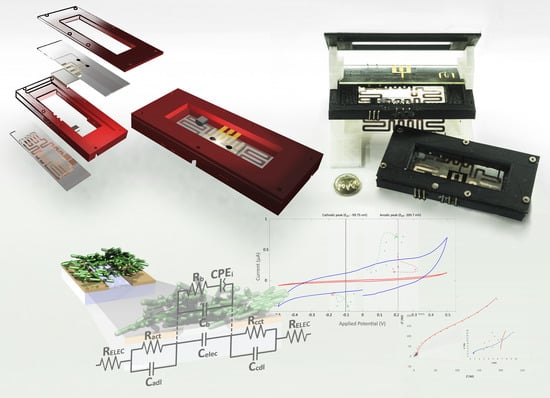
Fully Automated Microsystem for Unmediated Electrochemical Characterization, Visualization and Monitoring of Bacteria on Solid Media; E. coli K-12: A Case Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Equipment
2.2. Microorganism
2.3. Design of the Microsystem: Microelectrodes and Confinement Microstructure
2.3.1. Microelectrodes
2.3.2. Confinement Microstructures
2.4. Test Cell Design
2.5. Automated Monitoring System
2.6. Bacterial Microculture
2.7. Electrochemical Procedures
2.7.1. Cyclic Voltammetry
2.7.2. Electrochemical Impedance Spectroscopy
2.8. Data Processing
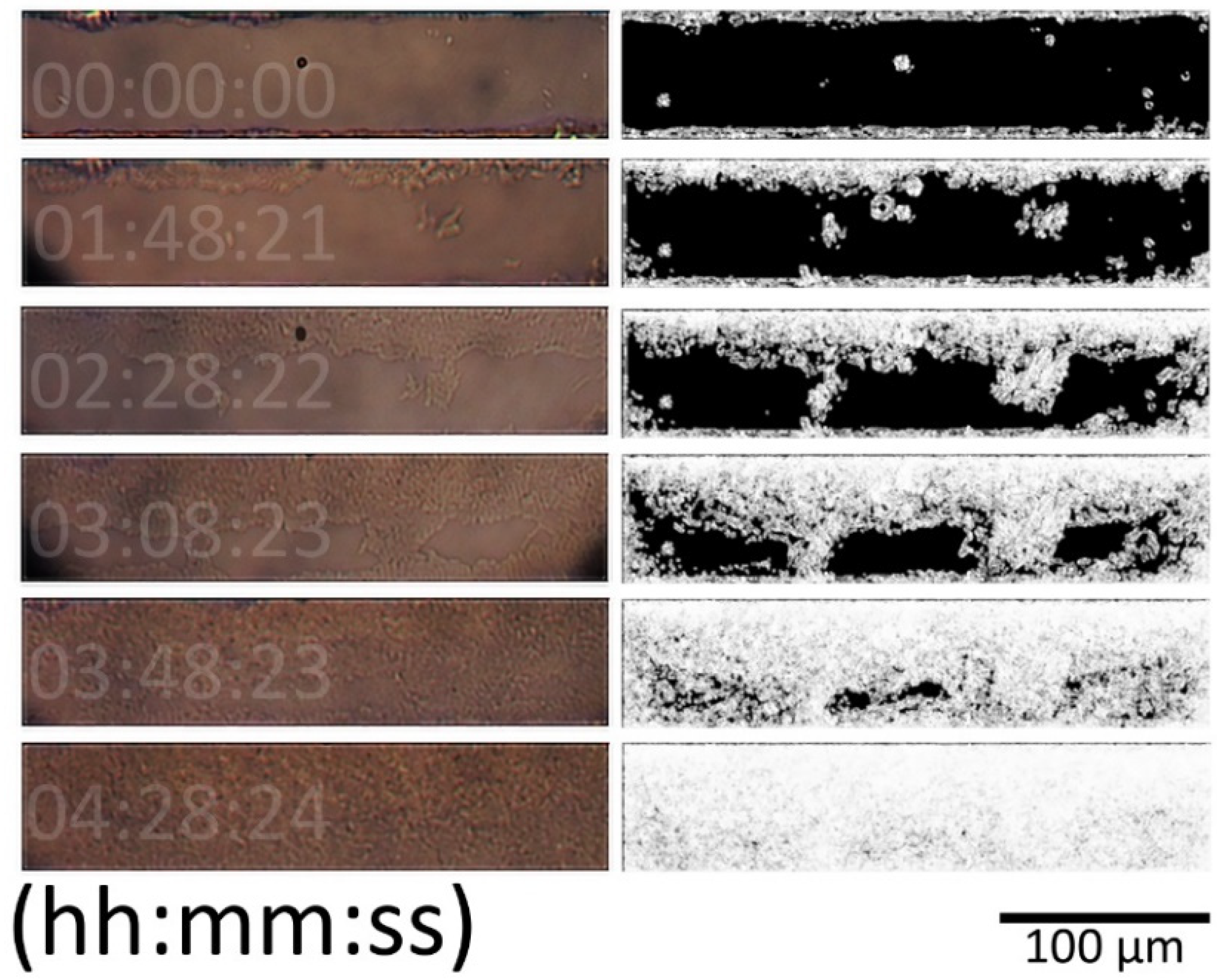
2.8.1. Image Analysis
2.8.2. Electrochemical Analysis
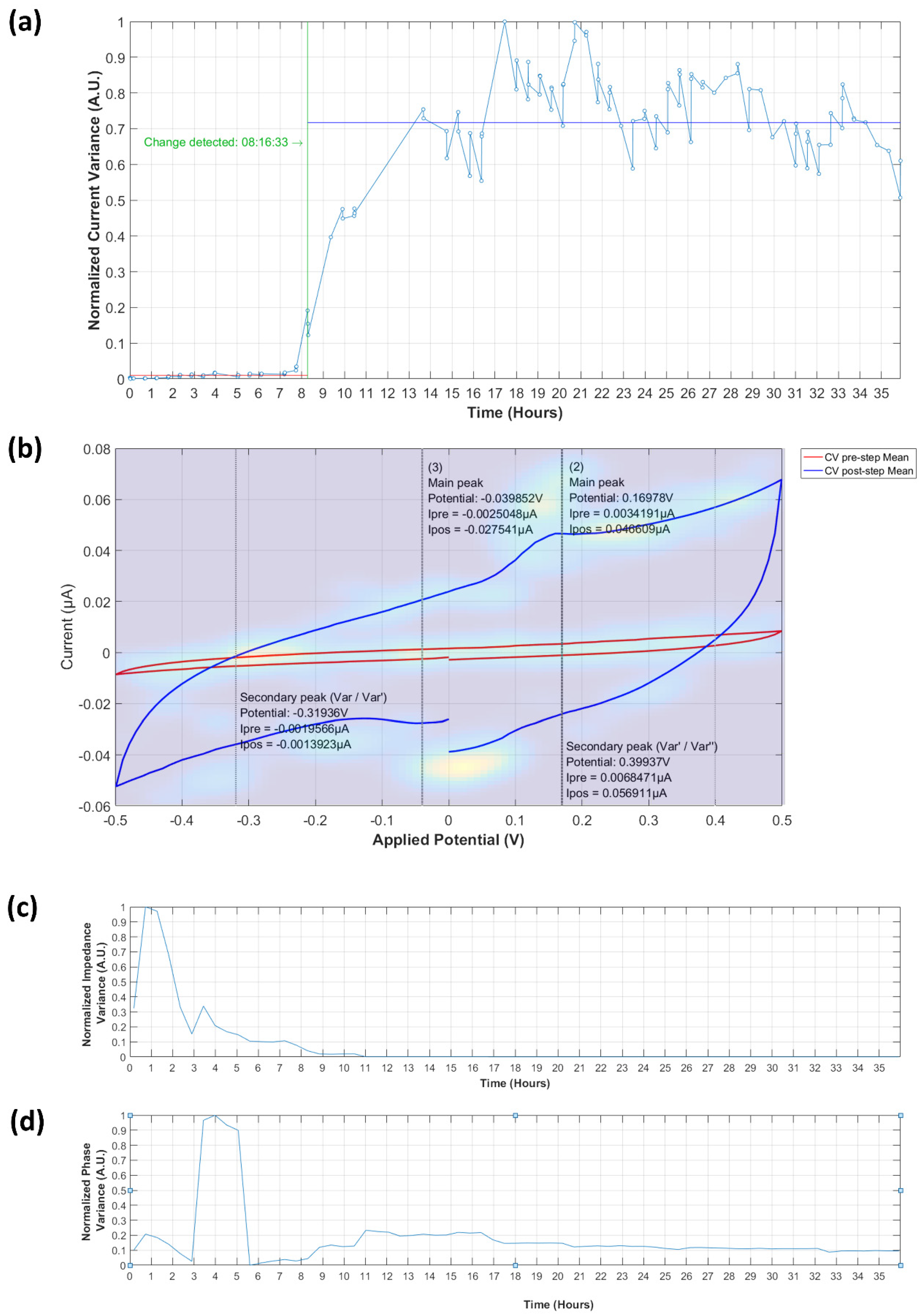
Cyclic Voltammetry Current Variance
Identification of Cyclic Voltammetry Current Peaks
Electrochemical Impedance Spectrometry Frequency and Phase Variance
Electrochemical Impedance Spectrometry Circuit Fitting
3. Results
3.1. Characterization of the Microsystems: Microelectrodes and Confinement Microstructure
3.2. Image Analysis
3.3. Cyclic Voltammetry
3.3.1. Analysis of Normalized Current Variance
3.3.2. Current Peaks
3.4. Electrochemical Impedance Spectroscopy
3.4.1. Normalized Impedance and Phase Variance
3.4.2. Equivalent Circuit Model Fitting
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A







References
- Gupta, V.K.; Zeilinger-Migsich, S.; Ferreira Filho, E.X.; del Durán Domínguez de Bazúa, M.C.; Purchase, D. Microbial Applications: Recent Advancements and Future Developments; Walter de Gruyter: Berlin, Germany, 2016; ISBN 3110412780. [Google Scholar]
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J.; Bhriain, N.N.; Dorman, M.J. The evolution of gene regulatory mechanisms in bacteria. In Molecular Mechanisms of Microbial Evolution. Grand Challenges in Biology and Biotechnology; Rampelotto, P.H., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 125–152. ISBN 9783319690780. [Google Scholar]
- Richardson, D.J. Bacterial respiration: A flexible process for a changing environment. Microbiology 2000, 146, 551–571. [Google Scholar] [CrossRef] [PubMed]
- Schoepp-Cothenet, B.; van Lis, R.; Atteia, A.; Baymann, F.; Capowiez, L.; Ducluzeau, A.-L.; Duval, S.; ten Brink, F.; Russell, M.J.; Nitschke, W. On the universal core of bioenergetics. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Gunsalus, R.P.; Cecchini, G.; Schröder, I. Bacterial respiration. In Methods for General and Molecular Microbiology, Third Edition; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G., Schmidt, T.M., Snyder, L.R., Eds.; American Society of Microbiology: Washington, DC, USA, 2014; pp. 539–557. [Google Scholar]
- Haddock, B.A.; Jones, C.W. Bacterial respiration. Bacteriol. Rev. 1977, 41, 47–99. [Google Scholar] [PubMed]
- Marreiros, B.C.; Calisto, F.; Castro, P.J.; Duarte, A.M.; Sena, F.V.; Silva, A.F.; Sousa, F.M.; Teixeira, M.; Refojo, P.N.; Pereira, M.M. Exploring membrane respiratory chains. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1039–1067. [Google Scholar] [CrossRef]
- Price, C.E.; Driessen, A.J.M. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 748–766. [Google Scholar] [CrossRef]
- Babauta, J.; Renslow, R.; Lewandowski, Z.; Beyenal, H. Electrochemically active biofilms: Facts and fiction. A review. Biofouling 2012, 28, 789–812. [Google Scholar] [CrossRef]
- Koch, C.; Harnisch, F. Is there a specific ecological niche for electroactive microorganisms? ChemElectroChem 2016, 3, 1282–1295. [Google Scholar] [CrossRef]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef]
- Lovley, D.R. Electromicrobiology. Annu. Rev. Microbiol. 2012, 66, 391–409. [Google Scholar] [CrossRef]
- Patil, S.A.; Hägerhäll, C.; Gorton, L. Electron transfer mechanisms between microorganisms and electrodes in bioelectrochemical systems. In Advances in Chemical Bioanalysis; Matysk, F.M., Ed.; Springer International Publishing: Cham, Switzerland, 2012; Volume 1, pp. 71–129. [Google Scholar]
- Lovley, D.R. Powering microbes with electricity: Direct electron transfer from electrodes to microbes. Environ. Microbiol. Rep. 2011, 3, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Schröder, U. Anodic electron transfer mechanisms in microbial fuel cells and their energy efficiency. Phys. Chem. Chem. Phys. 2007, 9, 2619–2629. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.; Aulenta, F.; Villano, M.; Angenent, L.T. Cathodes as electron donors for microbial metabolism: Which extracellular electron transfer mechanisms are involved? Bioresour. Technol. 2011, 102, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Green, J.; Paget, M.S. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Bajracharya, S.; Sharma, M.; Mohanakrishna, G.; Dominguez Benneton, X.; Strik, D.P.B.T.B.; Sarma, P.M.; Pant, D. An overview on emerging bioelectrochemical systems (BESs): Technology for sustainable electricity, waste remediation, resource recovery, chemical production and beyond. Renew. Energy 2016, 98, 153–170. [Google Scholar] [CrossRef]
- Agler-Rosenbaum, M.; Schröder, U.; Harnisch, F. Mikroben unter Strom. Biol. Unserer Zeit 2013, 43, 96–103. [Google Scholar] [CrossRef]
- Schröder, U.; Greiner, A.; Rosenbaum, M.A.; Harnisch, F. Wie Mikroorganismen und Elektroden interagieren. Nachrichten aus der Chemie 2016, 64, 732–737. [Google Scholar] [CrossRef]
- Ikeda, T.; Kano, K. An electrochemical approach to the studies of biological redox reactions and their applications to biosensors, bioreactors, and biofuel cells. J. Biosci. Bioeng. 2001, 92, 9–18. [Google Scholar] [CrossRef]
- Rabaey, K.; Rodríguez, J.; Blackall, L.L.; Keller, J.; Gross, P.; Batstone, D.; Verstraete, W.; Nealson, K.H. Microbial ecology meets electrochemistry: Electricity-driven and driving communities. ISME J. 2007, 1, 9–18. [Google Scholar] [CrossRef]
- Aulenta, F.; Puig, S.; Harnisch, F. Microbial electrochemical technologies: Maturing but not mature. Microb. Biotechnol. 2018, 11, 18–19. [Google Scholar] [CrossRef]
- Holtmann, D.; Harnisch, F. Electrification of biotechnology: Quo Vadis? In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2018; Volume 123, pp. 395–411. ISBN 0724-6145. [Google Scholar]
- Schröder, U.; Harnisch, F. Life electric—Nature as a blueprint for the development of microbial electrochemical technologies. Joule 2017, 1, 244–252. [Google Scholar] [CrossRef]
- Koch, C.; Harnisch, F. What is the essence of microbial electroactivity? Front. Microbiol. 2016, 7, 1–5. [Google Scholar] [CrossRef]
- Schröder, U.; Harnisch, F.; Angenent, L.T. Microbial electrochemistry and technology: Terminology and classification. Energy Environ. Sci. 2015, 8, 513–519. [Google Scholar] [CrossRef]
- Kracke, F.; Krömer, J.O. Identifying target processes for microbial electrosynthesis by elementary mode analysis. BMC Bioinformatics 2014, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, M.; Guo, J.; Sun, G. Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochem. 2012, 47, 1707–1714. [Google Scholar] [CrossRef]
- Patil, S.A.; Gildemyn, S.; Pant, D.; Zengler, K.; Logan, B.E.; Rabaey, K. A logical data representation framework for electricity-driven bioproduction processes. Biotechnol. Adv. 2015, 33, 736–744. [Google Scholar] [CrossRef]
- Harnisch, F.; Rosa, L.F.M.; Kracke, F.; Virdis, B.; Krömer, J.O. Electrifying white biotechnology: Engineering and economic potential of electricity-driven bio-production. ChemSusChem 2015, 8, 758–766. [Google Scholar] [CrossRef]
- Harnisch, F.; Rabaey, K. The diversity of techniques to study electrochemically active biofilms highlights the need for standardization. ChemSusChem 2012, 5, 1027–1038. [Google Scholar] [CrossRef]
- Sutherland, I.W. The biofilm matrix—An immobilized but dynamic microbial environment. Trends Microbiol. 2001, 9, 222–227. [Google Scholar] [CrossRef]
- Capdeville, B.; Nguyen, K.M.; Rols, J.L. Biofilms—Science and Technology; Melo, L.F., Bott, T.R., Fletcher, M., Capdeville, B., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 1992; Volume 223, ISBN 978-94-010-4805-7. [Google Scholar]
- Xie, X.-H.; Li, E.L.; Tang, Z.K. Real-time monitoring of induced adaptation of redox active Escherichia coli biofilm by EQCM-controlled extracellular redox environment. Electrochem. Commun. 2010, 12, 600–602. [Google Scholar] [CrossRef]
- Harnisch, F.; Freguia, S. A basic tutorial on Cyclic Voltammetry for the investigation of electroactive microbial biofilms. Chem. An Asian J. 2012, 7, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yoav, H.; Biran, A.; Sternheim, M.; Belkin, S.; Freeman, A.; Shacham-Diamand, Y. Functional modeling of electrochemical whole-cell biosensors. Sens. Actuators B Chem. 2013, 181, 479–485. [Google Scholar] [CrossRef]
- Tian, M.; Kanavillil, N.; Davey, L.; Leung, K.T.; Schraft, H.; Chen, A. Direct growth of biofilms on an electrode surface and its application in electrochemical biosensoring. J. Electroanal. Chem. 2007, 611, 133–139. [Google Scholar] [CrossRef]
- Devadas, D.; Young, E.W.K. Microfluidics for cell culture. In Microfluidic Methods for Molecular Biology; Lu, C., Verbridge, S.S., Eds.; Springer International Publishing: Blacksburg, VA, USA, 2016; pp. 323–347. [Google Scholar]
- Yi, C.; Li, C.-W.; Ji, S.; Yang, M. Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar] [CrossRef]
- Weibel, D.B.; DiLuzio, W.R.; Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007, 5, 209–218. [Google Scholar] [CrossRef]
- Wu, F.; Dekker, C. Nanofabricated structures and microfluidic devices for bacteria: From techniques to biology. Chem. Soc. Rev. 2016, 45, 268–280. [Google Scholar] [CrossRef]
- Hol, F.J.H.; Dekker, C. Zooming in to see the bigger picture: Microfluidic and nanofabrication tools to study bacteria. Science 2014, 346, 1251821. [Google Scholar] [CrossRef]
- Rusconi, R.; Garren, M.; Stocker, R. Microfluidics Expanding the Frontiers of Microbial Ecology. Annu. Rev. Biophys. 2014, 43, 65–91. [Google Scholar] [CrossRef] [Green Version]
- Fraiwan, A.; Choi, S. A biomicrosystem for simultaneous optical and electrochemical monitoring of electroactive microbial biofilm. In Proceedings of the 2015 IEEE Sensors, Busan, Korea, 1–4 November 2015; pp. 2–5. [Google Scholar]
- Pham, H.T.; Boon, N.; Aelterman, P.; Clauwaert, P.; De Schamphelaire, L.; van Oostveldt, P.; Verbeken, K.; Rabaey, K.; Verstraete, W. High shear enrichment improves the performance of the anodophilic microbial consortium in a microbial fuel cell. Microb. Biotechnol. 2008, 1, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, C.A.; Gaviria, L.N.; Segura, S.M.; Osma, J.F. Concept design for a novel confined-bacterial-based biosensor for water quality control. In Proceedings of the 2013 Pan American Health Care Exchanges (PAHCE), Medellin, Colombia, 4 May 2013; pp. 1–3. [Google Scholar]
- Hernandez, C.A.; Lopez-Barbosa, N.; Segura, C.C.; Osma, J.F. High definition method for imaging bacteria in microconfined environments on solid media. In Proceedings of the International Conference on Bioinformatics and Biomedical Engineering; Rojas, I., Ortuño, F., Eds.; Springer: Cham, Switzerland, 2017; Volume 10209 LNCS, pp. 726–736. [Google Scholar]
- Grant, M.A.A.; Wacław, B.; Allen, R.J.; Cicuta, P. The role of mechanical forces in the planar-to-bulk transition in growing Escherichia coli microcolonies. J. R. Soc. Interface 2014, 11, 20140400. [Google Scholar] [CrossRef]
- Volfson, D.; Cookson, S.; Hasty, J.; Tsimring, L.S. Biomechanical ordering of dense cell populations. Proc. Natl. Acad. Sci. USA 2008, 105, 15346–15351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, B.J. Derivations and genotypes of some mutant derivatives of Escherichia coli K12. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology; Neidhardt, F.C., Curtiss, R., Ingraham, J.L., Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W., Riley, M., Schaechter, M., et al., Eds.; ASM Press: Washington, DC, USA, 1996; pp. 2460–2488. [Google Scholar]
- Kracke, F. Understanding Extracellular Electron Transport of Industrial Microorganisms and Optimization for Production Application. Ph.D. Thesis, Advanced Water Management Centre, The University of Queensland, Brisbane, Australia, 2016. [Google Scholar]
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta Bioenerg. 1997, 1320, 217–234. [Google Scholar] [CrossRef]
- Xie, X.H.; Li, E.L.; Tang, Z.K. Sudden emergence of redox active Escherichia coli phenotype: Cyclic voltammetric evidence of the overlapping pathways. Int. J. Electrochem. Sci. 2010, 5, 1070–1081. [Google Scholar]
- Tran, Q.H.; Unden, G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 1998, 251, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Chalenko, Y.; Shumyantseva, V.; Ermolaeva, S.; Archakov, A. Electrochemistry of Escherichia coli JM109: Direct electron transfer and antibiotic resistance. Biosens. Bioelectron. 2012, 32, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Chaudhuri, B.B. A survey of Hough Transform. Pattern Recognit. 2015, 48, 993–1010. [Google Scholar] [CrossRef]
- Doyle, W. Operations Useful for Similarity-Invariant Pattern Recognition. J. ACM 1962, 9, 259–267. [Google Scholar] [CrossRef]
- Gagne, R.R.; Koval, C.A.; Lisensky, G.C. Ferrocene as an internal standard for electrochemical measurements. Inorg. Chem. 1980, 19, 2854–2855. [Google Scholar] [CrossRef]
- Trasatti, S. The absolute electrode potential: An explanatory note (Recommendations 1986). Pure Appl. Chem. 1986, 58, 955–966. [Google Scholar] [CrossRef]
- O’Reilly, J.E. Oxidation-reduction potential of the ferro-ferricyanide system in buffer solutions. Biochim. Biophys. Acta Bioenerg. 1973, 292, 509–515. [Google Scholar] [CrossRef]
- Kolthoff, I.M.; Tomsicek, W.J. The oxidation potential of the system potassium ferrocyanide–potassium ferricyanide at various ionic strengths. J. Phys. Chem. 1935, 39, 945–954. [Google Scholar] [CrossRef]
- Nordstrom, D.K. Thermochemical redox equilibria of ZoBell’s solution. Geochim. Cosmochim. Acta 1977, 41, 1835–1841. [Google Scholar] [CrossRef]
- Buerger, S.; Spoering, A.; Gavrish, E.; Leslin, C.; Ling, L.; Epstein, S.S. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 2012, 78, 3221–3228. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.S. Microbial awakenings. Nature 2009, 457, 1083. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Berbel, X.; Vigués, N.; Jenkins, A.T.A.; Mas, J.; Muñoz, F.J. Impedimetric approach for quantifying low bacteria concentrations based on the changes produced in the electrode–solution interface during the pre-attachment stage. Biosens. Bioelectron. 2008, 23, 1540–1546. [Google Scholar] [CrossRef]
- Varshney, M.; Li, Y. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle–antibody conjugates for detection of Escherichia coli O157:H7 in food samples. Biosens. Bioelectron. 2007, 22, 2408–2414. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y. AFM and impedance spectroscopy characterization of the immobilization of antibodies on indium–tin oxide electrode through self-assembled monolayer of epoxysilane and their capture of Escherichia coli O157:H7. Biosens. Bioelectron. 2005, 20, 1407–1416. [Google Scholar] [CrossRef]
- Muñoz-Berbel, X.; Vigués, N.; Mas, J.; Jenkins, A.T.A.; Muñoz, F.J. Impedimetric characterization of the changes produced in the electrode–solution interface by bacterial attachment. Electrochem. Commun. 2007, 9, 2654–2660. [Google Scholar] [CrossRef]
- Kracke, F.; Vassilev, I.; Krömer, J.O. Microbial electron transport and energy conservation – the foundation for optimizing bioelectrochemical systems. Front. Microbiol. 2015, 6, 1–18. [Google Scholar] [CrossRef]
- Liu, C.-G.; Xue, C.; Lin, Y.-H.; Bai, F.-W. Redox potential control and applications in microaerobic and anaerobic fermentations. Biotechnol. Adv. 2013, 31, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Schon, E.A. Bioenergetics through thick and thin. Science 2018, 362, 1114–1115. [Google Scholar] [CrossRef] [PubMed]















| Electric Component | Value before Colonization Event | Value after Colonization Event | Unit | |
|---|---|---|---|---|
| Cbelec | 0.919 | 6.72 | nF | |
| Relec | 15.96 | 3.904 | KΩ | |
| Rb | 5.71 | 0.107 | MΩ | |
| CPEi | Qi | 31.37848 × 10−9 | 69.78041 × 10−9 | T |
| ni | 0.77428 | 0.865045 | φ | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez, C.A.; Beni, V.; Osma, J.F. Fully Automated Microsystem for Unmediated Electrochemical Characterization, Visualization and Monitoring of Bacteria on Solid Media; E. coli K-12: A Case Study. Biosensors 2019, 9, 131. https://doi.org/10.3390/bios9040131
Hernandez CA, Beni V, Osma JF. Fully Automated Microsystem for Unmediated Electrochemical Characterization, Visualization and Monitoring of Bacteria on Solid Media; E. coli K-12: A Case Study. Biosensors. 2019; 9(4):131. https://doi.org/10.3390/bios9040131
Chicago/Turabian StyleHernandez, Cesar A., Valerio Beni, and Johann F. Osma. 2019. "Fully Automated Microsystem for Unmediated Electrochemical Characterization, Visualization and Monitoring of Bacteria on Solid Media; E. coli K-12: A Case Study" Biosensors 9, no. 4: 131. https://doi.org/10.3390/bios9040131
APA StyleHernandez, C. A., Beni, V., & Osma, J. F. (2019). Fully Automated Microsystem for Unmediated Electrochemical Characterization, Visualization and Monitoring of Bacteria on Solid Media; E. coli K-12: A Case Study. Biosensors, 9(4), 131. https://doi.org/10.3390/bios9040131