Label-Free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials and Reagents
2.2. Preparation and Characterization of Flat Graphite Surfaces
2.3. Modification of AFM Probes

2.4. AFM-Based Force Spectroscopy Experiments
2.5. Detection Limit and Selectivity Tests
2.6. Effects of Loading Rates and Solutions
2.7. Statistic Analysis
3. Results and Discussion
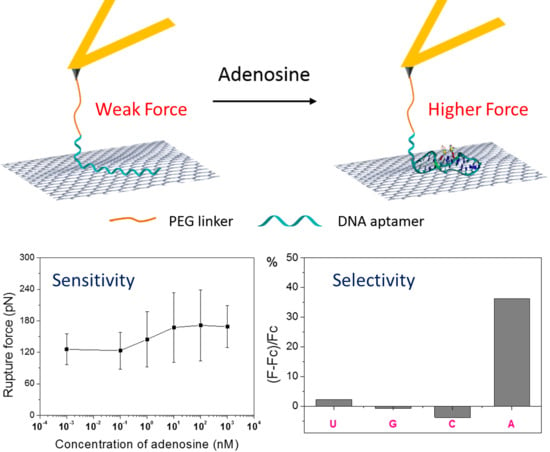
3.1. Sensing Principle

3.2. Typical FD Curves before and After Adding Adenosine


3.3. Detection Limit Test

3.4. Selectivity of the Fabricated Sensor Architecture

3.5. Factors Affecting the Test


4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hasko, G.; Linden, J.; Cronstein, B.; Pacher, P. Adenosine receptor: Therapeutic aspects for inflammatory and immune diseases. Nat. Rev. Drug. Discov. 2008, 7, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2005, 45, 90–94. [Google Scholar] [CrossRef]
- Xue, X.F.; Zhou, J.H.; Wu, L.M.; Fu, L.H.; Zhao, J. HPLC determination of adenosine in royal jelly. Food Chem. 2009, 115, 715–719. [Google Scholar] [CrossRef]
- Bock, R.M.; Ling, N.S.; Morell, S.A.; Lipton, S.H. Ultraviolet absorption spectra of adenosine-5'-triphosphate and related 5'-ribonucleotides. Arch. Biochem. Biophys. 1956, 62, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Rimai, L.; Cole, T.; Parsons, J.L.; Hickmott, J.T.; Carew, E.B. Studies of Raman spectra of water solutions of adenosine tri-, di- and monophosphate and some related compounds. Biophys. J. 1969, 9, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sharon, E.; Freeman, R.; Liu, X.; Willner, I. Fluorescence detection of DNA, adenosine-5'-triphosphate (ATP), and telomerase activity by Zinc(II)-protoporphyrin IX/G-quadruplex labels. Anal. Chem. 2012, 84, 4789–4797. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.Y.; Shih, Y.C.; Tseng, W.L. Tween 20-stabilized gold nanoparticles combined with adenosine triphosphate-BODIPY conjugates for the fluorescence detection of adenosine with more than 1000-fold selectivity. Anal. Chim. Acta 2015, 857, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Tseng, W.L. A method for fluorescence sensing of adenosine and alkaline phosphatase based on the inhibition of S-adenosylhomocysteine hydrolase activity. Biosens. Bioelectron. 2013, 41, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Llaudet, E.; Botting, N.P.; Crayston, J.A.; Dale, N. A three-enzyme microelectrode sensor for detecting purine release from central nervous system. Biosens. Bioelectron. 2003, 18, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xia, J.; Li, X. Electrochemical biosensor for detection of adenosine based on structure-switching aptamer and amplification with reporter probe DNA modified Au nanoparticles. Anal. Chem. 2008, 80, 8382–8388. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, J.; Suri, S.; Sooter, L.J.; Ma, D.; Wu, N. Detection of adenosine triphosphate with an aptamer biosensor based on surface-enhanced Raman scattering. Anal. Chem. 2012, 84, 2837–2842. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Sun, C.; Kang, Y.; Chen, J.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. Label-free electrochemical detection of nanomolar adenosine based on target-induced aptamer displacement. Electrochem. Commun. 2008, 10, 531–535. [Google Scholar] [CrossRef]
- Liu, H.; Xiang, Y.; Lu, Y.; Crooks, R.M. Aptamer-based origami paper analytical device for electrochemical detection of adenosine. Angew. Chem. 2012, 124, 7031–7034. [Google Scholar] [CrossRef]
- Yan, X.; Cao, Z.; Kai, M.; Lu, J. Label-free aptamer-based chemiluminescence detection of adenosine. Talanta 2009, 79, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Schwesinger, F.; Ros, R.; Strunz, T.; Anselmetti, D.; Güntherodt, H.J.; Honegger, A.; Jermutus, L.; Tiefenauer, L.; Pluckthun, A. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA 2000, 97, 9972–9977. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhu, C.; Ling, L.; Wan, L.; Fang, X.; Bai, C. Specific aptamer-protein interaction studied by atomic force microscopy. Anal. Chem. 2003, 75, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Merkel, R.; Nassoy, P.; Leung, A.; Ritchie, K.; Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bard, A.J. Monitoring DNA immobilization and hybridization on surfaces by atomic force microscopy force measurements. Anal. Chem. 2001, 73, 2207–2212. [Google Scholar] [CrossRef] [PubMed]
- Monohar, S.; Mantz, A.R.; Bancroft, K.E.; Hui, C.Y.; Jagota, A.; Vezenov, D.V. Peeling single-stranded DNA from graphite surface to determine oligonucleotide binding energy by force spectroscopy. Nano Lett. 2008, 8, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Iliafar, S.; Wagner, K.; Manohar, S.; Jagota, A.; Vezenov, D. Quantifying interactions between DNA oligomers and graphite surface using single molecule force spectroscopy. J. Phys. Chem. C 2012, 116, 13896–13903. [Google Scholar] [CrossRef]
- Lulevich, V.; Kim, S.; Grigoropoulos, C.P.; Noy, A. Frictionless sliding of single-stranded DNA in a carbon nanotube pore observed by single molecule force spectroscopy. Nano Lett. 2011, 11, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Li, Q.; Steckbeck, S.; Colombi Ciacchi, L. Direct force measurement on peeling heteropolymeric DNA on a graphite surface with single-molecule force spectroscopy. Phys. Chem. Chem. Phys. 2014, 16, 3995–4001. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, Y.; Yu, Y.; Wang, Z.; Zhang, X. Study on intercalations between double-stranded DNA and pyrene by single-molecule force spectroscopy: Toward the detection of mismatch in DNA. Langmuir 2010, 26, 13773–13777. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Steinbock, L.J.; Butt, H.J.; Helm, M.; Berger, R. Measuring single small molecule binding via rupture forces of a split aptamer. J. Am. Chem. Soc. 2011, 133, 2025–2027. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Steckbeck, S.; Köppen, S.; Colombi Ciacchi, L. Label-free biosensing with single-molecule force spectroscopy. Chem. Commun. 2013, 49, 3239–3241. [Google Scholar] [CrossRef]
- Jung, Y.J.; Albrecht, J.A.; Kwak, J.W.; Park, J.W. Direct quantitative analysis of HCV RNA by atomic force microscopy without labeling or amplification. Nucleic Acids Res. 2012, 40, 11728–11736. [Google Scholar] [CrossRef] [PubMed]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef]
- Lin, C.H.; Patel, D.J. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: Distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997, 4, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Huizenga, D.E.; Szostak, J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, W.; Cui, S.; Wang, Z.; Zhang, X. Force spectroscopy of polymers: Beyond single chain mechanics. Curr. Opin. Solid State Mater. Sci. 2005, 9, 140–148. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, Q.; Ciacchi, L.C.; Wei, G. Label-Free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition. Biosensors 2015, 5, 85-97. https://doi.org/10.3390/bios5010085
Li J, Li Q, Ciacchi LC, Wei G. Label-Free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition. Biosensors. 2015; 5(1):85-97. https://doi.org/10.3390/bios5010085
Chicago/Turabian StyleLi, Jingfeng, Qing Li, Lucio Colombi Ciacchi, and Gang Wei. 2015. "Label-Free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition" Biosensors 5, no. 1: 85-97. https://doi.org/10.3390/bios5010085
APA StyleLi, J., Li, Q., Ciacchi, L. C., & Wei, G. (2015). Label-Free Sensing of Adenosine Based on Force Variations Induced by Molecular Recognition. Biosensors, 5(1), 85-97. https://doi.org/10.3390/bios5010085