FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme
Abstract
:1. Introduction
2. Results
2.1. FtsH Sensitizes Methicillin-Resistant S. aureus to Oxacillin
2.2. The Sensitization Effect of ftsH Is Specific to β-Lactams
2.3. FtsH Does Not Degrade PBP2a and PBP2
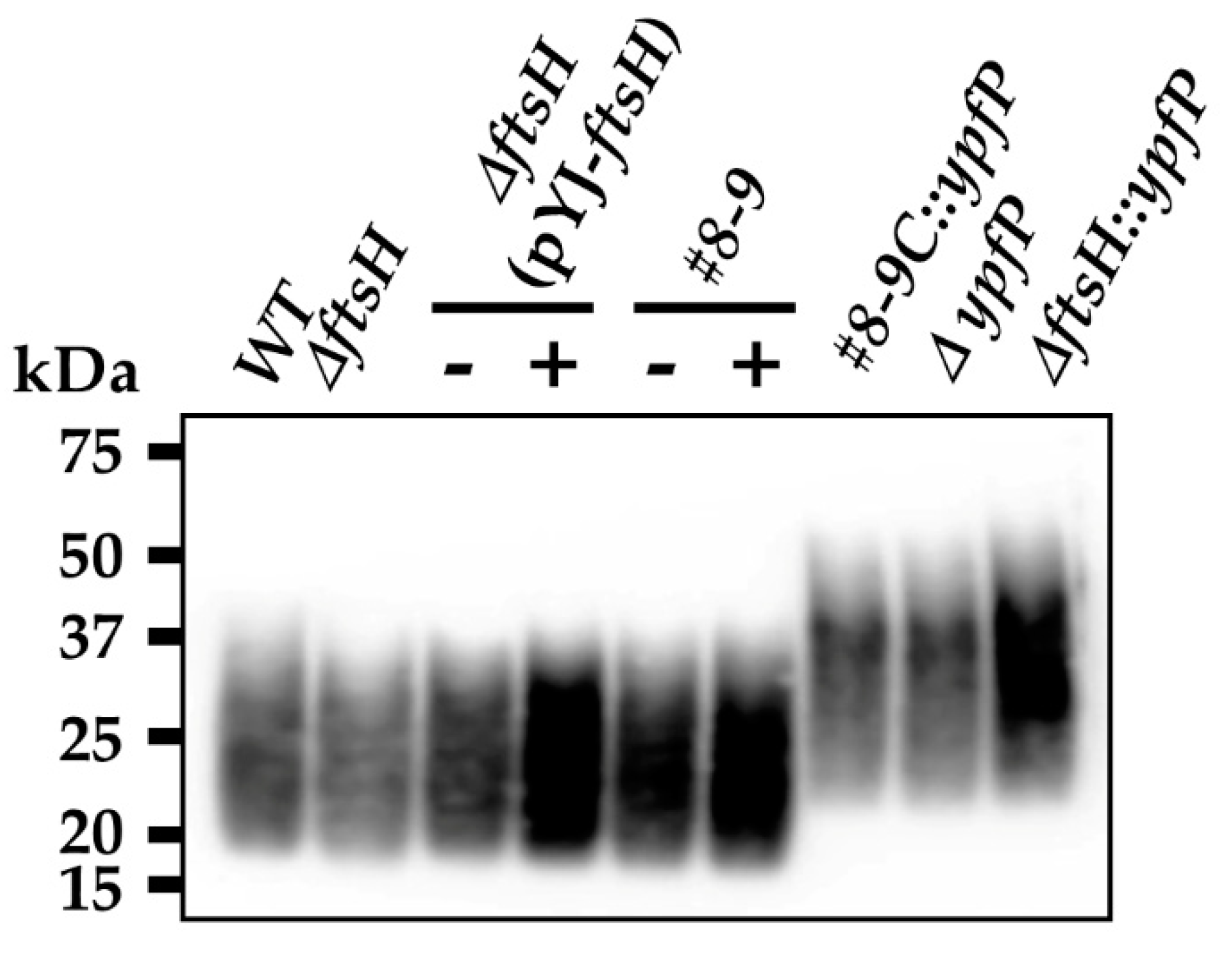
2.4. The Deletion of the FtsH Substrate Gene ypfP Sensitizes the USA300 Strain to Oxacillin
2.5. The Production of Aberrantly Large LTA Molecules Coincides with Increased Sensitivity to Oxacillin
2.6. The ftsH and ypfP Mutations Do Not Affect Autolysis Activity But Increase the Cell Wall Thickness
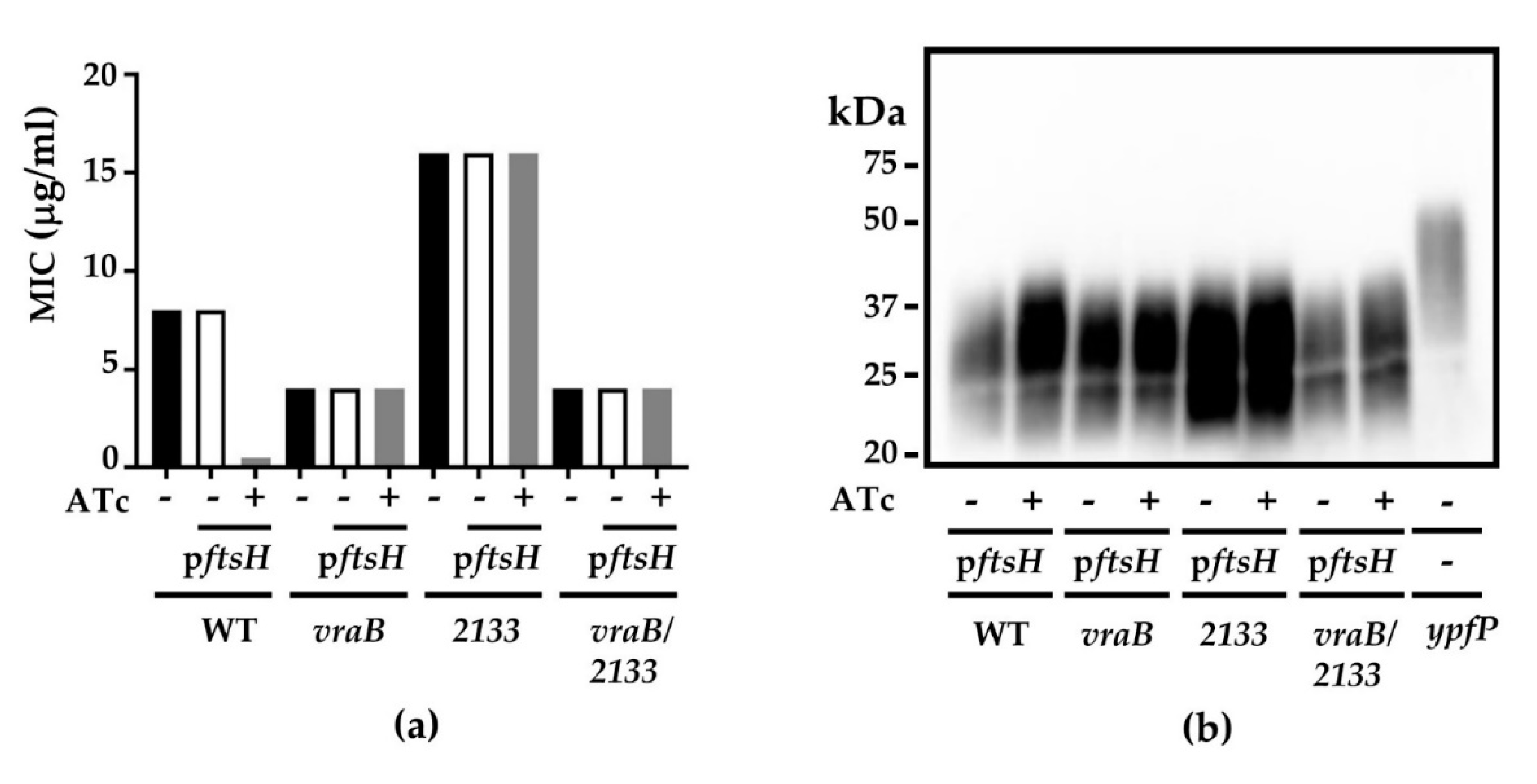
2.7. Identification of Suppressor Mutations
2.8. The Suppressor Mutations in vraB and SAUSA300_2133 Confer S. aureus Insensitivity to FtsH
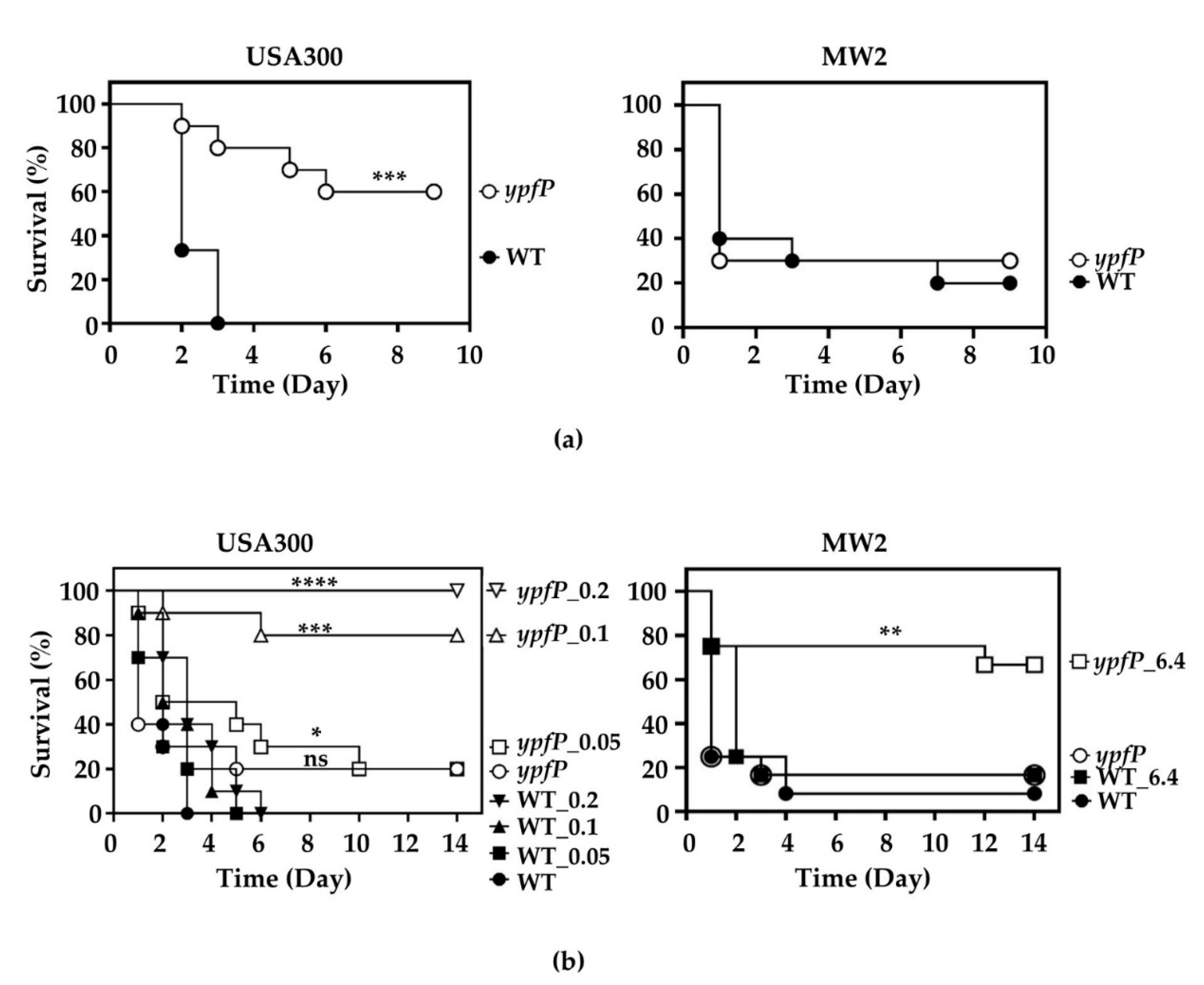
2.9. The Infection by the ypfP Mutant Could Be Treated with Oxacillin
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Culture Conditions
4.2. Plasmid Constructions and Mutagenesis
4.3. Determination of MIC
4.4. Isolation of Suppressor Mutants in the Strain USA Overexpressing FtsH
4.5. Western Blot Analysis
4.6. Labeling of Penicillin-binding Proteins (PBPs) Using Bocillin-FL
4.7. Autolysis Assay
4.8. Electron Microscopy
4.9. Animal Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Gordon, R.J.; Lowy, F.D. Pathogenesis of Methicillin-Resistant Staphylococcus aureus Infection. Clin. Infect. Dis. 2008, 46, S350–S359. [Google Scholar] [CrossRef]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef] [PubMed]
- Utsui, Y.; Yokota, T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1985, 28, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef]
- Otero, L.; Rojas-Altuve, A.; Llarrull, L.; Carrasco-Lopez, C.; Kumarasiri, M.; Lastochkin, E.; Fishovitz, J.; Dawley, M.; Hesek, D.; Lee, M.; et al. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl. Acad. Sci. USA 2013, 110, 16808–16813. [Google Scholar] [CrossRef]
- Fisher, J.F.; Mobashery, S. Beta-Lactams against the Fortress of the Gram-Positive Staphylococcus aureus Bacterium. Chem. Rev. 2021, 121, 3412–3463. [Google Scholar] [CrossRef]
- Pinho, M.; de Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef]
- Hamilton, S.M.; Alexander, J.A.N.; Choo, E.J.; Basuino, L.; da Costa, T.M.; Severin, A.; Chung, M.; Aedo, S.; Strynadka, N.C.J.; Tomasz, A.; et al. High-Level Resistance of Staphylococcus aureus to beta-Lactam Antibiotics Mediated by Penicillin-Binding Protein 4 (PBP4). Antimicrob. Agents Chemother. 2017, 61, e02727-16. [Google Scholar] [CrossRef]
- Basuino, L.; Jousselin, A.; Alexander, J.A.N.; Strynadka, N.C.; Pinho, M.G.; Chambers, H.F.; Chatterjee, S.S. PBP4 activity and its overexpression are necessary for PBP4-mediated high-level beta-lactam resistance. J. Antimicrob. Chemother. 2018, 73, 1177–1180. [Google Scholar] [CrossRef]
- Lee, S.H.; Jarantow, L.W.; Wang, H.; Sillaots, S.; Cheng, H.; Meredith, T.C.; Roemer, T. Antagonism of chemical genetic interaction networks resensitize MRSA to beta-lactam antibiotics. Chem. Biol. 2011, 18, 1379–1389. [Google Scholar] [CrossRef]
- Qamar, A.; Golemi-Kotra, D. Dual Roles of FmtA in Staphylococcus aureus Cell Wall Biosynthesis and Autolysis. Antimicrob. Agents Chemother. 2012, 56, 3797–3805. [Google Scholar] [CrossRef]
- Brown, S.; Xia, G.; Luhachack, L.G.; Campbell, J.; Meredith, T.C.; Chen, C.; Winstel, V.; Gekeler, C.; Irazoqui, J.; Peschel, A.; et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 18909–18914. [Google Scholar] [CrossRef]
- Meredith, T.C.; Wang, H.; Beaulieu, P.; Gründling, A.; Roemer, T. Harnessing the power of transposon mutagenesis for antibacterial target identification and evaluation. Mob. Genet. Elem. 2012, 2, 171–178. [Google Scholar] [CrossRef]
- Karinou, E.; Schuster, C.F.; Pazos, M.; Vollmer, W.; Gründling, A. Inactivation of the Monofunctional Peptidoglycan Glycosyltransferase SgtB Allows Staphylococcus aureus To Survive in the Absence of Lipoteichoic Acid. J. Bacteriol. 2019, 201, e00574-18. [Google Scholar] [CrossRef]
- Hesser, A.R.; Schaefer, K.; Lee, W.; Walker, S. Lipoteichoic acid polymer length is determined by competition between free starter units. Proc. Natl. Acad. Sci. USA 2020, 117, 29669–29676. [Google Scholar] [CrossRef]
- Hesser, A.R.; Matano, L.M.; Vickery, C.R.; Wood, B.M.; Santiago, A.G.; Morris, H.G.; Do, T.; Losick, R.; Walker, S. The Length of Lipoteichoic Acid Polymers Controls Staphylococcus aureus Cell Size and Envelope Integrity. J. Bacteriol. 2020, 202, e00149-20. [Google Scholar] [CrossRef]
- Dordel, J.; Kim, C.; Chung, M.; Pardos de la Gándara, M.; Holden, M.T.; Parkhill, J.; Tomasz, A. Novel determinants of antibiotic resistance: Identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio 2014, 5, e01000. [Google Scholar] [CrossRef]
- Kim, C.K.; Milheiriço, C.; de Lencastre, H.; Tomasz, A. Antibiotic Resistance as a Stress Response: Recovery of High-Level Oxacillin Resistance in Methicillin-Resistant Staphylococcus aureus “Auxiliary” (fem) Mutants by Induction of the Stringent Stress Response. Antimicrob. Agents Chemother. 2017, 61, e00313-17. [Google Scholar] [CrossRef]
- Ito, K.; Akiyama, Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 2005, 59, 211–231. [Google Scholar] [CrossRef]
- Lithgow, J.K.; Ingham, E.; Foster, S.J. Role of the hprT-ftsH locus in Staphylococcus aureus. Microbiology 2004, 150, 373–381. [Google Scholar] [CrossRef]
- Yeo, W.S.; Anokwute, C.; Marcadis, P.; Levitan, M.; Ahmed, M.; Bae, Y.; Kim, K.; Kostrominova, T.; Liu, Q.; Bae, T. A Membrane-Bound Transcription Factor is Proteolytically Regulated by the AAA+ Protease FtsH in Staphylococcus aureus. J. Bacteriol. 2020, 202, e00019-20. [Google Scholar] [CrossRef]
- Liu, Q.; Hu, M.; Yeo, W.-S.; He, L.; Li, T.; Zhu, Y.; Meng, H.; Wang, Y.; Lee, H.; Liu, X.; et al. Rewiring of the FtsH regulatory network by a single nucleotide change in saeS of Staphylococcus aureus. Sci. Rep. 2017, 7, 8456. [Google Scholar] [CrossRef]
- Beltrame, C.O.; Cortes, M.F.; Bonelli, R.R.; Côrrea, A.B.D.A.; Botelho, A.M.N.; Americo, M.A.; Figueiredo, A.M.S. Inactivation of the Autolysis-Related Genes lrgB and yycI in Staphylococcus aureus Increases Cell Lysis-Dependent eDNA Release and Enhances Biofilm Development In Vitro and In Vivo. PLoS ONE 2015, 10, e0138924. [Google Scholar] [CrossRef]
- Brunskill, E.W.; Bayles, K.W. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1996, 178, 5810–5812. [Google Scholar] [CrossRef]
- Jeong, D.-W.; Cho, H.; Jones, M.B.; Shatzkes, K.; Sun, F.; Ji, Q.; Liu, Q.; Peterson, S.N.; He, C.; Bae, T. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 2012, 86, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Fedtke, I.; Mader, D.; Kohler, T.; Moll, H.; Nicholson, G.; Biswas, R.; Henseler, K.; Götz, F.; Zähringer, U.; Peschel, A. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol. Microbiol. 2007, 65, 1078–1091. [Google Scholar] [CrossRef]
- Kiriukhin, M.Y.; Debabov, D.V.; Shinabarger, D.L.; Neuhaus, F.C. Biosynthesis of the Glycolipid Anchor in Lipoteichoic Acid of Staphylococcus aureus RN4220: Role of YpfP, the Diglucosyldiacylglycerol Synthase. J. Bacteriol. 2001, 183, 3506–3514. [Google Scholar] [CrossRef] [PubMed]
- Voggu, L.; Schlag, S.; Biswas, R.; Rosenstein, R.; Rausch, C.; Gotz, F. Microevolution of Cytochrome bd Oxidase in Staphylococci and Its Implication in Resistance to Respiratory Toxins Released by Pseudomonas. J. Bacteriol. 2006, 188, 8079–8086. [Google Scholar] [CrossRef] [PubMed]
- Stevens, E.; Laabei, M.; Gardner, S.; Somerville, G.A.; Massey, R.C. Cytolytic toxin production by Staphylococcus aureus is dependent upon the activity of the protoheme IX farnesyltransferase. Sci. Rep. 2017, 7, 13744. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.J.; Silhavy, T. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 1992, 359, 744–746. [Google Scholar] [CrossRef] [PubMed]
- Wessler, S.; Schneider, G.; Backert, S. Bacterial serine protease HtrA as a promising new target for antimicrobial therapy? Cell Commun. Signal 2017, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.B.; Stauff, D.L.; Pishchany, G.; Whitwell, C.W.; Torres, V.; Skaar, E.P. Staphylococcus aureus Redirects Central Metabolism to Increase Iron Availability. PLOS Pathog. 2006, 2, e87. [Google Scholar] [CrossRef]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Charleston, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on Evolution of Virulence and Resistance from the Complete Genome Analysis of an Early Methicillin-Resistant Staphylococcus aureus Strain and a Biofilm-Producing Methicillin-Resistant Staphylococcus epidermidis Strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef]
- Baba, T.; Takeuchi, F.; Kuroda, M.; Yuzawa, H.; Aoki, K.-I.; Oguchi, A.; Nagai, Y.; Iwama, N.; Asano, K.; Naimi, T.; et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2002, 359, 1819–1827. [Google Scholar] [CrossRef]
- Pinho, M.; Filipe, S.R.; de Lencastre, H.; Tomasz, A. Complementation of the Essential Peptidoglycan Transpeptidase Function of Penicillin-Binding Protein 2 (PBP2) by the Drug Resistance Protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef]
- Peacock, S.; Paterson, G. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Zhao, G.; Meier, T.I.; Kahl, S.; Gee, K.R.; Blaszczak, L.C. BOCILLIN FL, a Sensitive and Commercially Available Reagent for Detection of Penicillin-Binding Proteins. Antimicrob. Agents Chemother. 1999, 43, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Jorasch, P.; Warnecke, D.C.; Lindner, B.; Zähringer, U.; Heinz, E. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. JBIC J. Biol. Inorg. Chem. 2000, 267, 3770–3783. [Google Scholar] [CrossRef]
- Gründling, A.; Schneewind, O. Genes Required for Glycolipid Synthesis and Lipoteichoic Acid Anchoring in Staphylococcus aureus. J. Bacteriol. 2007, 189, 2521–2530. [Google Scholar] [CrossRef]
- Bae, T.; Schneewind, O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006, 55, 58–63. [Google Scholar] [CrossRef]
- Coe, K.A.; Lee, W.; Stone, M.C.; Komazin-Meredith, G.; Meredith, T.C.; Grad, Y.H.; Walker, S. Multi-strain Tn-Seq reveals common daptomycin resistance determinants in Staphylococcus aureus. PLoS Pathog. 2019, 15, e1007862. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.U.; Haas, R.; Fischer, W. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staph-ylococcus aureus. Eur. J. Biochem. 1984, 138, 357–363. [Google Scholar] [CrossRef]
- Panchal, V.V.; Griffiths, C.; Mosaei, H.; Bilyk, B.; Sutton, J.A.; Carnell, O.T.; Hornby, D.P.; Green, J.; Hobbs, J.K.; Kelley, W.L.; et al. Evolving MRSA: High-level beta-lactam resistance in Staphylococcus aureus is associated with RNA Polymerase alterations and fine tuning of gene expression. PLoS Pathog. 2020, 16, e1008672. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- A Kelley, L.; Mezulis, S.; Yates, C.M.; Wass, M.; Sternberg, E.M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Johnson, Z.L.; Cheong, C.-G.; Lee, S.-Y. Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4 Å. Nature 2012, 483, 489–493. [Google Scholar] [CrossRef]
- Ryuno, H.; Nigo, F.; Naguro, I.; Sekimizu, K.; Kaito, C. Staphylococcus aureus aggregation in the plasma fraction of silkworm hemolymph. PLoS ONE 2019, 14, e0217517. [Google Scholar] [CrossRef] [PubMed]
- Kreiswirth, B.N.; Löfdahl, S.; Betley, M.J.; O’Reilly, M.; Schlievert, P.; Bergdoll, M.S.; Novick, R. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983, 305, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.-W.; Cho, H.; Lee, H.; Li, C.; Garza, J.; Fried, M.; Bae, T. Identification of the P3 Promoter and Distinct Roles of the Two Promoters of the SaeRS Two-Component System in Staphylococcus aureus. J. Bacteriol. 2011, 193, 4672–4684. [Google Scholar] [CrossRef]
- Ji, Y.; Marra, A.; Rosenberg, M.; Woodnutt, G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 1999, 181, 6585–6590. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. MBio 2013, 4, e00537-12. [Google Scholar] [CrossRef] [PubMed]
- Bae, T.; Banger, A.K.; Wallace, A.; Glass, E.M.; Aslund, F.; Schneewind, O.; Missiakas, D.M. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 2004, 101, 12312–12317. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Suspceptability Testing; CLSI approved standard M100-S15; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2005. [Google Scholar]
- Liu, Q.; Cho, H.; Yeo, W.-S.; Bae, T. The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals. PLoS Pathog. 2015, 11, e1004799. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.L.; Lehman, M.K.; Fey, P.D.; Bayles, K.W. Contribution of the Staphylococcus aureus Atl AM and GL Murein Hydrolase Activities in Cell Division, Autolysis, and Biofilm Formation. PLoS ONE 2012, 7, e42244. [Google Scholar] [CrossRef]
- Yeo, W.-S.; Arya, R.; Kim, K.K.; Jeong, H.; Cho, K.H.; Bae, T. The FDA-approved anti-cancer drugs, streptozotocin and floxuridine, reduce the virulence of Staphylococcus aureus. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]


| Strains | MIC (µg/mL) | |
|---|---|---|
| USA300 | WT * | 8 |
| ΔftsH | 64 | |
| ΔmecA | 0.25 | |
| ΔftsHΔmecA | 0.25 | |
| WT (pYJ335) + ATc ** | 8 | |
| WT (pYJ-ftsH-His6) | 8 | |
| WT (pYJ-ftsH-His6) + ATc | 0.5 | |
| COL | WT | 256 |
| WT (pYJ335) + ATc | 256 | |
| WT (pYJ-ftsH-His6) | 128 | |
| WT (pYJ-ftsH-His6) + ATc | 8 | |
| MW2 | WT | 8 |
| WT (pYJ335) + ATc | 8 | |
| WT (pYJ-ftsH-His6) | 4 | |
| WT (pYJ-ftsH-His6) + ATc | 2 | |
| MIC (µg/mL) | ||||
|---|---|---|---|---|
| Antibiotics | WT | ΔftsH | ΔftsH(pYJ-ftsH-His6) | |
| −ATc | +ATc | |||
| Oxacillin | 8 | 64 | 16 | 0.5 |
| Cefotaxime | 32 | 128 | 16 | 8 |
| Cefazolin | 32 | 128 | 32 | 4 |
| Vancomycin | 2 | 1 | 1 | 2 |
| Dalbavancin | 0.2 | 0.4 | 0.4 | 0.2 |
| Teicoplanin | 1 | 0.5 | 0.5 | 1 |
| Linezolid | 4 | 2 | 2 | 2 |
| Daptomycin ** | 0.5 | 0.25 | 0.5 | 0.5 |
| Antibiotics | WT | ypfP |
|---|---|---|
| Oxacillin | 8 | 0.5 |
| Cefotaxime | 32 | 2 |
| Cefazolin | 32 | 0.5 |
| Vancomycin | 2 | 2 |
| Delbavancin | 0.2 | 0.4 |
| Teicoplanin | 1 | 2 |
| Linezolid | 4 | 1 |
| Daptomycin * | 0.5 | 0.5 |
| No. | Gene | Function | Changes in DNA | Changes in Protein | Number of Mutants (Total 18) |
|---|---|---|---|---|---|
| 1 | rpoC | RNA polymerase β’ subunit | T591408A | S723T | 18 |
| 2 | vraB | Acetyl-CoA c-acetyltransferase | G635085T | A144S | 18 |
| 3 | 2133 | Hypothetical protein | C2306775A | V220L | 18 |
| 4 | 0606 | Hypothetical protein | Insertion A680372 (2 nt) T680375 (4 nt) T680375 (7 nt) T680376 (10 nt) | Frame-shift | 3 2 2 2 |
| 5 | clpP | ATP-dependent Clp protease, proteolytic subunit | G838600A C838711A A838825T C839105A A839135G T839161C | E9K A53E I84F A177E D187G Stop196Q | 1 1 5 1 2 1 |
| 6 | clpX | ATP-dependent Clp protease, ATP-binding subunit | A1775135C C1775137T | F267V G266D | 1 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeo, W.-S.; Jeong, B.; Ullah, N.; Shah, M.A.; Ali, A.; Kim, K.K.; Bae, T. FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme. Antibiotics 2021, 10, 1198. https://doi.org/10.3390/antibiotics10101198
Yeo W-S, Jeong B, Ullah N, Shah MA, Ali A, Kim KK, Bae T. FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme. Antibiotics. 2021; 10(10):1198. https://doi.org/10.3390/antibiotics10101198
Chicago/Turabian StyleYeo, Won-Sik, Bohyun Jeong, Nimat Ullah, Majid Ali Shah, Amjad Ali, Kyeong Kyu Kim, and Taeok Bae. 2021. "FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme" Antibiotics 10, no. 10: 1198. https://doi.org/10.3390/antibiotics10101198
APA StyleYeo, W.-S., Jeong, B., Ullah, N., Shah, M. A., Ali, A., Kim, K. K., & Bae, T. (2021). FtsH Sensitizes Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics by Degrading YpfP, a Lipoteichoic Acid Synthesis Enzyme. Antibiotics, 10(10), 1198. https://doi.org/10.3390/antibiotics10101198










