Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine
Abstract
:1. Introduction
2. Results
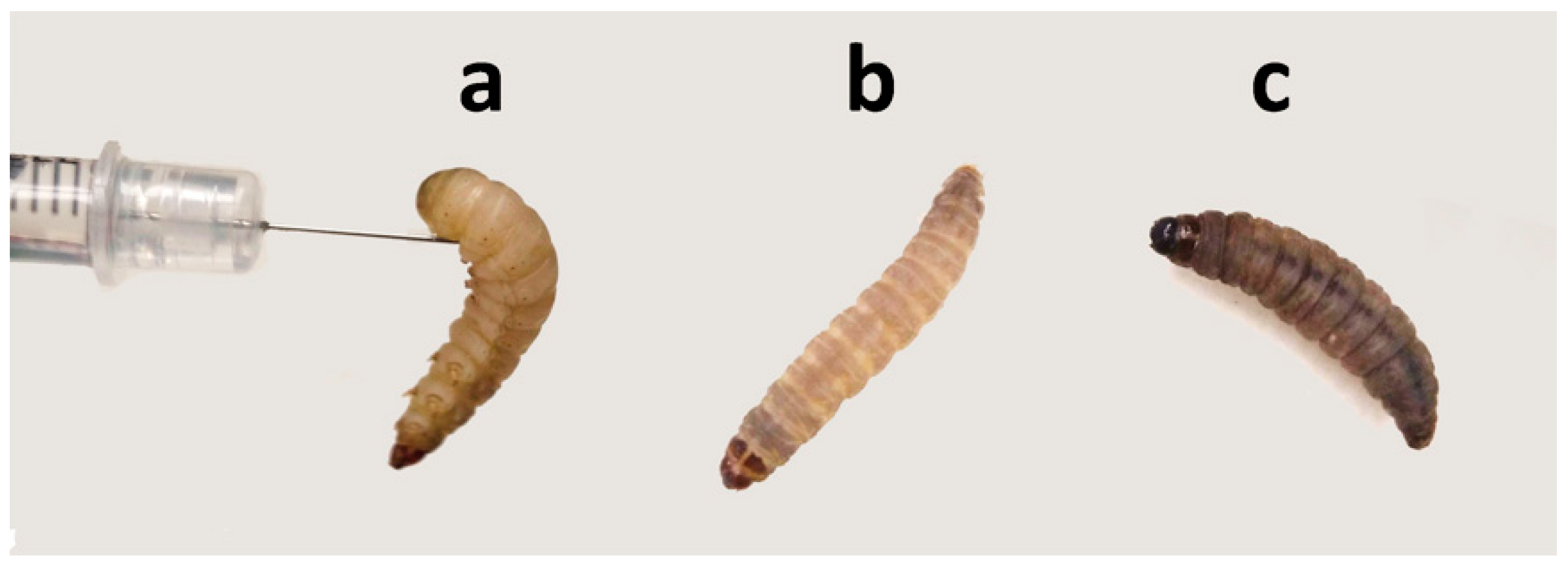
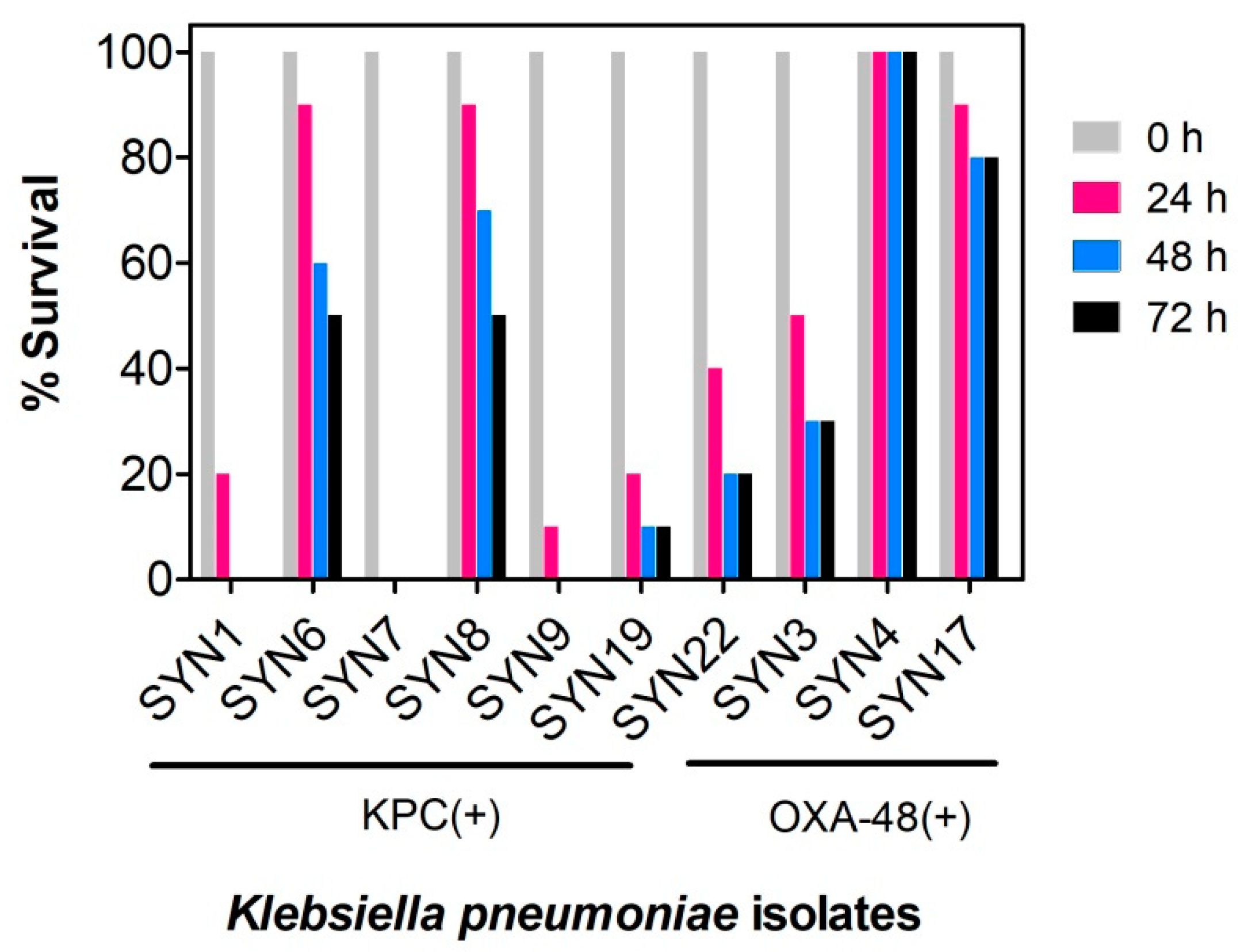
2.1. Evaluation of K. pneumoniae Virulance in G. mellonella Infection Model
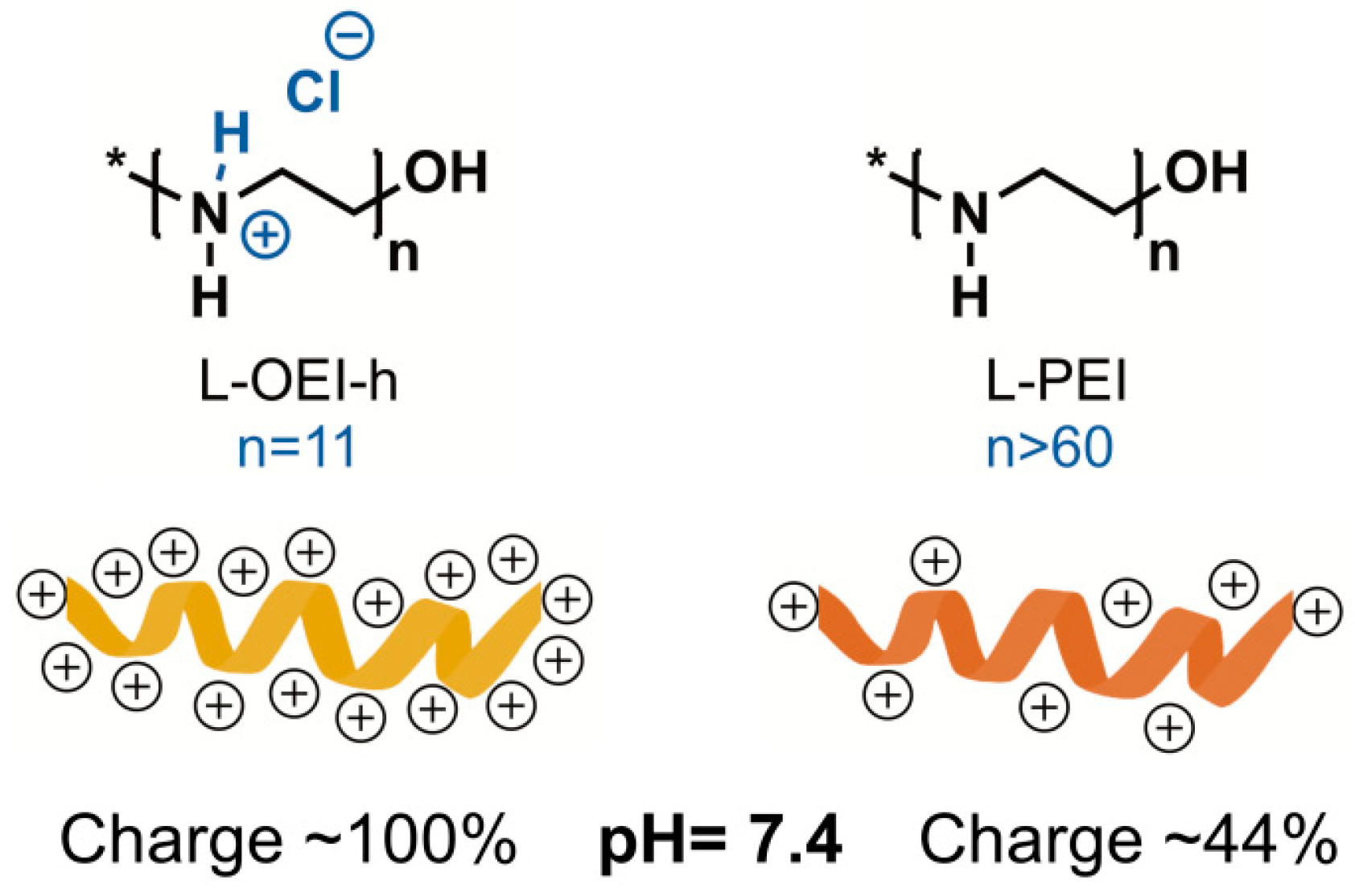
2.2. Antimicrobial Activity of L-OEI-h
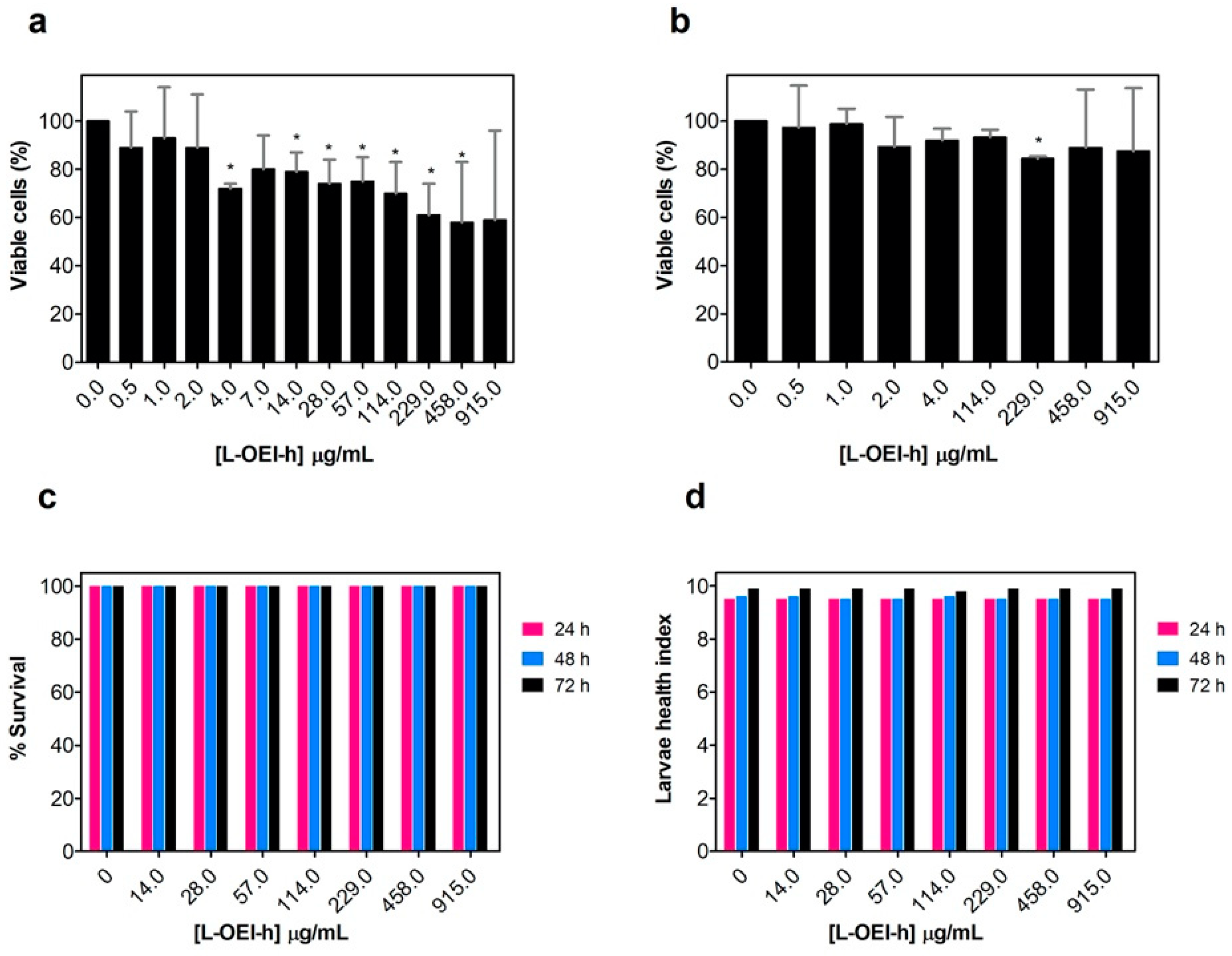
2.3. Biocompatibility Studies
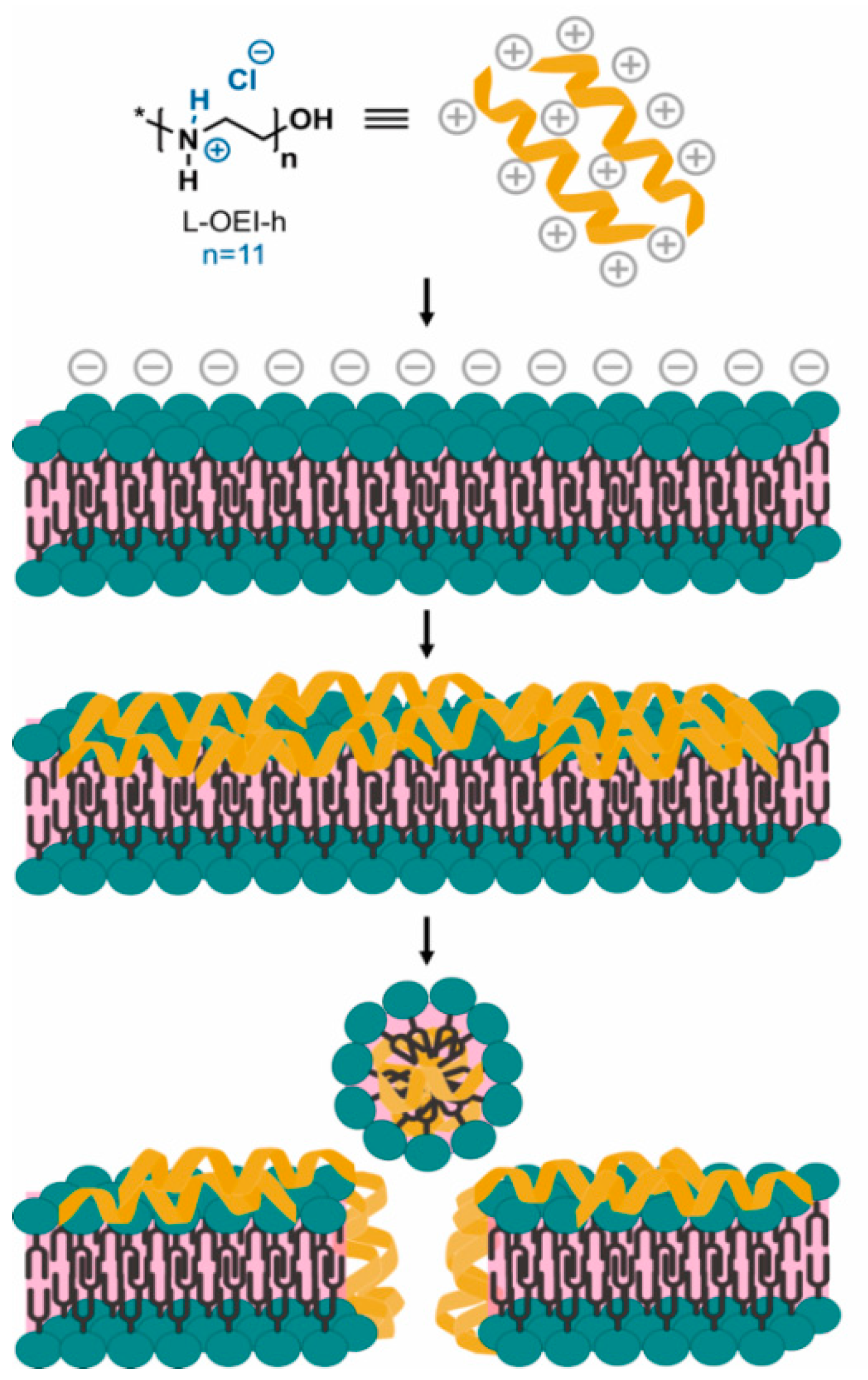
2.4. Exploring the L-OEI-h Mechanism of Action
3. Discussion
4. Materials and Methods
4.1. Synthesis of Linear Oligoethyleneimine Hydrochloride (L-OEI-h)
4.2. Clinical Isolates Collection and Identification
4.3. Galleria mellonella Infection Model
4.4. Antimicrobial Activity
4.4.1. Minimum Inhibitory Concentration (MIC) Determination
4.4.2. Minimum Bactericidal Concentration (MBC) Determination
4.4.3. In Vitro Time-Kill Curves
4.5. Biocompatibility Assays
4.5.1. MTT Viability Assay
4.5.2. Galleria mellonella Toxicity Assay
4.6. Exploring the L-OEI-h Mechanism of Action
Liposome Preparation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Coates, A.R.; Halls, G.; Hu, Y. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 2011, 163, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. PT 2015, 40, 277–283. [Google Scholar]
- Nordmann, P.; Cuzon, G.; Naas, T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009, 9, 228–236. [Google Scholar] [CrossRef]
- Molton, J.S.; Tambyah, P.A.; Ang, B.S.; Ling, M.L.; Fisher, D.A. The global spread of healthcare-associated multidrug-resistant bacteria: A perspective from Asia. Clin. Infect. Dis. 2013, 56, 1310–1318. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to Gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The current burden of carbapenemases: Review of significant properties and dissemination among Gram-negative bacteria. Antibiotics (Basel) 2020, 9, 186. [Google Scholar] [CrossRef]
- Mil-Homens, D.; Bernardes, N.; Fialho, A.M. The antibacterial properties of docosahexaenoic omega-3 fatty acid against the cystic fibrosis multiresistant pathogen Burkholderia cenocepacia. FEMS Microbiol. Lett. 2012, 328, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutuli, M.A.; Petronio, G.P.; Vergalito, F.; Magnifico, I.; Pietrangelo, L.; Venditti, N.; Di Marco, R. Galleria mellonella as a consolidated in vivo model hosts: New developments in antibacterial strategies and novel drug testing. Virulence 2019, 10, 527–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jander, G.; Rahme, L.G.; Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000, 182, 3843–3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mil-Homens, D.; Barahona, S.; Moreira, R.N.; Silva, I.J.; Pinto, S.N.; Fialho, A.M.; Arraiano, C.M. Stress response protein BolA influences fitness and promotes Salmonella enterica serovar Typhimurium virulence. Appl. Environ. Microbiol. 2018, 84, e02850-17. [Google Scholar] [CrossRef] [Green Version]
- Vilmos, P.; Kurucz, E. Insect immunity: Evolutionary roots of the mammalian innate immune system. Immunol. Lett. 1998, 62, 59–66. [Google Scholar] [CrossRef]
- Hoffmann, J.A. Innate immunity of insects. Curr. Opin. Immunol. 1995, 7, 4–10. [Google Scholar] [CrossRef]
- Papst, L.; Beović, B.; Pulcini, C.; Durante-Mangoni, E.; Rodríguez-Baño, J.; Kaye, K.S.; Daikos, G.L.; Raka, L.; Paul, M. Antibiotic treatment of infections caused by carbapenem-resistant Gram-negative bacilli: An international ESCMID cross-sectional survey among infectious diseases specialists practicing in large hospitals. Clin. Microbiol. Infect. 2018, 24, 1070–1076. [Google Scholar] [CrossRef] [Green Version]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Correia, V.G.; Bonifácio, V.D.B.; Raje, V.P.; Casimiro, T.; Moutinho, G.; da Silva, C.L.; Pinho, M.G.; Aguiar-Ricardo, A. Oxazoline-based antimicrobial oligomers: Synthesis by CROP using supercritical CO2. Macromol. Biosci. 2011, 11, 1128–1137. [Google Scholar] [CrossRef]
- McLaughlin, M.M.; Advincula, M.R.; Malczynski, M.; Barajas, G.; Qi, C.; Scheetz, M.H. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect. Dis. 2014, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Malanovic, N.; Lohner, K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals (Basel) 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohner, K. New strategies for novel antibiotics: Peptides targeting bacterial cell membranes. Gen. Physiol. Biophys. 2009, 28, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.M.; Adenwalla, N.; Wiles, S.; Proft, T. Galleria mellonella larvae as an infection model for group A streptococcus. Virulence 2013, 4, 419–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmona-Ribeiro, A.M.; de Melo Carrasco, L.D. Cationic antimicrobial polymers and their assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front Cell Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, J.B.; Velliyagounder, K.; Ragunath, C.; Rohde, H.; Mack, D.; Knobloch, J.K.; Ramasubbu, N. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 2004, 186, 8213–8220. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochimica Biophysica Acta (BBA) Biomembranes 1999, 1462, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, C.W.; Sim, J.H.; Shah, K.R.; Kolesnikova-Kaplan, A.; Shi, W.; Eckert, R. Selective membrane disruption: Mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob. Agents Chemother. 2011, 55, 3446–3452. [Google Scholar] [CrossRef] [Green Version]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en (accessed on 6 January 2021).
- Palermo, E.F.; Lienkamp, K.; Gillies, E.R.; Ragogna, P.J. Antibacterial activity of polymers: Discussions on the nature of amphiphilic balance. Angew. Chem. Int. Ed. 2019, 58, 3690–3693. [Google Scholar] [CrossRef]
- Kenawy, E.-R.; Worley, S.D.; Broughton, R. The chemistry and applications of antimicrobial polymers: A state-of-the-art review. Biomacromolecules 2007, 8, 1359–1384. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, K.; Caputo, G.A.; DeGrado, W.F. The role of hydrophobicity in the antimicrobial and hemolytic activities of polymethacrylate derivatives. Chemistry 2009, 15, 1123–1133. [Google Scholar] [CrossRef] [Green Version]
- Palermo, E.F.; Kuroda, K. Chemical structure of cationic groups in amphiphilic polymethacrylates modulates the antimicrobial and hemolytic activities. Biomacromolecules 2009, 10, 1416–1428. [Google Scholar] [CrossRef]
- Palermo, E.F.; Lee, D.K.; Ramamoorthy, A.; Kuroda, K. Role of cationic group structure in membrane binding and disruption by amphiphilic copolymers. J. Phys. Chem. B 2011, 115, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Al-Badri, Z.M.; Som, A.; Lyon, S.; Nelson, C.F.; Nusslein, K.; Tew, G.N. Investigating the effect of increasing charge density on the hemolytic activity of synthetic antimicrobial polymers. Biomacromolecules 2008, 9, 2805–2810. [Google Scholar] [CrossRef]
- Lin, J.; Qiu, S.; Lewis, K.; Klibanov, A.M. Bactericidal properties of flat surfaces and nanoparticles derivatized with alkylated polyethylenimines. Biotechnol. Prog. 2002, 18, 1082–1086. [Google Scholar] [CrossRef]
- Curtis, K.A.; Miller, D.; Millard, P.; Basu, S.; Horkay, F.; Chandran, P.L. Unusual salt and pH induced changes in polyethylenimine solutions. PLoS ONE 2016, 11, e0158147. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xu, K.; Wang, H.; Tan, P.K.; Fan, W.; Venkatraman, S.S.; Li, L.; Yang, Y.Y. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotechnol. 2009, 4, 457–463. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 7063. [Google Scholar] [CrossRef] [Green Version]
- Kwon, J.Y.; Kim, M.K.; Mereuta, L.; Seo, C.H.; Luchian, T.; Park, Y. Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB Express. 2019, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Bengoechea, J.A.; Sa Pessoa, J. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019, 43, 123–144. [Google Scholar] [CrossRef] [Green Version]
- Fleeman, R.M.; Macias, L.A.; Brodbelt, J.S.; Davies, B.W. Defining principles that influence antimicrobial peptide activity against capsulated Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2020, 117, 27620. [Google Scholar] [CrossRef]
- Liu, X.; Tang, M.; Zhang, T.; Hu, Y.; Zhang, S.; Kong, L.; Xue, Y. Determination of a threshold dose to reduce or eliminate CdTe-induced toxicity in L929 cells by controlling the exposure dose. PLoS ONE 2013, 8, e59359. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, M.; Barathi, V.A.; Goh, E.T.L.; Anggara, R.; Fazil, M.; Ng, A.J.Y.; Harini, S.; Aung, T.T.; Fox, S.J.; Liu, S.; et al. Antimicrobial activity and cell selectivity of synthetic and biosynthetic cationic polymers. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Aguiar-Ricardo, A.; Bonifácio, V.D.B.; Casimiro, T.; Correia, V.G. Supercritical carbon dioxide design strategies: From drug carriers to soft killers. Philos. Trans. A Math Phys. Eng. Sci. 2015, 373. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Susceptibility testing of bacteria using Maldi-Tof Mass Spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef] [Green Version]
- Edwards-Jones, V.; Claydon, M.A.; Evason, D.J.; Walker, J.; Fox, A.J.; Gordon, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Pankey, G.A.; Sabath, L.D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 2004, 38, 864–870. [Google Scholar] [CrossRef] [Green Version]
- García-Armesto, M.R.; Prieto, M.; García-López, M.L.; Otero, A.; Moreno, B. Modern microbiological methods for foods: Colony count and direct count methods. A review. Microbiologia 1993, 9, 1–13. [Google Scholar]
- Mangoni, M.L.; Papo, N.; Barra, D.; Simmaco, M.; Bozzi, A.; Di Giulio, A.; Rinaldi, A.C. Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochem. J. 2004, 380, 859–865. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; McCanna, D.J.; Sivak, J.G. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J. Pharmacol. Toxicol. Methods 2015, 71, 1–7. [Google Scholar] [CrossRef]
- Pinheiro, M.; Lúcio, M.; Lima, J.L.; Reis, S. Liposomes as drug delivery systems for the treatment of TB. Nanomedicine (Lond.) 2011, 6, 1413–1428. [Google Scholar] [CrossRef]


| Clinical Isolate | MIC (μg/mL) | MBC (μg/mL) |
|---|---|---|
| SYN1 KPC | 458 | 458 |
| SYN3 OXA-48 | 915 | >915 |
| SYN4 OXA-48 | 458 | 915 |
| SYN6 KPC | 915 | 915 |
| SYN7 KPC | >915 | >915 |
| SYN8 KPC | 458 | 458 |
| SYN9 KPC | 229 | 229–458 |
| SYN17 OXA-48 | 915 | 915 |
| SYN19 KPC | 915 | 915 |
| SYN22 KPC | 915 | >915 |
| P. aeruginosa PAO | 114 | 114 |
| E. coli AB1157 * | 90 | 90 |
| S. aureus JE2 | >915 | >915 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mil-Homens, D.; Martins, M.; Barbosa, J.; Serafim, G.; Sarmento, M.J.; Pires, R.F.; Rodrigues, V.; Bonifácio, V.D.B.; Pinto, S.N. Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine. Antibiotics 2021, 10, 56. https://doi.org/10.3390/antibiotics10010056
Mil-Homens D, Martins M, Barbosa J, Serafim G, Sarmento MJ, Pires RF, Rodrigues V, Bonifácio VDB, Pinto SN. Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine. Antibiotics. 2021; 10(1):56. https://doi.org/10.3390/antibiotics10010056
Chicago/Turabian StyleMil-Homens, Dalila, Maria Martins, José Barbosa, Gabriel Serafim, Maria J. Sarmento, Rita F. Pires, Vitória Rodrigues, Vasco D.B. Bonifácio, and Sandra N. Pinto. 2021. "Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine" Antibiotics 10, no. 1: 56. https://doi.org/10.3390/antibiotics10010056
APA StyleMil-Homens, D., Martins, M., Barbosa, J., Serafim, G., Sarmento, M. J., Pires, R. F., Rodrigues, V., Bonifácio, V. D. B., & Pinto, S. N. (2021). Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates: In Vivo Virulence Assessment in Galleria mellonella and Potential Therapeutics by Polycationic Oligoethyleneimine. Antibiotics, 10(1), 56. https://doi.org/10.3390/antibiotics10010056








