for Clinical Utility. Clin. Chem. Lab. Med. (CCLM) 2024, 62, 2094–2110. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.A.; Guttery, D.S.; Hills, A.; Fernandez-Garcia, D.; Page, K.; Rosales, B.M.; Goddard, K.S.; Hastings, R.K.; Luo, J.; Ogle, O. Mutation Analysis of Cell-free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin. Cancer Res. 2017, 23, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Onidani, K.; Shoji, H.; Kakizaki, T.; Yoshimoto, S.; Okaya, S.; Miura, N.; Sekikawa, S.; Furuta, K.; Lim, C.T.; Shibahara, T. Monitoring of Cancer Patients via Next-generation Sequencing of Patient-derived Circulating Tumor Cells and Tumor DNA. Cancer Sci. 2019, 110, 2590–2599. [Google Scholar] [CrossRef]
- Peneder, P.; Stütz, A.M.; Surdez, D.; Krumbholz, M.; Semper, S.; Chicard, M.; Sheffield, N.C.; Pierron, G.; Lapouble, E.; Tötzl, M. Multimodal Analysis of Cell-free DNA Whole-genome Sequencing for Pediatric Cancers with Low Mutational Burden. Nat. Commun. 2021, 12, 3230. [Google Scholar] [CrossRef]
- Xie, J.; Hu, B.; Gong, Y.; He, S.; Lin, J.; Huang, Q.; Cheng, J. A Comparative Study on Ctdna and Tumor DNA Mutations in Lung Cancer and Benign Cases with a High Number of Ctcs and Ctecs. J. Transl. Med. 2023, 21, 873. [Google Scholar] [CrossRef]
- Gerratana, L.; Davis, A.A.; Foffano, L.; Reduzzi, C.; Rossi, T.; Medford, A.; Clifton, K.; Shah, A.N.; Bucheit, L.; Cristofanilli, M. Integrating Machine Learning-Predicted Circulating Tumor Cells (CTCs) and circulating tumor DNA (ctDNA) in Metastatic Breast Cancer: A proof of principle study on endocrine resistance profiling. Cancer Lett. 2024, 609, 217325. [Google Scholar] [CrossRef]
- Sørlie, T. Introducing Molecular Subtyping of Breast Cancer into the Clinic? J. Clin. Oncol. 2009, 27, 1153–1154. [Google Scholar] [CrossRef]
- Norum, J.H.; Andersen, K.; Sørlie, T. Lessons learned from the intrinsic subtypes of breast cancer in the quest for precision therapy. J. Br. Surg. 2014, 101, 925–938. [Google Scholar] [CrossRef]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast Cancer Heterogeneity and Its Implication in Personalized Precision Therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Bagaria, S.P.; Ray, P.S.; Sim, M.S.; Ye, X.; Shamonki, J.M.; Cui, X.; Giuliano, A.E. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg. 2014, 149, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.D.; Migliaccio, I.; Malorni, L.; Guarducci, C.; Biganzoli, L.; Leo, A.D. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Palmieri, C.; Tilley, W.D. Renewed Interest in the Progesterone Receptor in Breast Cancer. Br. J. Cancer 2016, 115, 909–911. [Google Scholar] [CrossRef] [PubMed]
- Gerratana, L.; Davis, A.; Medford, A.; Reduzzi, C.; Clifton, K.; Podany, E.; Hensing, W.; Velimirovic, M.; Shah, A.M.; Cristofanilli, M. Abstract PO4-15-04: Differential genomic profiling of progesterone receptor (PR) negative, estrogen receptor (ER) positive HER2-negative metastatic breast cancer (MBC) through circulating tumor DNA. Cancer Res. 2024, 84 (Suppl. S9), PO4–PO15. [Google Scholar] [CrossRef]
- Fernandez-Garcia, D.; Hills, A.; Page, K.; Hastings, R.K.; Toghill, B.; Goddard, K.S.; Ion, C.; Ogle, O.; Boydell, A.R.; Gleason, K. Plasma Cell-free DNA (cfdna) as a Predictive and Prognostic Marker in Patients with Metastatic Breast Cancer. Breast Cancer Res. 2019, 21, 149. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, C.; Wan, S.; Mu, Z.; Zhang, Z.; Abu-Khalaf, M.M.; Fellin, F.M.; Silver, D.P.; Neupane, M.; Jaslow, R.J. Association of Clinical Outcomes in Metastatic Breast Cancer Patients with Circulating Tumour Cell and Circulating Cell-free DNA. Eur. J. Cancer 2019, 106, 133–143. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Schneider, B.P. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: Preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef]
- Keup, C.; Storbeck, M.; Hauch, S.; Hahn, P.; Sprenger-Haussels, M.; Hoffmann, O.; Kimmig, R.; Kasimir-Bauer, S. Multimodal Targeted Deep Sequencing of Circulating Tumor Cells and Matched Cell-free DNA Provides a More Comprehensive Tool to Identify Therapeutic Targets in Metastatic Breast Cancer Patients. Cancers 2020, 12, 1084. [Google Scholar] [CrossRef]
- Chimonidou, M.; Strati, A.; Malamos, N.; Georgoulias, V.; Lianidou, E.S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin. Chem. 2013, 59, 270–279. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Adjei, A.A.; Gridelli, C.; Reck, M.; Kerr, K.; Felip, E. Metastatic Non-small-cell Lung Cancer (NSCLC): ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2012, 23, vii56–vii64. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, E.; Coco, S.; Genova, C.; Rossi, G.; Longo, L.; Grossi, F. Liquid Biopsy in Non-small Cell Lung Cancer: Highlights and Challenges. Cancers 2019, 12, 17. [Google Scholar] [CrossRef]
- Raez, L.E.; Brice, K.; Dumais, K.; Lopez-Cohen, A.; Wietecha, D.; Izquierdo, P.A.; Santos, E.S.; Powery, H.W. Liquid biopsy versus tissue biopsy to determine front line therapy in metastatic non-small cell lung cancer (NSCLC). Clin. Lung Cancer 2023, 24, 120–129. [Google Scholar] [CrossRef]
- Moon, S.M.; Kim, J.-H.; Kim, S.K.; Kim, S.; Kwon, H.-J.; Bae, J.-S.; Lee, S.; Lee, H.S.; Choi, M.-Y.; Jeon, B.H. Clinical Utility of Combined Circulating Tumor Cell and Circulating Tumor DNA Assays for Diagnosis of Primary Lung Cancer. Anticancer Res. 2020, 40, 3435–3444. [Google Scholar] [CrossRef]
- Kong, S.L.; Liu, X.; Tan, S.J.; Tai, J.A.; Phua, L.Y.; Poh, H.M.; Yeo, T.; Chua, Y.W.; Haw, Y.X.; Ling, W.H. Complementary Sequential Circulating Tumor Cell (CTC) and Cell-free Tumor DNA (ctdna) Profiling Reveals Metastatic Heterogeneity and Genomic Changes in Lung Cancer and Breast Cancer. Front. Oncol. 2021, 11, 698551. [Google Scholar] [CrossRef]
- Markou, A.N.; Londra, D.; Stergiopoulou, D.; Vamvakaris, I.; Potaris, K.; Pateras, I.S.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Preoperative Mutational Analysis of Circulating Tumor Cells (ctcs) and Plasma-cfdna Provides Complementary Information for Early Prediction of Relapse: A Pilot Study in Early-stage Non-small Cell Lung Cancer. Cancers 2023, 15, 1877. [Google Scholar] [CrossRef]
- Liu, H.E.; Vuppalapaty, M.; Wilkerson, C.; Renier, C.; Chiu, M.; Lemaire, C.; Che, J.; Matsumoto, M.; Carroll, J.; Crouse, S. Detection of EGFR Mutations in Cfdna and Ctcs, and Comparison to Tumor Tissue in Non-small-cell-lung-cancer (NSCLC) Patients. Front. Oncol. 2020, 10, 572895. [Google Scholar] [CrossRef]
- Jemal, A.; Center, M.M.; Desantis, C.; Ward, E.M. Global Patterns of Cancer Incidence and Mortality Rates and Trends. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1893–1907. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Vega, P.; Valentin, F.; Cubiella, J. Colorectal cancer diagnosis: Pitfalls and opportunities. World J. Gastrointest. Oncol. 2015, 7, 422. [Google Scholar] [CrossRef] [PubMed]
- Namløs, H.M.; Boye, K.; Mishkin, S.J.; Barøy, T.; Lorenz, S.; Bjerkehagen, B.; Stratford, E.W.; Munthe, E.; Kudlow, B.A.; Myklebost, O. Noninvasive Detection of Ctdna Reveals Intratumor Heterogeneity and Is Associated with Tumor Burden in Gastrointestinal Stromal Tumor. Mol. Cancer Ther. 2018, 17, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Salvianti, F.; Gelmini, S.; Mancini, I.; Pazzagli, M.; Pillozzi, S.; Giommoni, E.; Brugia, M.; Costanzo, F.D.; Galardi, F.; Antonuzzo, L. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: The OMITERC prospective study. Br. J. Cancer 2021, 125, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Yamada, T.; Takahashi, G.; Iwai, T.; Ueda, K.; Kuriyama, S.; Koizumi, M.; Matsuda, A.; Shinji, S.; Ohta, R. Analysis of Colorectal Cancer-related Mutations by Liquid Biopsy: Utility of Circulating Cell-free DNA and Circulating Tumor Cells. Cancer Sci. 2019, 110, 3497–3509. [Google Scholar] [CrossRef]
- Kidess-Sigal, E.; Liu, H.E.; Triboulet, M.M.; Che, J.; Ramani, V.C.; Visser, B.C.; Poultsides, G.A.; Longacre, T.A.; Marziali, A.; Vysotskaia, V. Enumeration and Targeted Analysis of KRAS, BRAF and PIK3CA Mutations in Ctcs Captured by a Label-free Platform: Comparison to Ctdna and Tissue in Metastatic Colorectal Cancer. Oncotarget 2016, 7, 85349–85364. [Google Scholar] [CrossRef]
- Yu, P.; Zhu, S.; Luo, Y.; Li, G.; Pu, Y.; Cai, B.; Zhang, C. Application of Circulating Tumor Cells and Circulating Free DNA from Peripheral Blood in the Prognosis of Advanced Gastric Cancer. J. Oncol. 2022, 2022, 9635218. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodríguez-Garrote, M.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N. Circulating Tumor Cells (CTC) and KRAS Mutant Circulating Free DNA (cfdna) Detection in Peripheral Blood as Biomarkers in Patients Diagnosed with Exocrine Pancreatic Cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.C. Whole-exome Sequencing of Cell-free DNA and Circulating Tumor Cells in Multiple Myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Alix-PanabièRes, C.; MüLler, I.; Letang, N.; Vendrell, J.-P.; Rebillard, X.; Pantel, K. Cell-free Tumor DNA in Blood Plasma as a Marker for Circulating Tumor Cells in Prostate Cancer. Clin. Cancer Res. 2009, 15, 1032–1038. [Google Scholar] [CrossRef]
- Kobayashi, M.; Abe, H.; Arai, K.; Murakami, S.; Kamai, T. Circulating tumor cells and cell-free tumor DNA analyses in urothelial cancer using the LiquidBiopsy platform. Curr. Urol. 2022, 16, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Muraro, E.; Del Ben, F.; Turetta, M.; Cesselli, D.; Bulfoni, M.; Zamarchi, R.; Rossi, E.; Spazzapan, S.; Dolcetti, R.; Steffan, A. Clinical Relevance of the Combined Analysis of Circulating Tumor Cells and Anti-tumor T-cell Immunity in Metastatic Breast Cancer Patients. Front. Oncol. 2022, 12, 983887. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Rawding, P.A.; Bu, J.; Hyun, S.; Rou, W.; Jeon, H.; Kim, S.; Lee, B.; Kubiatowicz, L.J.; Kim, D. Machine-learning-based Clinical Biomarker Using Cell-free DNA for Hepatocellular Carcinoma (HCC). Cancers 2022, 14, 2061. [Google Scholar] [CrossRef] [PubMed]
- Shaker, F.; Razi, S.; Rezaei, N. Circulating miRNA and circulating tumor DNA application as liquid biopsy markers in gastric cancer. Clin. Biochem. 2024, 129, 110767. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Hadlock, T.; Lo, T.; Purcell, E.; Mutukuri, A.; Fouladdel, S.; Raguera, M.D.S.; Fairbairn, H.; Murlidhar, V.; Durham, A. Dual-isolation and Profiling of Circulating Tumor Cells and Cancer Exosomes from Blood Samples with Melanoma Using Immunoaffinity-based Microfluidic Interfaces. Adv. Sci. 2020, 7, 2001581. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Lee, T.H.; Poellmann, M.J.; Rawding, P.A.; Jeong, W.; Hong, R.S.; Hyun, S.H.; Eun, H.S.; Hong, S. Tri-modal Liquid Biopsy: Combinational Analysis of Circulating Tumor Cells, Exosomes, and Cell-free DNA Using Machine Learning Algorithm. Clin. Transl. Med. 2021, 11, e499. [Google Scholar] [CrossRef]
- Kumar, K.; Kim, E.; Alhammadi, M.; Reddicherla, U.; Aliya, S.; Tiwari, J.N.; Park, H.S.; Choi, J.H.; Son, C.Y.; Huh, Y.S. Recent advances in microfluidic approaches for the isolation and detection of exosomes. TrAC Trends Anal. Chem. 2023, 159, 116912. [Google Scholar] [CrossRef]
- Yang, S.; Kim, S.H.; Intisar, A.; Shin, H.Y.; Kanf, H.G.; Kim, M.Y.; Kim, J.M.; Roh, H.R.; Kim, M. Fully Automated Continuous Centrifugal Microfluidics Isolates Natural Killer Cells with High Performance and Minimal Stress. Anal. Chem. 2023, 95, 9949–9958. [Google Scholar] [CrossRef]

| Type | Samples | CTC | ctDNA | Summary | Ref. | ||
|---|---|---|---|---|---|---|---|
| Isolation | Analysis | Extraction | Analysis | ||||
| LC | 111 individuals (99 LC, 12 benign) | Cyogen CTC isolation system (Size-based filtration) | IF staining (EpCAM+, CK+, DAPI+, CD45−) | Chemagic cfDNA 5k kit special H24 | Targeted NGS for somatic mutations using a 54-gene panel | Combined analysis achieved a sensitivity of 95%, outperforming CTCs (66%), ctDNA (73%), and other serum markers (CEA, CYFRA 21-1 at 66.7%). | [56] |
| LC, BC | 12 LC patients and 12 BC patients | ClearCell FX1 (Size-based Isolation) followed by single-cell isolation using DropCell platform (CD45−negative enrichment) | Targeted amplicon sequencing of gDNA from CTCs using QIAseq DNA Panel for CD45-negative cells with specific morphology | QIAamp Circulating Nucleic Acid Kit | Targeted NGS for somatic mutations using a 45-gene panel for LC and 58 gene panel for BC | Significant mutational heterogeneity observed between the paired CTCs and ctDNA samples. Variants in TP53, PIK3CA, and ESR1 identified. | [57] |
| Early-stage NSCLC | 28 SCC, 18 ADC, 3 NOS carcinoma patients | Parsortix System (size-based isolation) followed by single-cell isolation using DropCell platform (CD45−negative enrichment) | Mutational analysis of CTC-derived DNA using ddPCR | Qiagen Circulating Nucleic Acids Kit | Hotspot mutations in BRAF, EGFR, KRAS, and PIK3CA genes analyzed using ddPCR | Mutations detected in 53% of patients when both CTC and ctDNA were assessed. Detection of mutations in either biomarkers associated with higher recurrence rates | [58] |
| NSCLC | 24 NSCLC patients and 6 HDs | Vortex microfluidic platform (size-based isolation) | IF staining (CK+, DAPI+, CD45−) EGFR mutations analyzed using EntroGen ctEGFR assay. | QIAamp Circulating Nucleic Acid Kit | EGFR mutation profiling using qPCR-based EntroGen ctEGFR assay targeting Exon 19 del, L858R, and T790M mutations. | Combining ctDNA and CTCs provided complementary information for EGFR mutation detection. | [58] |
| Type | Samples | CTC | ctDNA | Summary | Ref. | ||
|---|---|---|---|---|---|---|---|
| Isolation | Analysis | Extraction | Analysis | ||||
| mCRC | 20 mCRC patients with KRAS mutations | CellSearch System (EpCAM-positive selection) followed by single-cell isolation using DEPArray™ | Mutational analysis performed via WGA using the Ampli1 WGA kit followed by NGS targeting hotspot regions in 50 oncogenes and tumor suppressor genes | QIAsymphony Circulating DNA Kit. | Targeted NGS for detecting SNVs and indels in 14 CRC-relevant genes using Oncomine Colon cfDNA Assay | CTC positivity at baseline predicted worse OS, while cfDNA enabled reliable detection of KRAS mutations. Increases in cfDNA and CTC counts associated with disease progression. | [64] |
| CRC | 56 CRC patients (34 untreated, 22 stage IV with RAS mutations) | Cynvenio Biosystems LiquidBiopsy Platform (targeting EpCAM, Her2, and Trop2) | IF staining (CK+, CD45−, DAPI+) RAS mutations analyzed using ddPCR NGS for the mutation analysis on 50 oncogenes (Ion AmpliSeq Cancer Hotspot Panel v2) | QIAamp Circulating Nucleic Acid Kit | RAS mutations analyzed using ddPCR NGS for the mutation analysis on 50 oncogenes (Ion AmpliSeq Cancer Hotspot Panel v2) | Combining cfDNA and CTC improved sensitivity for detecting mutations, identifying mutations not found in tumor tissue. | [65] |
| mCRC | 15 mCRC patients undergoing liver metastasectomy (41 blood samples) | Vortex Microfluidic Platform (size-based isolation) | IF staining (CK+, CD45−, DAPI+) Mutation analysis using PCR targeting hotspot mutations in the KRAS, BRAF, and PIK3CA | QIAamp Circulating Nucleic Acid Kit | Mutations analyzed using SCODA mutation enrichment followed by targeted sequencing for KRAS, BRAF, and PIK3CA mutations | Mutations were detected in 77.8% of patients. Concordance rates were 78.2% for KRAS, 73.9% for BRAF, and 91.3% for PIK3CA between CTCs and ctDNA. | [66] |
| AGC | 45 AGC patients undergoing neoadjuvant chemotherapy and surgery | CanPatrol system (nanomembrane filtration) | RNA in situ hybridization to detect epithelial (EpCAM, CK8/18/19), mesenchymal (vimentin, twist), and mixed CTCs. | KminTrak plasma extractor | Quantitatively measured using Qbit fluorescence method. | Mesenchymal CTC levels correlated with advanced N stage and poor chemotherapy response, while higher baseline cfDNA levels predicted better sensitivity to chemotherapy. | [67] |
| PDAC | 45 PDAC patients (31 cfDNA, 35 CTCs) | CellSearch System (EpCAM-positive selection) followed by Immunomagentic depletion of CD45+ cells | IF staining (CK+, CD45−, DAPI+) KRAS mutation analysis Sanger sequencing (G12D, G12R, and G12V) | QIAamp Circulating Nucleic Acid Kit | KRAS mutations (G12D, G12V, G12R) detected via ddPCR | cfDNA was detected across all disease stages, while CTC detection was limited to advanced stages. | [68] |
| Type | Samples | CTC | ctDNA | Summary | Ref. | ||
|---|---|---|---|---|---|---|---|
| Isolation | Analysis | Extraction | Analysis | ||||
| MM | 139 MM patients (107 cfDNA, 56 CTC) | Anti-CD138 magnetic bead-based positive selection | Clonal somatic mutations and CNAs detected using ULP-WGS and WES | Qiagen Circulating Nucleic Acids Kit | Clonal somatic mutations and CNAs detected using ULP-WGS and WES | CTCs and cfDNA demonstrated complementary profiles, with some mutations unique to each. Sequential cfDNA monitoring correlated with disease progression and therapeutic responses. | [69] |
| PCa | 81 PCa patients (69 localized, 12 metastatic) | RosetteSep negative enrichment | PSA-EPISPOT assay (PSA-secreting tumor cells) | QIAamp Circulating Nucleic Acid Kit | Microsatellite analysis using PCR | cfDNA levels were significantly higher in metastatic patients compared to localized cases. Allelic imbalances in cfDNA and CTC presence showed significant associations. | [70] |
| UC | 16 UC patients (6 blood samples from UTUC and 10 urine samples from bladder cancer patients) | Cynvenio LiquidBiopsy Blood Collection Kit (Targeting EpCAM, HER2, EGFR, and Trop2) | IF staining (CK+, CD45−, DAPI+) NGS using Ion AmpliSeq Cancer Hotspot Panel v2 (50 cancer-related genes) | MagMAX Cell-Free DNA Isolation Kit | NGS using Ion AmpliSeq Cancer Hotspot Panel v2 (50 cancer-related genes) | Combined NGS analysis of CTCs and cfDNA identified actionable mutations, with cfDNA revealing additional mutations not found in CTCs | [71] |
| Type | Advantages | Limitations | Ref. |
|---|---|---|---|
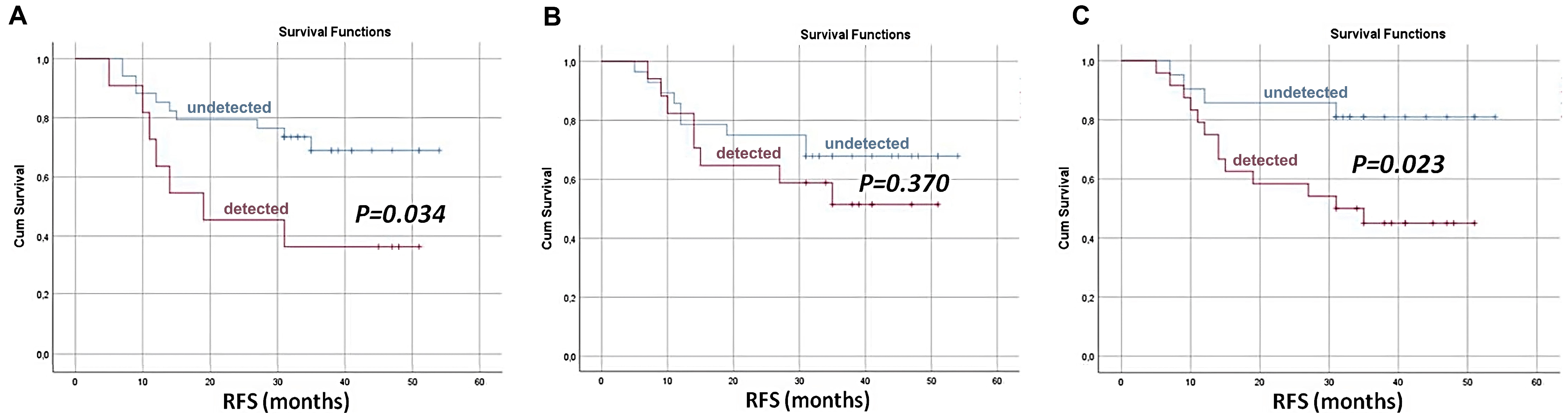
| TNBC | Co-analyzing CTCs and ctDNA provides enhanced predictive accuracy for DFS and DDFS outcomes, as demonstrated by the combined presence of these biomarkers after neoadjuvant chemotherapy being strongly associated with an increased risk of recurrence, which cannot be reliably detected by analyzing either biomarker alone. | The inconsistent detection of CTCs and ctDNA in patients has been described, as individual biomarkers often fail to capture all cases of recurrence; for instance, not all patients with disease relapse had detectable ctDNA or CTCs. | [48] |
| MBC | Co-analyzing CTCs and ctDNA provides a comprehensive view of tumor heterogeneity, as cfDNA captures a broader range of mutations that reflect both individual CTC profiles and additional mutations potentially acquired during disease progression. | The method is constrained by technical challenges, such as the limited detection of low-frequency mutations in individual CTCs due to allelic distortion or dropout during sequencing. | [34] |
| mCRC | Co-analyzing CTCs and ctDNA enhances prognostic accuracy in mCRC patients, as the presence of both biomarkers correlates with a poorer PFS and OS, providing a more comprehensive assessment of patient prognosis than either biomarker alone | The detection rates of CTCs and cfDNA can vary among patients, and the absence of one biomarker does not necessarily predict a better prognosis. | [64] |
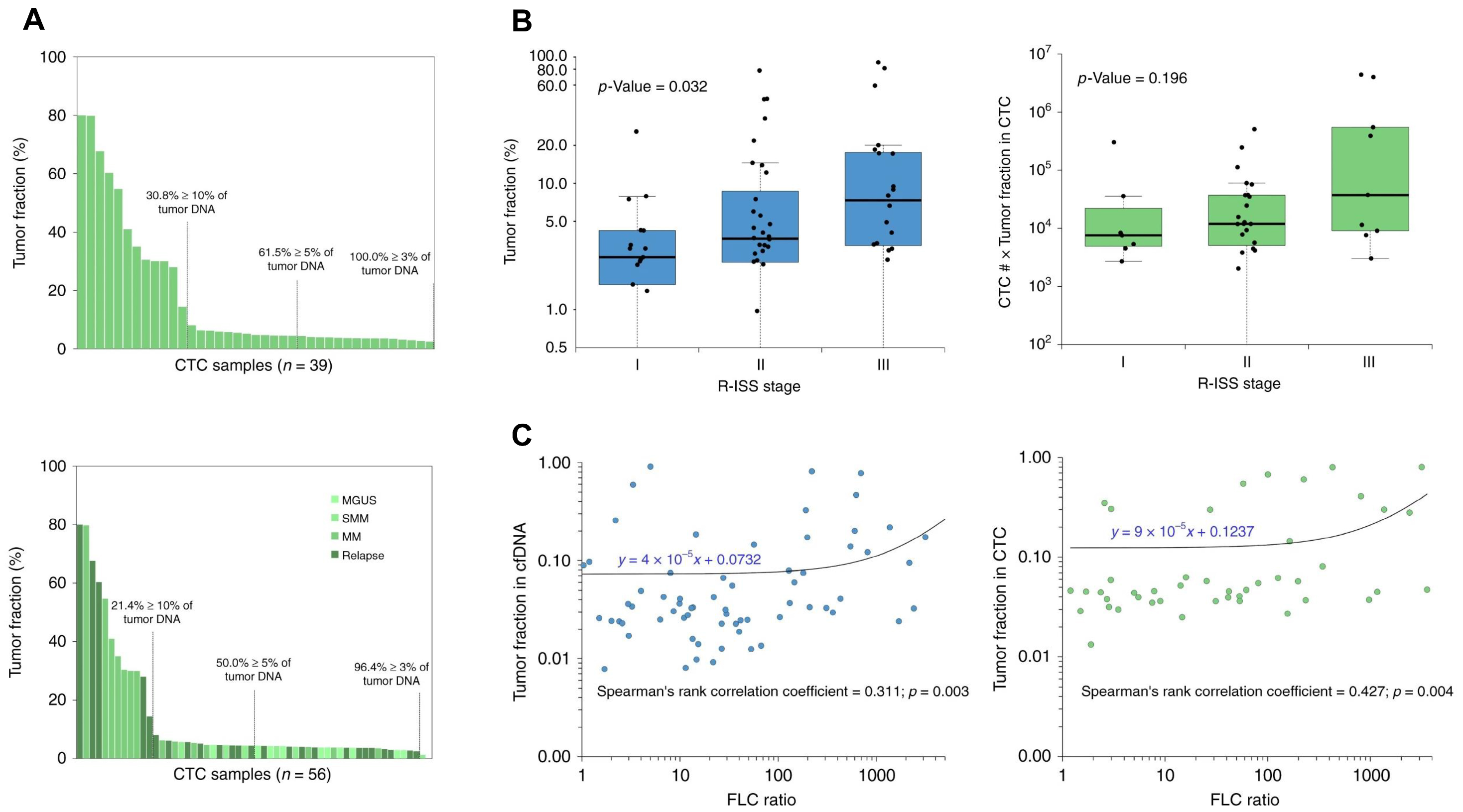
| MM | The presence of both biomarkers is associated with a poorer PFS and OS, suggesting that their combined detection can enhance the accuracy of patient prognosis compared to evaluating each biomarker individually. | The approach is limited by variable tumor fractions in CTCs and cfDNA across patients, which can impact detectability. | [69] |
| PCa | cfDNA provides a broader view of tumor-specific genomic aberrations, such as allelic imbalances, while CTCs offer direct evidence of tumor cell dissemination, together enabling more accurate assessment of tumor progression and metastatic potential. | cfDNA not always correlating with CTC presence due to differences in tumor shedding. | [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, G.Y.; Dalkiran, B.; Park, H.S.; Shin, D.; Son, C.; Choi, J.H.; Bang, S.; Lee, H.; Doh, I.; Kim, D.H.; et al. Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring. Biosensors 2025, 15, 74. https://doi.org/10.3390/bios15020074
Moon GY, Dalkiran B, Park HS, Shin D, Son C, Choi JH, Bang S, Lee H, Doh I, Kim DH, et al. Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring. Biosensors. 2025; 15(2):74. https://doi.org/10.3390/bios15020074
Chicago/Turabian StyleMoon, Ga Young, Basak Dalkiran, Hyun Sung Park, Dongjun Shin, Chaeyeon Son, Jung Hyun Choi, Seha Bang, Hosu Lee, Il Doh, Dong Hyung Kim, and et al. 2025. "Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring" Biosensors 15, no. 2: 74. https://doi.org/10.3390/bios15020074
APA StyleMoon, G. Y., Dalkiran, B., Park, H. S., Shin, D., Son, C., Choi, J. H., Bang, S., Lee, H., Doh, I., Kim, D. H., Jeong, W.-j., & Bu, J. (2025). Dual Biomarker Strategies for Liquid Biopsy: Integrating Circulating Tumor Cells and Circulating Tumor DNA for Enhanced Tumor Monitoring. Biosensors, 15(2), 74. https://doi.org/10.3390/bios15020074





