A Molecular Interaction Analysis Reveals the Possible Roles of Graphene Oxide in a Glucose Biosensor
Abstract
:1. Introduction
2. Materials and Methods
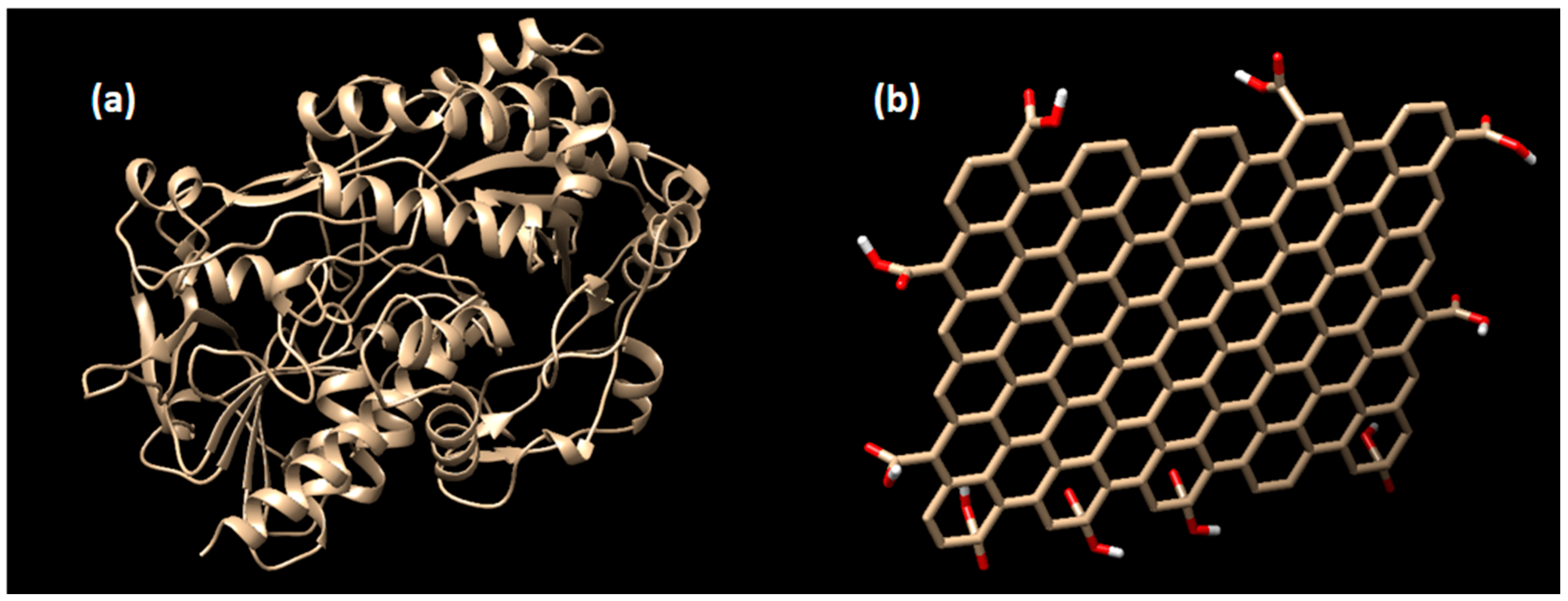
2.1. Enzyme Preparation
2.2. Ligand Preparation
2.3. Blind Docking Simulation
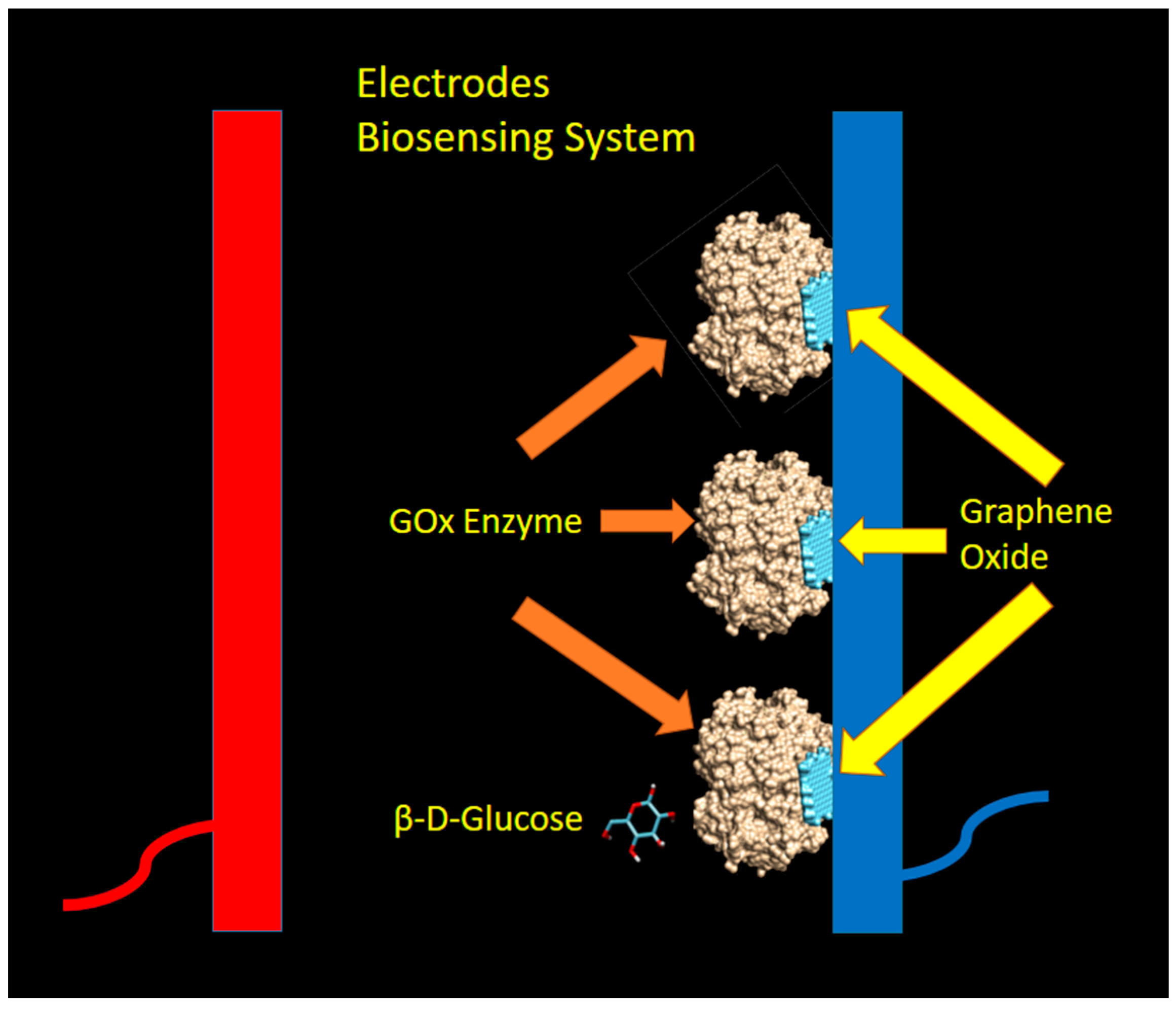
2.4. Biosensing Mechanism and Model
3. Results and Discussion
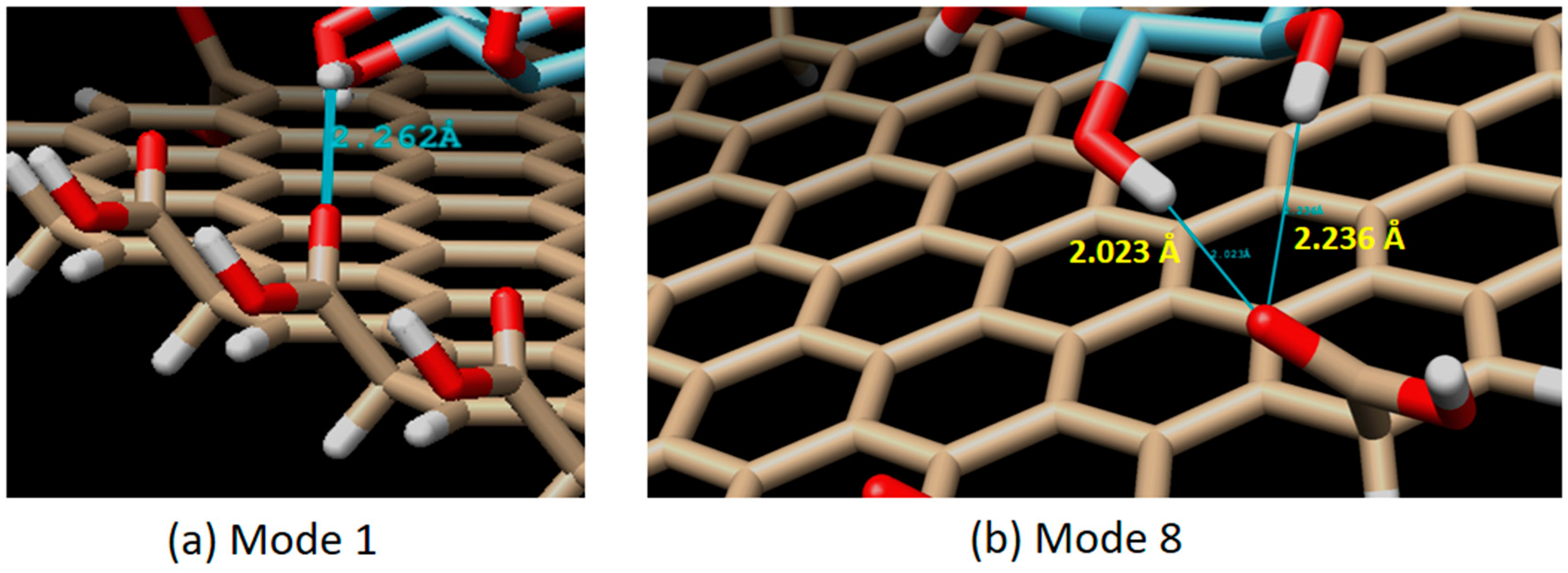
3.1. Molecular Interaction Analysis of Graphene Oxide and GOx
3.2. Direct Sensing of Glucose by Graphene Oxide: A Non-Enzymatic Mechanism
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Novoselov 2004. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.C.; Geim, A.K.; Katsnelson, M.I.; Novoselov, K.S.; Booth, T.J.; Roth, S. The structure of suspended graphene sheets. Nature 2007, 446, 60–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, C.H.; Liu, L.; Mak, K.F.; Flynn, G.W.; Heinz, T.F. Ultraflat graphene. Nature 2009, 462, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Acik, M.; Chabal, Y.J. Nature of graphene edges: A review. Jpn. J. Appl. Phys. 2011, 50, 070101. [Google Scholar] [CrossRef]
- Afsahi, S.; Lerner, M.B.; Goldstein, J.M.; Lee, J.; Tang, X.; Bagarozzi, D.A.; Pan, D.; Locascio, L.; Walker, A.; Barron, F.; et al. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018, 100, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Pumera, M. Graphene in biosensing. Mater. Today 2011, 14, 308–315. [Google Scholar] [CrossRef]
- Megawati, M.; Chua, C.K.; Sofer, Z.; Klímová, K.; Pumera, M. Nitrogen-doped graphene: Effect of graphite oxide precursors and nitrogen content on the electrochemical sensing properties. Phys. Chem. Chem. Phys. 2017, 19, 15914–15923. [Google Scholar] [CrossRef] [PubMed]
- Kybert, N.J.; Han, G.H.; Lerner, M.B.; Dattoli, E.N.; Esfandiar, A.; Charlie Johnson, A.T. Scalable arrays of chemical vapor sensors based on DNA-decorated graphene. Nano Res. 2014, 7, 95–103. [Google Scholar] [CrossRef]
- Lerner, M.B.; Matsunaga, F.; Han, G.H.; Hong, S.J.; Xi, J.; Crook, A.; Perez-Aguilar, J.M.; Park, Y.W.; Saven, J.G.; Liu, R.; et al. Scalable production of highly sensitive nanosensors based on graphene functionalized with a designed G protein-coupled receptor. Nano Lett. 2014, 14, 2709–2714. [Google Scholar] [CrossRef]
- Lerner, M.B.; Goldsmith, B.; Rockway, J.; Perez, I. Towards a Carbon Nanotube Intermodulation Product Sensor for Nonlinear Energy Harvesting. J. Sens. 2015, 2015, 983697. [Google Scholar] [CrossRef]
- Esfandiar, A.; Kybert, N.J.; Dattoli, E.N.; Hee Han, G.; Lerner, M.B.; Akhavan, O.; Irajizad, A.; Charlie Johnson, A.T. DNA-decorated graphene nanomesh for detection of chemical vapors. Appl. Phys. Lett. 2013, 103, 183110. [Google Scholar] [CrossRef]
- Fujimoto, Y. First-principles theoretical investigation of graphene layers for sensor applications: A review. Nanomater. Nanotechnol. 2017, 7. [Google Scholar] [CrossRef]
- Lu, Y.; Lerner, M.B.; John Qi, Z.; Mitala, J.J.; Hsien Lim, J.; Discher, B.M.; Charlie Johnson, A.T. Graphene-protein bioelectronic devices with wavelength-dependent photoresponse. Appl. Phys. Lett. 2012, 100, 033110. [Google Scholar] [CrossRef] [Green Version]
- Dey, R.S.; Raj, C.R. Development of an amperometric cholesterol biosensor based on graphene-Pt nanoparticle hybrid material. J. Phys. Chem. C 2010, 114, 21427–21433. [Google Scholar] [CrossRef]
- Dey, R.S.; Raj, C.R. Enzyme-integrated cholesterol biosensing scaffold based on in situ synthesized reduced graphene oxide and dendritic Pd nanostructure. Biosens. Bioelectron. 2014, 62, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sinsinbar, G.; Choudhary, M.; Kumar, V.; Pasricha, R.; Verma, H.N.; Singh, S.P.; Arora, K. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens. Actuators B Chem. 2013, 185, 675–684. [Google Scholar] [CrossRef]
- Zhu, L.; Luo, L.; Wang, Z. DNA electrochemical biosensor based on thionine-graphene nanocomposite. Biosens. Bioelectron. 2012, 35, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Hernández, R.; Vallés, C.; Benito, A.M.; Maser, W.K.; Xavier Rius, F.; Riu, J. Graphene-based potentiometric biosensor for the immediate detection of living bacteria. Biosens. Bioelectron. 2014, 54, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, H.; Huang, M.; Zhou, J. One-step hydrothermal synthesis of nitrogen doping graphene based cobalt oxide and its supercapacitive properties. J. Alloys Compd. 2017, 705, 801–805. [Google Scholar] [CrossRef]
- Zhang, M.; Liao, C.; Mak, C.H.; You, P.; Mak, C.L.; Yan, F. Highly sensitive glucose sensors based on enzyme-modified whole-graphene solution-gated transistors. Sci. Rep. 2015, 5, 8311. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.D.; Kim, S.K.; Chang, H.; Roh, K.M.; Choi, J.W.; Huang, J. A glucose biosensor based on TiO2-Graphene composite. Biosens. Bioelectron. 2012, 38, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, D.; Zeng, C.; Miao, Z.; Dai, L. Biocompatible graphene oxide-based glucose biosensors. Langmuir 2010, 26, 6158–6160. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, Q.; Li, F.; Niu, L. Glucose oxidase and graphene bionanocomposite bridged by ionic liquid unit for glucose biosensing application. Sens. Actuators B Chem. 2012, 161, 728–733. [Google Scholar] [CrossRef]
- Wu, H.; Wang, J.; Kang, X.; Wang, C.; Wang, D.; Liu, J.; Aksay, I.A.; Lin, Y. Glucose biosensor based on immobilization of glucose oxidase in platinum nanoparticles/graphene/chitosan nanocomposite film. Talanta 2009, 80, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shao, Y.; Matson, D.W.; Li, J.; Lin, Y. Nitrogen-doped graphene and its application in electrochemical biosensing. ACS Nano 2010, 4, 1790–1798. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Shao, Q.; Hu, Y.; Jin, J.; Yin, Y.; Zhang, H.; Cai, C. Direct electrochemistry of glucose oxidase assembled on graphene and application to glucose detection. Electrochim. Acta 2010, 55, 8606–8614. [Google Scholar] [CrossRef]
- Fu, C.; Yang, W.; Chen, X.; Evans, D.G. Direct electrochemistry of glucose oxidase on a graphite nanosheet-Nafion composite film modified electrode. Electrochem. Commun. 2009, 11, 997–1000. [Google Scholar] [CrossRef]
- Shan, C.; Yang, H.; Han, D.; Zhang, Q.; Ivaska, A.; Niu, L. Graphene/AuNPs/chitosan nanocomposites film for glucose biosensing. Biosens. Bioelectron. 2010, 25, 1070–1074. [Google Scholar] [CrossRef]
- Ye, Y.; Ding, S.; Ye, Y.; Xu, H.; Cao, X.; Liu, S.; Sun, H. Enzyme-based sensing of glucose using a glassy carbon electrode modified with a one-pot synthesized nanocomposite consisting of chitosan, reduced graphene oxide and gold nanoparticles. Microchim. Acta 2015, 182, 1783–1789. [Google Scholar] [CrossRef]
- Xu, D.S.; Chang, J.P.; Li, J.; Yang, R.; Li, D.; Yip, S. Dislocation slip or deformation twinning: Confining pressure makes a difference. Mater. Sci. Eng. A 2004, 387–389, 840–844. [Google Scholar] [CrossRef]
- Liu, H.P.; Zhan, G.Y.; Dong, Q.Z.; Lv, Y.A.; Wang, J.F.; Tao, C.-A.; Hu, Z.H. Glucose biosensor based on Pt nanoparticles/graphene-chitosan bionanocomposites. Appl. Mech. Mater. 2013, 328, 695–699. [Google Scholar] [CrossRef]
- Wang, G.; He, X.; Wang, L.; Gu, A.; Huang, Y.; Fang, B.; Geng, B.; Zhang, X. Non-enzymatic electrochemical sensing of glucose. Microchim. Acta 2013, 180, 161–186. [Google Scholar] [CrossRef]
- Ma, R.; Wang, B.; Liu, Y.; Li, J.; Zhao, Q.; Wang, G.; Jia, W.; Wang, H. Direct electrochemistry of glucose oxidase on the hydroxyapatite/Nafion composite film modified electrode and its application for glucose biosensing. Sci. China Ser. B Chem. 2009, 52, 2013–2019. [Google Scholar] [CrossRef]
- Demming, A.; Hierold, C. Sensing at the nanoscale. Nanotechnology 2013, 24, 440201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, P.; Huang, Y.; Wang, T.; Ma, J. Nanomaterials for electrochemical non-enzymatic glucose biosensors. RSC Adv. 2013, 3, 3487–3502. [Google Scholar] [CrossRef]
- Alexander, S.; Baraneedharan, P.; Balasubrahmanyan, S.; Ramaprabhu, S. Highly sensitive and selective non enzymatic electrochemical glucose sensors based on Graphene Oxide-Molecular Imprinted Polymer. Mater. Sci. Eng. C 2017, 78, 124–129. [Google Scholar] [CrossRef]
- Moozarm Nia, P.; Meng, W.P.; Lorestani, F.; Mahmoudian, M.R.; Alias, Y. Electrodeposition of copper oxide/polypyrrole/reduced graphene oxide as a nonenzymatic glucose biosensor. Sens. Actuators B Chem. 2015, 209, 100–108. [Google Scholar] [CrossRef]
- Hughes, Z.E.; Walsh, T.R. What makes a good graphene-binding peptide? Adsorption of amino acids and peptides at aqueous graphene interfaces. J. Mater. Chem. B 2015, 3, 3211–3221. [Google Scholar] [CrossRef] [Green Version]
- Wohlfahrt, G.; Witt, S.; Hendle, J.; Schomburg, D.; Kalisz, H.M.; Hecht, H.J. 1.8 and 1.9 Å resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 969–977. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Morris, G.; Huey, R. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-based materials for biosensors: A review. Sensors (Switzerland) 2017, 17, 2161. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, M.; Ptitsyn, A.; McLamore, E.S.; Claussen, J.C. Nanomaterial-mediated biosensors for monitoring glucose. J. Diabetes Sci. Technol. 2014, 8, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Wohlfahrt, G.; Knäblein, J.; Schomburg, D. Aspects of the mechanism of catalysis of glucose oxidase: A docking, molecular mechanics and quantum chemical study. J. Comput. Aided. Mol. Des. 1998, 12, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.K.; Fillat, M.F.; Gémez-Moreno, C.; Tollin, G. Electrostatic and hydrophobic interactions during complex formation and electron transfer in the ferredoxin/ferredoxin:NADP+reductase system from Anabaena. J. Am. Chem. Soc. 1996, 118, 5526–5531. [Google Scholar] [CrossRef]
- Viswanathan, S.; Narayanan, T.N.; Aran, K.; Fink, K.D.; Paredes, J.; Ajayan, P.M.; Filipek, S.; Miszta, P.; Tekin, H.C.; Inci, F.; et al. Graphene-protein field effect biosensors: Glucose sensing. Mater. Today 2015, 18, 513–522. [Google Scholar] [CrossRef]
- Sakr, M.A.; Serry, M. Non-enzymatic graphene-based biosensors for continous glucose monitoring. In Proceedings of the 2015 IEEE SENSORS, Busan, Korea, 1–4 November 2015. [Google Scholar]
- Chaiyo, S.; Mehmeti, E.; Siangproh, W.; Hoang, T.L.; Nguyen, H.P.; Chailapakul, O.; Kalcher, K. Non-enzymatic electrochemical detection of glucose with a disposable paper-based sensor using a cobalt phthalocyanine–ionic liquid–graphene composite. Biosens. Bioelectron. 2018, 102, 113–120. [Google Scholar] [CrossRef]
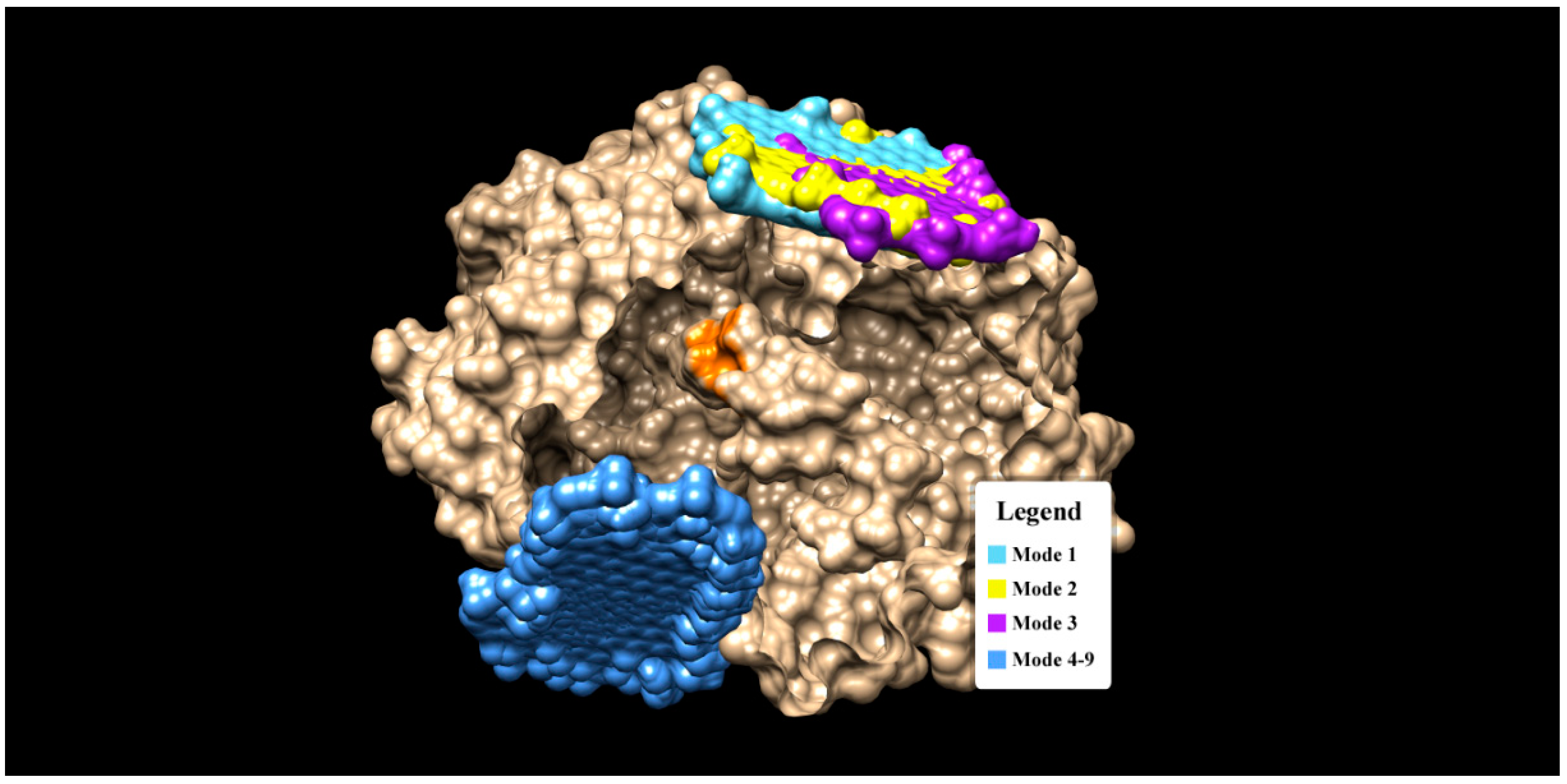



| Mode | ΔG (kCal/mol) | Residue Involved in H-Bond | H-Bond Distance | Residue Involved in Hydrophobic Interaction |
|---|---|---|---|---|
| 1 | −11.6 | - | - | ARG 37, GLU 40, ASN 41, ASP 134, ASN 135, ALA 138, TYR 139, LEU 141, GLN 142, ARG 145, SER 163, GLY 166, VAL 167, ASN 168, GLY 169, ARG 239, ASP 573, LEU 576, GLU 577 |
| 2 | −11.0 | - | - | ARG 37, GLU 40, ASN 41, PRO 42, ASP 134, ASN 135, ALA 138, LEU 141, GLN 142, ALA 162, SER 163, CYS 164, HIS 165, GLY 166, VAL 167, ASN 168, GLY 169, ARG 239, ASP 573, LEU 576, GLU 577, TYR 579, ALA 580 |
| 3 | −10.5 | GLY 169 ASN 168 | 3.149 2.859 | ARG 37, GLU 40, ASN 41, PRO 42, ALA 138, TYR 139, LEU 141, GLN 142, ALA 143, GLU 144, ARG 145, GLY 166, VAL 167, THR 170, ASP 573, GLU 577, ALA 580, SER 581 |
| 4 | −10.1 | GLU 374 GLU 378 | 2.770 3.143 | MET 305, SER 307, ILE 308, ASP 319, LEU 320, PRO 321, LEU 324, VAL 381, ALA 382, GLY 384, PHE 386, HIS 387, ASN 388, THR 389, THR 390, LYS 526, GLU 527 |
| 5 | −9.80 | ALA 382 ALA 382 | 2.832 2.782 | MET 305, SER 307, ASP 319, LEU 320, PRO 321, LEU 324, GLU 378, VAL 381, ARG 383, GLY 384, PHE 386, HIS 387, ASN 388, THR 389, THR 390, LYS 526, GLU 527 |
| 6 | −9.70 | - | - | MET 305, LYS 306, SER 307, ASP 319, GLU 374, GLU 378, VAL 381, ALA 382, HIS 387, ASN 388, THR 389, THR 390, LYS 526, GLU 527 |
| 7 | −9.50 | - | MET 305, LYS 306, SER 307, ILE 308, ASP 319, LEU 320, GLU 374, GLU 378, VAL 381, ALA 382, ARG 383, HIS 387, ASN 388, THR 389, THR 390, LYS 526, GLU 527 | |
| 8 | −9.40 | - | - | MET 305, SER 307, ASP 319, GLU 378, VAL 381, ALA 382, HIS 387, ASN 388, THR 389, THR 390, LYS 526, GLU 527 |
| 9 | −9.30 | SER 307 ASP 319 | 2.959 2.703 | MET 305, LYS 306, ILE 308, LEU 320, GLU 378, VAL 381, ALA 382, HIS 387, ASN 388, THR 389, THR 390, LYS 526 |
| Mode | ΔG (kCal/mol) | RMSD l.b (Å) * | RMSD u.b (Å) * |
|---|---|---|---|
| 1 | −4.20 | 0.000 | 0.000 |
| 2 | −4.10 | 1.234 | 3.649 |
| 3 | −4.10 | 1.059 | 4.093 |
| 4 | −4.00 | 2.818 | 5.098 |
| 5 | −4.00 | 1.272 | 2.009 |
| 6 | −4.00 | 2.362 | 3.661 |
| 7 | −4.00 | 1.254 | 2.314 |
| 8 | −3.90 | 2.211 | 4.563 |
| 9 | −3.90 | 1.502 | 3.750 |
| 10 | −3.90 | 1.535 | 2.754 |
| 11 | −3.90 | 3.528 | 5.889 |
| 12 | −3.80 | 2.321 | 4.232 |
| 13 | −3.80 | 2.426 | 4.128 |
| 14 | −3.80 | 3.587 | 4.876 |
| 15 | −3.80 | 2.106 | 4.095 |
| 16 | −3.80 | 1.113 | 3.851 |
| 17 | −3.80 | 2.604 | 3.853 |
| 18 | −3.80 | 1.680 | 2.305 |
| 19 | −3.80 | 1.789 | 3.733 |
| 20 | −3.70 | 5.241 | 7.421 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumaryada, T.; Sandy Gunawan, M.; Perdana, S.; Arjo, S.; Maddu, A. A Molecular Interaction Analysis Reveals the Possible Roles of Graphene Oxide in a Glucose Biosensor. Biosensors 2019, 9, 18. https://doi.org/10.3390/bios9010018
Sumaryada T, Sandy Gunawan M, Perdana S, Arjo S, Maddu A. A Molecular Interaction Analysis Reveals the Possible Roles of Graphene Oxide in a Glucose Biosensor. Biosensors. 2019; 9(1):18. https://doi.org/10.3390/bios9010018
Chicago/Turabian StyleSumaryada, Tony, Muhammad Sandy Gunawan, Salahuddin Perdana, Sugianto Arjo, and Akhiruddin Maddu. 2019. "A Molecular Interaction Analysis Reveals the Possible Roles of Graphene Oxide in a Glucose Biosensor" Biosensors 9, no. 1: 18. https://doi.org/10.3390/bios9010018
APA StyleSumaryada, T., Sandy Gunawan, M., Perdana, S., Arjo, S., & Maddu, A. (2019). A Molecular Interaction Analysis Reveals the Possible Roles of Graphene Oxide in a Glucose Biosensor. Biosensors, 9(1), 18. https://doi.org/10.3390/bios9010018





