Label-Free Imaging and Biochemical Characterization of Bovine Sperm Cells
Abstract
:1. Introduction
2. Theoretical Background
2.1. Digital Holography
2.2. Raman Spectroscopy
3. Experimental Set Up

Sample Preparation
4. Results and Discussion

4.1. Morphological Characterization of Bull Sperm Cells



| Raman peak assignment | |||
|---|---|---|---|
| Raman band (cm−1) | Nucleic acids | Protein | Lipids |
| 751 | U, C | ||
| 785 | T, C, bk | ||
| 831 | OPO | Tyr | |
| 1005 | Phe | ||
| 1095 | PO2- | C–C | |
| 1255 | Amide III | =CH | |
| 1290 | A, T | Amide III | =CH |
| 1320–1380 | A, G | CH def | CH3, CH2 |
| 1450–1480 | A, G | CH2 | CH2 |
| 1575–1585 | A, G | ||
| 1600–1700 | T | Amide I | C=C |

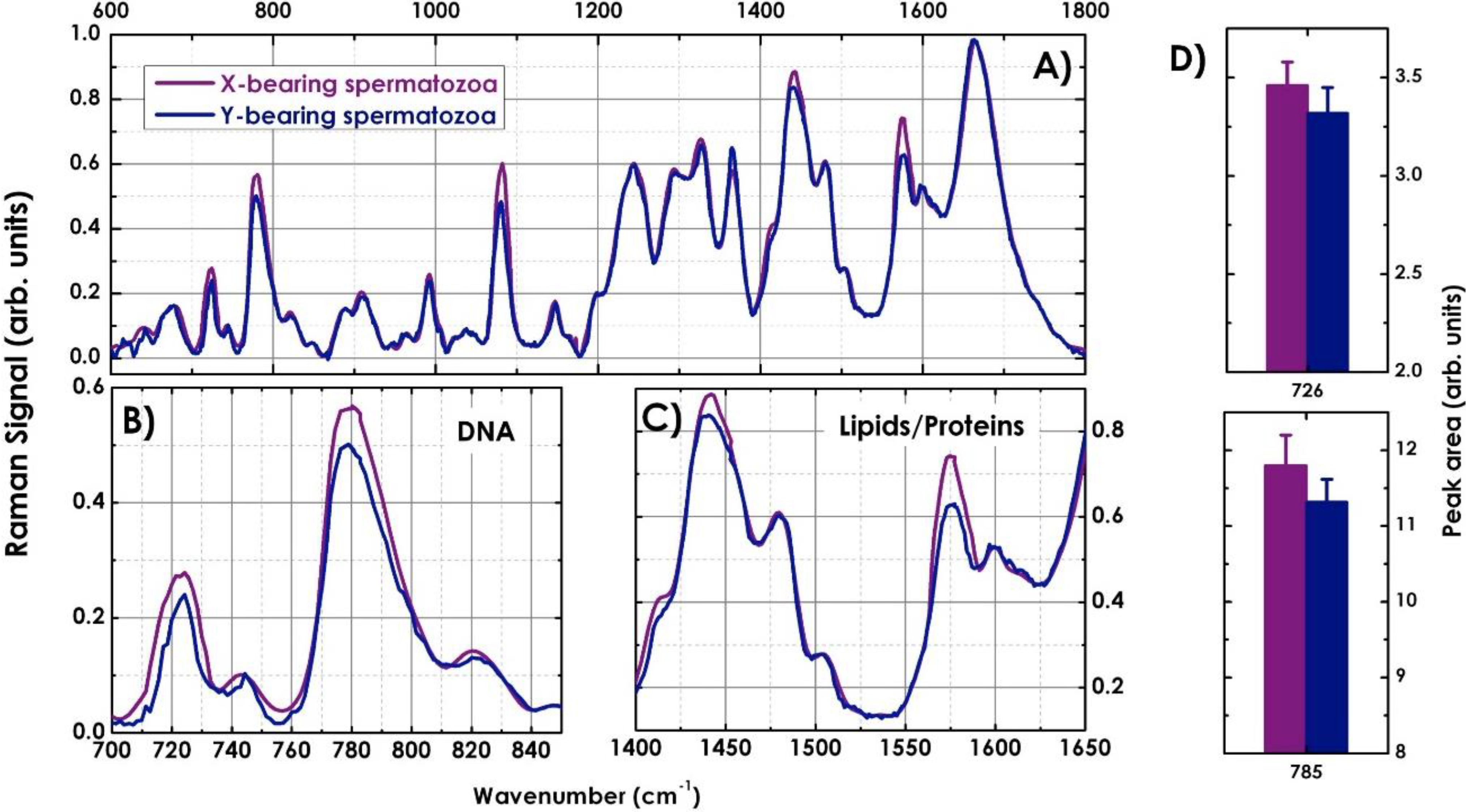
4.2. Characterization of X-and Y-Bearing Sperm Cells



5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Al-Makhzoomi, A.; Lundeheim, N.; Håård, M.; Rodríguez-Martínez, H. Sperm morphology and fertility of progeny-tested AI dairy bulls in Sweden. Theriogenology 2008, 70, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, G.; Maree, L. SpermBlue: A new universal stain for human and animal sperm which is also amenable to automated sperm morphology analysis. Biotech. Histochem. 2009, 84, 1–10. [Google Scholar]
- De Luca, A.C.; Managó, S.; Ferrara, M.A.; Rendina, I.; Sirleto, L.; Puglisi, R.; Balduzzi, D.; Galli, A.; Ferraro, P.; Coppola, G. Non-invasive sex assessment in bovine semen by Raman spectroscopy. Laser Phys. Lett. 2014, 11. [Google Scholar] [CrossRef]
- Soggiu, S.; Piras, C.; Hussein, H.A.; de Canio, M.; Gaviraghi, A.; Galli, A.; Urbani, A.; Binizzi, L.; Roncada, P. Unravelling the bull fertility proteome. Mol. BioSyst. 2013, 9, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Pace, M.M.; Sullivan, J.J.; Elliott, F.I.; Graham, E.F.; Coulter, G.H. Effects of thawing temperature, number of spermatozoa and spermatozoa quality on fertility of bovine spermatozoa packaged in 0.5-mL french straws. J. Anim. Sci. 1981, 53, 693–701. [Google Scholar]
- Amann, R.P.; Hammerstedt, R.H. In vitro evaluation of sperm quality: An Opinion. J. Androl. 1993, 14, 397–406. [Google Scholar] [PubMed]
- Graham, J.K. In vitro assays of bull fertility. In Proceedings of the 15th Technical Conference on Artificial Insemination and Reproduction National Association of Animal Breeders, Milwaukee, WI, USA, April 1994; pp. 74–81.
- Ostermeier, G.C.; Sartor-Bergfelt, R.; Susko-Parrish, J.L.; Parrish, J.J. Bull fertility and sperm nuclear shape. AgBiotechNet 2000, 2, ABN 055:1–ABN 055:6. [Google Scholar]
- Di Caprio, G.; Ferrara, M.A.; Miccio, L.; Merola, F.; Memmolo, P.; Ferraro, P.; Coppola, G. Holographic imaging of unlabelled sperm cells for semen analysis: A review. J. Biophotonics 2014, 9999, 1–11. [Google Scholar]
- Kumar, S.; Chaudhury, K.; Sen, P.; Guha, S.K. Atomic force microscopy: A powerful tool for high-resolution imaging of spermatozoa. J. Nanobiotechnol. 2005, 3, 3–9. [Google Scholar] [CrossRef]
- Allen, M.J.; Bradbury, E.M.; Balhorn, R. The natural subcellular surface structure of the bovine sperm cell. J. Struct. Biol. 1995, 114, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Di Caprio, G.; Gioffrè, M.; Saffioti, N.; Grilli, S.; Ferraro, P.; Puglisi, R.; Balduzzi, D.; Galli, A.; Coppola, G. Quantitative label-free animal sperm imaging by means of digital holographic microscopy. IEEE J. Quant. Electron. 2010, 16, 833–840. [Google Scholar] [CrossRef]
- Memmolo, P.; Di Caprio, G.; Distante, C.; Paturzo, M.; Puglisi, R.; Balduzzi, D.; Galli, A.; Coppola, G.; Ferraro, P. Identification of bovine sperm head for morphometry analysis in quantitative phase-contrast holographic microscopy. Opt. Express 2011, 19, 23215–23226. [Google Scholar]
- Merola, F.; Miccio, L.; Memmolo, P.; Di Caprio, G.; Galli, A.; Puglisi, R.; Balduzzi, D.; Coppola, G.; Netti, P.; Ferraro, P. Digital holography as a method for 3D imaging and estimating biovolume of motile cells. Lab Chip 2013, 13, 4512–4516. [Google Scholar] [CrossRef] [PubMed]
- Seidel, E.G.; Garner, D.L. Current status of sexing mammalian spermatozoa. Reproduction 2002, 124, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Garner, D.L. Flow cytometric sexing of mammalian sperm. Theriogenology 2006, 65, 943–957. [Google Scholar]
- Howes, E.A.; Miller, N.G.A.; Dolby, C.; Hutchings, A.; Butcher, G.W.; Jones, R. A search for sex-specific antigens on bovine spermatozoa using immunological and biochemical techniques to compare the protein profiles of X and Y chromosome-bearing sperm populations separated by fluorescence-activated cell sorting. J. Reprod. Fertil. 1997, 110, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, P.J.M. Do X and Y spermatozoa differ in proteins? Theriogenology 1999, 52, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Zavaczki, Z.; Celik-Ozenci, C.; Ovari, L.; Jakab, A.; Sati, G.L.; Ward, D.C.; Huszar, G. Dimensional assessment of X-bearing and Y-bearing haploid and disomic human sperm with the use of fluorescence in situ hybridization and objective morphometry. Fertil. Steril. 2006, 85, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.O.; Silva, L.P.; Sartori, R.; Dode, M.A.N. Nanoscale Differences in the Shape and Size of X and Y Chromosome-Bearing Bovine Sperm Heads Assessed by Atomic Force Microscopy. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Kaneko, S.; Oshio, S.; Kobayashi, T.; Iizuka, R.; Mohri, H. Human X- and Y-bearing sperm differ in cell surface sialic acid content. Biochem. Biophys. Res. Commun. 1984, 124, 950–955. [Google Scholar] [CrossRef] [PubMed]
- Canetta, E.; Mazilu, M.; de Luca, A.C.; Currethers, A.E.; Dholakia, K.; Neilson, S.; Sargeant, H.; Briscoe, T.; Herrington, C.S.; Riches, A.C. Modulated Raman spectroscopy for enhanced identification of bladder tumor cells in urine samples. J. Biomed. Opt. 2011, 16. [Google Scholar] [CrossRef] [Green Version]
- Bräutigam, K.; Bocklitz, T.; Silge, A.; Dierker, C.; Ossig, R.; Schnekenburger, J.; Cialla, D.; Rösch, P.; Popp, J. Comparative two- and three-dimensional analysis of nanoparticle localization in different cell types by Raman spectroscopic imaging. J. Mol. Struct. 2014, 1073, 44–50. [Google Scholar] [CrossRef]
- Adar, F.; Mamedov, S.; Whitely, A. Limits of Spatial Resolution of a Raman Microscope. Microscopy Microanal. 2005, 11, S728–S729. [Google Scholar] [CrossRef]
- Notingher, I. Raman Spectroscopy Cell-based Biosensors. Sensors 2007, 7, 1343–1358. [Google Scholar] [CrossRef]
- De Luca, A.C.; Render-Harris, P.; Mazilu, M.; Mariggiò, S.; Corda, D.; Di Falco, A. Reproducible Surface-Enhanced Raman quantification of biomarkers in multicomponent mixtures. ACS Nano 2014, 8, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Rusciano, G.; De Luca, A.C.; Pesce, G.; Sasso, A. Enhancing Raman Tweezers by phase-sensitive detection. Anal. Chem. 2007, 79, 3708–3715. [Google Scholar] [CrossRef] [PubMed]
- Samek, O.; Jonáš, A.; Pilát, Z.; Zemánek, P.; Nedbal, L.; Tříska, J.; Kotas, P.; Trtílek, M. Raman Microspectroscopy of Individual Algal Cells: Sensing Unsaturation of Storage Lipids in vivo. Sensors 2010, 10, 8635–8651. [Google Scholar] [CrossRef]
- Schmälzlin, E.; Moralejo, B.; Rutowska, M.; Monreal-Ibero, A.; Sandin, C.; Tarcea, N.; Popp, J.; Roth, M.M. Raman Imaging with a Fiber-Coupled Multichannel Spectrograph. Sensors 2014, 14, 21968–21980. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of tumors during tissue-conserving surgery with integrated auto fluorescence and Raman scattering microscopy. PNAS 2013, 110, 15189–15194. [Google Scholar] [CrossRef] [PubMed]
- Uzunbajakava, N.; Lenferink, A.; Kraan, Y.; Volokhina, E.; Vrensen, G.; Greve, J.; Otto, C. Nonresonant Confocal Raman Imaging of DNA and Protein Distribution in Apoptotic Cells. Biophys. J. 2003, 84, 3968–3981. [Google Scholar] [CrossRef] [PubMed]
- Kosmeier, S.; Zolotovskaya, S.; De Luca, A.C.; Riches, A.; Herrington, C.S.; Dholakia, K.; Mazilu, M. Nonredundant Raman imaging using optical eigenmodes. Optica 2014, 1, 257–263. [Google Scholar] [CrossRef]
- Marquet, P.; Rappaz, B.; Magistretti, P.J.; Cuche, E.; Emery, Y.; Colomb, T.; Depeursinge, C. Digital holographic microscopy: A noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt. Lett. 2005, 30, 468–470. [Google Scholar] [CrossRef] [PubMed]
- Takaki, Y.; Kawai, H.; Ohzu, H. Hybrid holographic microscopy free of conjugate and zero-order images. Appl. Opt. 1999, 38, 4990–4996. [Google Scholar] [CrossRef] [PubMed]
- Pedrini, G.; Schedin, S.; Tiziani, H.J. Aberration compensation in digital holographic reconstruction of microscopic objects. J. Mod. Opt. 2001, 48, 1035–1041. [Google Scholar] [CrossRef]
- Schnars, U.; Jüptner, W.P.O. Digital recording and numerical reconstruction of holograms. Meas. Sci. Technol. 2002, 13, R85–R101. [Google Scholar] [CrossRef]
- Yu, L.; Cai, L. Iterative algorithm with a constraint condition for numerical reconstruction of threedimensional object from its hologram. J. Opt. Soc. Am. 2001, 18, 1033–1045. [Google Scholar] [CrossRef]
- Goodman, J.W. Introduction to Fourier Optics, 2nd ed.; Mc Graw-Hill: New York, NY, USA, 1996. [Google Scholar]
- Ashok, P.C.; De Luca, A.C.; Mazilu, M.; Dholakia, K. Enhanced bioanalyte detection in waveguide confined Raman spectroscopy using modulation techniques. J. Biophot. 2011, 4, 514–518. [Google Scholar] [CrossRef]
- Bell, S.E.; Burns, D.T.; Dennis, A.C.; Matchett, L.J.; Speers, J.S. Composition profiling of seized ecstasy tablets by Raman spectroscopy. Analyst. 2000, 125, 1811–1815. [Google Scholar] [CrossRef] [PubMed]
- Rusciano, G.; De Luca, A.C.; Pesce, G.; Sasso, A. On the interaction of nano-sized organic carbon particles with model lipid membranes. Carbon 2009, 47, 2950–2957. [Google Scholar] [CrossRef]
- Klein, K. Label-Free Live-Cell Imaging with Confocal Raman Microscopy. Biophys. J. 2012, 102, 360–368. [Google Scholar] [CrossRef]
- Rusciano, G.; De Luca, A.C.; Pesce, G.; Sasso, A. Raman Tweezers as a Diagnostic Tool of Hemoglobin-Related Blood Disorders. Sensors 2008, 8, 7818–7832. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. Automatica 1975, 11, 285–296. [Google Scholar] [CrossRef]
- De Canio, M.; Soggiu, A.; Piras, C.; Bonizzi, L.; Galli, A.; Urbani, A.; Roncada, P. Diffrential protein profile in sexed bovine semen: Shotgun proteomics investigation. Mol. BioSyst. 2014, 10, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Kosmeier, S.; De Luca, A.C.; Zolotovskaya, S.; Di Falco, A.; Dholakia, K.; Mazilu, M. Coherent control of plasmonic nanoantennas using optical eigenmodes. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrara, M.A.; Di Caprio, G.; Managò, S.; De Angelis, A.; Sirleto, L.; Coppola, G.; De Luca, A.C. Label-Free Imaging and Biochemical Characterization of Bovine Sperm Cells. Biosensors 2015, 5, 141-157. https://doi.org/10.3390/bios5020141
Ferrara MA, Di Caprio G, Managò S, De Angelis A, Sirleto L, Coppola G, De Luca AC. Label-Free Imaging and Biochemical Characterization of Bovine Sperm Cells. Biosensors. 2015; 5(2):141-157. https://doi.org/10.3390/bios5020141
Chicago/Turabian StyleFerrara, Maria Antonietta, Giuseppe Di Caprio, Stefano Managò, Annalisa De Angelis, Luigi Sirleto, Giuseppe Coppola, and Anna Chiara De Luca. 2015. "Label-Free Imaging and Biochemical Characterization of Bovine Sperm Cells" Biosensors 5, no. 2: 141-157. https://doi.org/10.3390/bios5020141
APA StyleFerrara, M. A., Di Caprio, G., Managò, S., De Angelis, A., Sirleto, L., Coppola, G., & De Luca, A. C. (2015). Label-Free Imaging and Biochemical Characterization of Bovine Sperm Cells. Biosensors, 5(2), 141-157. https://doi.org/10.3390/bios5020141