A Bioelectronic System to Measure the Glycolytic Metabolism of Activated CD4+ T Cells
Abstract
:1. Introduction
2. Methods
2.1. Metabolite Biosensor Device
2.1.1. The Probing Electrodes
2.1.2. The Microcontroller-Based Electronic Interface
2.1.3. The Algorithm for Data Acquisition and Signal Processing
2.1.4. The Labview Interface
3. Preparation, Storage, and Thawing of Human Peripheral Blood Mononuclear Cells (PBMCs)
3.1. Isolation and Activation of the CD4+ T Cells
3.2. Biosensor Measurements of the Cell-Free Culture Media
3.3. Biosensor Measurements of Lactate Standards
3.4. Glucose Uptake Assays
3.4.1. GlucMeter Reading
3.4.2. 2-NBDG Assay
3.4.3. L-Lactate Assay
3.5. Statistical Analysis
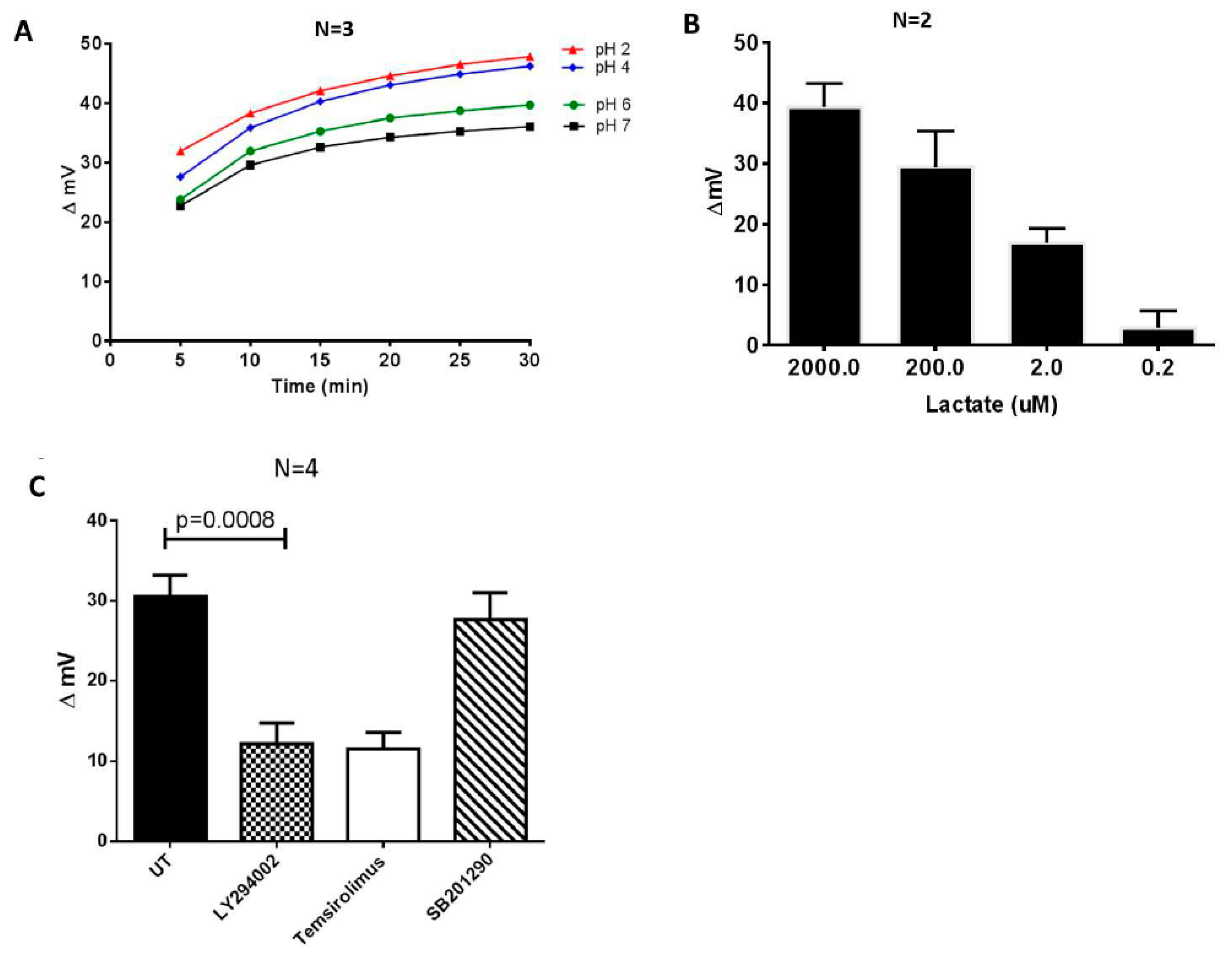
4. Results and Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Frauwirth, K.A.; Thompson, C.B. Regulation of T Lymphocyte Metabolism. J. Immunol. 2004, 172, 4661–4665. [Google Scholar] [CrossRef] [Green Version]
- Pearce, E.L.; Poffenberger, M.C.; Chang, C.-H.; Jones, R.G. Fueling immunity: Insights into metabolism and lymphocyte function. Science 2013, 342, 1242454. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.S.; Ostrowski, M.; Balderson, B.; Christian, N.; Crowe, S.W. Glucose metabolism regulates T cell activation, differentiation, and functions. Front. Immunol. 2015, 6, 1. [Google Scholar] [CrossRef]
- Ricciardi, S.; Manfrini, N.; Alfieri, R.; Calamita, P.; Crosti, M.C.; Gallo, S.; Müller, R.; Pagani, M.; Abrignani, S.; Biffo, S. The Translational Machinery of Human CD4+ T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metab. 2018, 28, 895–906.e5. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, X.; Sanders, K.L.; Edwards, J.L.; Ye, J.; Si, F.; Gao, A.; Huang, L.; Hsueh, E.C.; Ford, D.A.; et al. TLR8-Mediated Metabolic Control of Human Treg Function: A Mechanistic Target for Cancer Immunotherapy. Cell Metab. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.M.; Murphy, A.J.; Lee, M.K.S.; Gardiner, C.M.; Crowe, S.M.; Sanjabi, S.; Finlay, D.K.; Palmer, C.S. Sugar or Fat?—Metabolic Requirements for Immunity to Viral Infections. Front. Immunol. 2017, 8, 1311. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.S.; Ostrowski, M.; Gouillou, M.; Tsai, L.; Yu, D.; Zhou, J.; Henstridge, D.C.; Maisa, A.; Hearps, A.C.; Lewin, S.R.; et al. Increased glucose metabolic activity is associated with CD4+ T-cell activation and depletion during chronic HIV infection. AIDS 2014, 28, 297–309. [Google Scholar] [CrossRef]
- Maratou, E.; Dimitriadis, G.; Kollias, A.; Boutati, E.; Lambadiari, V.; Mitrou, P.; Raptics, S.A. Glucose transporter expression on the plasma membrane of resting and activated white blood cells. Eur. J. Clin. Invest. 2007, 37, 282–290. [Google Scholar] [CrossRef]
- Barata, J.T.; Boussiotis, V.A.; Yunes, J.A.; Ferrando, A.A.; Moreau, L.A.; Veiga, J.P.; Sallan, S.E.; Look, A.T.; Nadler, L.M.; Cardoso, A.A. IL-7-dependent human leukemia T-cell line as a valuable tool for drug discovery in T-ALL. Blood 2004, 103, 1891–1900. [Google Scholar] [CrossRef] [Green Version]
- Duette, G.; Gerber, P.P.; Rubione, J.; Perez, P.S.; Landay, A.L.; Crowe, S.M.; Liao, Z.; Witwer, K.W.; Holgado, M.P.; Salido, J.; et al. Induction of HIF-1α by HIV-1 Infection in CD4+ T Cells Promotes Viral Replication and Drives Extracellular Vesicle-Mediated Inflammation. mBio 2018, 9, e00757-18. [Google Scholar] [CrossRef]
- Palmer, C.S.; Duette, G.A.; Wagner, M.C.E.; Henstridge, D.C.; Saleh, S.; Pereira, C.; Zhou, J.; Simar, D.; Lewin, S.R.; Ostrowski, M.; et al. Metabolically active CD4+ T cells expressing Glut1 and OX40 preferentially harbor HIV during in vitro infection. FEBS Lett. 2017, 591, 3319–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanagh Williamson, M.; Coombes, N.; Juszczak, F.; Athanasopoulos, M.; Khan, M.B.; Eykyn, T.R.; Srenathan, U.; Taams, L.S.; Dias Zeidler, J.; Da Poian, A.T.; et al. Upregulation of Glucose Uptake and Hexokinase Activity of Primary Human CD4+ T Cells in Response to Infection with HIV-1. Viruses 2018, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Loisel-Meyer, S.; Swainson, L.; Craveiro, M.; Oburoglu, L.; Mongellaz, C.; Costa, C.; Martinez, M.; Cosset, F.-L.; Battini, J.-L.; Herzenberg, L.A.; et al. Glut1-mediated glucose transport regulates HIV infection. Proc. Natl. Acad. Sci. USA 2012, 109, 2549–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porras, O.H.; Ruminot, I.; Loaiza, A.; Barros, L.F. Na+-Ca2+ cosignaling in the stimulation of the glucose transporter GLUT1 in cultured astrocytes. Glia 2008, 56, 59–68. [Google Scholar] [CrossRef]
- Lang, F.; Strutz-Seebohm, N.; Seebohm, G.; Lang, U.E. Significance of sgk1 in the regulation of neuronal function. J. Physiol. 2010, 588, 3349–3354. [Google Scholar] [CrossRef] [PubMed]
- van der Windt, G.J.W.; Everts, B.; Chang, C.-H.; Curtis, J.D.; Freitas, T.C.; Amiel, E.; Pearce, E.J.; Pearce, E.L. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 2012, 36, 68–78. [Google Scholar] [CrossRef]
- Perry, C.G.R.; Kane, D.A.; Lanza, I.R.; Neufer, P.D. Methods for assessing mitochondrial function in diabetes. Diabetes 2013, 62, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B., Jr.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.-C.; van der Windt, G.J.W.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013, 153, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Yang, K.; Cloer, C.; Neale, G.; Vogel, P.; Chi, H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 2013, 499, 485–490. [Google Scholar] [CrossRef]
- Gubser, P.M.; Bantug, G.R.; Razik, L.; Fischer, M.; Dimeloe, S.; Hoenger, G.; Durovic, B.; Jauch, A.; Hess, C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 2013, 14, 1064–1072. [Google Scholar] [CrossRef]
- Billiard, J.; Dennison, J.B.; Briand, J.; Annan, R.S.; Chai, D.; Colón, M.; Dodson, C.S.; Gilbert, S.A.; Greshock, J.; Jing, J.; et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013, 1, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kintzios, S.; Pistola, E.; Konstas, J.; Bem, F.; Matakiadis, T.; Alexandropoulos, N.; Biselis, I.; Levin, R. The application of the bioelectric recognition assay for the detection of human and plant viruses: Definition of operational parameters. Biosens. Bioelectron. 2001, 16, 467–480. [Google Scholar] [CrossRef]
- Banerjee, P.; Kintzios, S.; Prabhakarpandian, B. Biotoxin detection using cell-based sensors. Toxins 2013, 5, 2366–2383. [Google Scholar] [CrossRef] [PubMed]
- Apostolou, T.; Pascual, N.; Marco, M.P.; Moschos, A.; Petropoulos, A.; Kaltsas, G.; Kintzios, S. Extraction-less, rapid assay for the direct detection of 2,4,6-trichloroanisole (TCA) in cork samples. Talanta 2014, 125, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.; Hampartzoumian, T.; Lloyd, A.; Zekry, A. A novel role for adiponectin in regulating the immune responses in chronic hepatitis c virus infection. Hepatology 2008, 48, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Pachow, D.; Andrae, N.; Kliese, N.; Angenstein, F.; Stork, O.; Wilisch-Neumann, A.; Kirches, E.; Mawrin, C. mTORC1 inhibitors suppress meningioma growth in mouse models. Clin. Cancer Res. 2013, 19, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Chiang, K.-C.; Yang, S.-W.; Chang, K.-P.; Feng, T.-H.; Chang, K.-S.; Tsui, K.-H.; Shin, Y.-S.; Chen, C.-C.; Chao, M.; Juang, H.-H. Caffeic Acid Phenethyl Ester Induces N-myc Downstream Regulated Gene 1 to Inhibit Cell Proliferation and Invasion of Human Nasopharyngeal Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1397. [Google Scholar] [CrossRef]
- Man, K.; Miasari, M.; Shi, W.; Xin, A.; Henstridge, D.C.; Preston, S.; Pellegrini, M.; Belz, G.T.; Smyth, G.K.; Febbraio, M.A.; et al. The transcription factor irf4 is essential for tcr affinity-mediated metabolic programming and clonal expansion of t cells. Nat. Immunol. 2013, 14, 1155–1165. [Google Scholar] [CrossRef]
- Kiss, L.; Bennett, P.B.; Uebele, V.N.; Koblan, K.S.; Kane, S.A.; Neagle, B.; Schroeder, K. High throughput ion-channel pharmacology: Planar-array-based voltage clamp. Assay Drug. Dev. Technol. 2003, 1, 127–135. [Google Scholar] [CrossRef]
- Gill, S.; Gill, R.; Lee, S.S.; Hesketh, J.C.; Fedida, D.; Rezazadeh, S.; Stankovich, L.; Liang, D. Flux assays in high throughput screening of ion channels in drug discovery. Assay Drug. Dev. Technol. 2003, 1, 709–717. [Google Scholar] [CrossRef]
- Moschopoulou, G.; Dourou, A.-M.; Fidaki, A.; Kintzios, S.E. Assessment of pesticides cytoxicity by means of bioelectric profiling of mammalian cells. Environ. Nanotechnol. Monit. Manag. 2017, 8, 254–260. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crowe, S.M.; Kintzios, S.; Kaltsas, G.; Palmer, C.S. A Bioelectronic System to Measure the Glycolytic Metabolism of Activated CD4+ T Cells. Biosensors 2019, 9, 10. https://doi.org/10.3390/bios9010010
Crowe SM, Kintzios S, Kaltsas G, Palmer CS. A Bioelectronic System to Measure the Glycolytic Metabolism of Activated CD4+ T Cells. Biosensors. 2019; 9(1):10. https://doi.org/10.3390/bios9010010
Chicago/Turabian StyleCrowe, Suzanne M., Spyridon Kintzios, Grigoris Kaltsas, and Clovis S. Palmer. 2019. "A Bioelectronic System to Measure the Glycolytic Metabolism of Activated CD4+ T Cells" Biosensors 9, no. 1: 10. https://doi.org/10.3390/bios9010010
APA StyleCrowe, S. M., Kintzios, S., Kaltsas, G., & Palmer, C. S. (2019). A Bioelectronic System to Measure the Glycolytic Metabolism of Activated CD4+ T Cells. Biosensors, 9(1), 10. https://doi.org/10.3390/bios9010010






