Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus)
Abstract
:1. Introduction
2. Results
2.1. Fish Performance
2.2. In Vitro Antimicrobial Testing of Prepared Diets
2.3. Sequence Data and Diversity Analysis
2.4. Microbiome Community Dynamics in Response to OTC
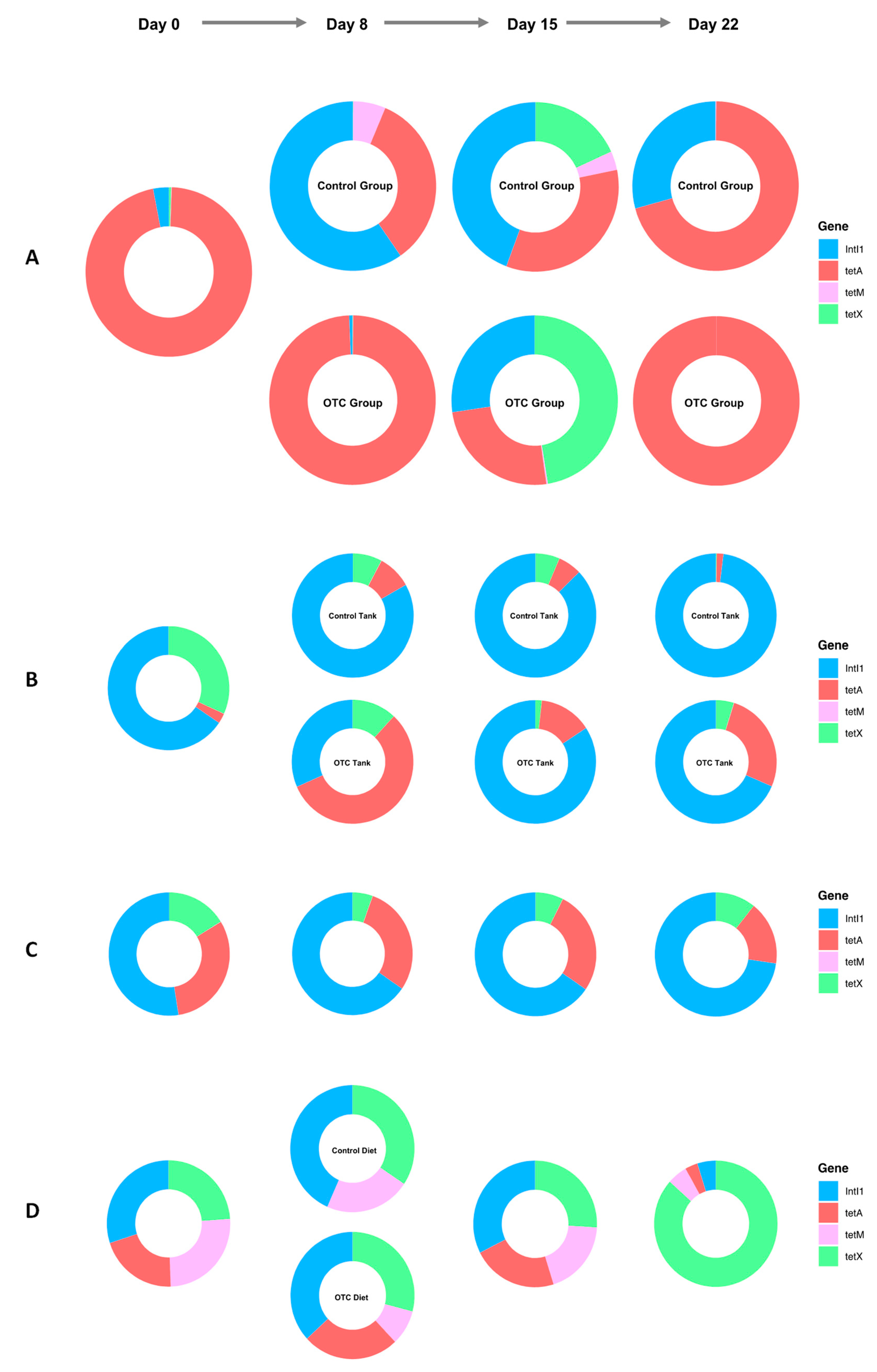
2.5. ARG Dynamics
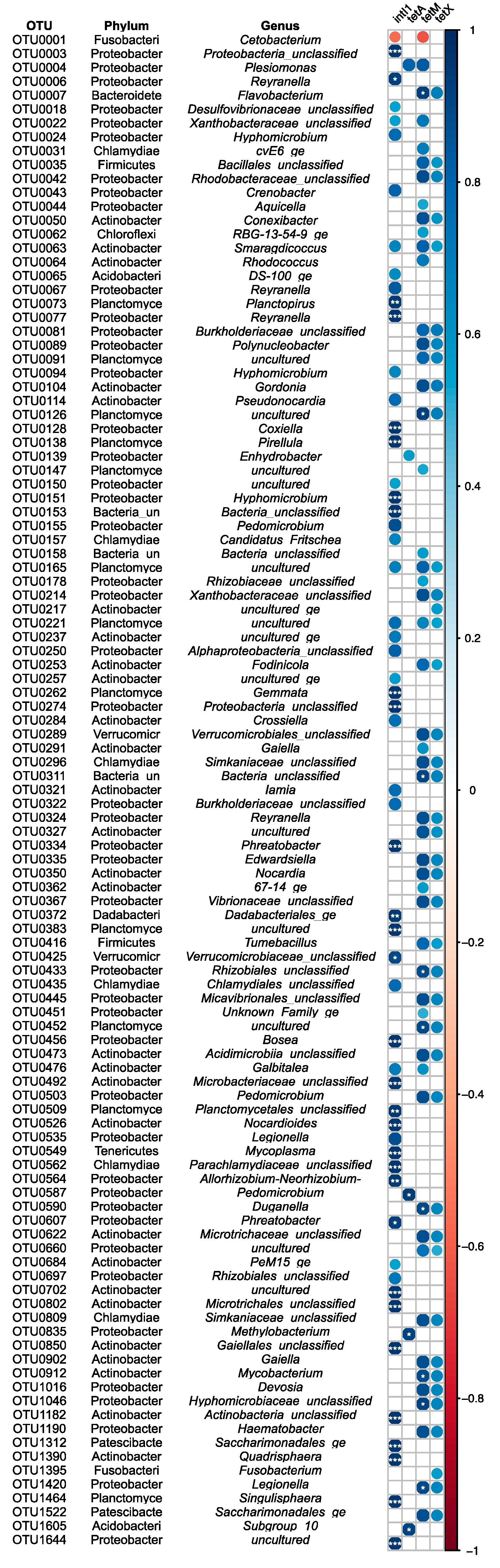
2.6. Correlation Analysis of ARGs and the Gut Microbiome in Fish
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Diet Preparation and In Vitro Antimicrobial Testing
4.3. Sample Collection
4.4. Library Preparation and Illumina Miseq Sequencing
4.5. Bioinformatic Analysis of Illumina Miseq Data
4.6. Quantification of ARGs
4.7. Data Visualisation and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO—The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020; p. 21. ISBN 9789251326923.
- FAO—The State of World Fisheries and Aquaculture 2018. In Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018; p. 23. ISBN 9789251305621.
- Kobayashi, M.; Msangi, S.; Batka, M.; Vannuccini, S.; Dey, M.M.; Anderson, J.L. Fish to 2030: The Role and Opportunity for Aquaculture. Aquac. Econ. Manag. 2015, 19, 282–300. [Google Scholar] [CrossRef] [Green Version]
- Ibrahem, M.; Mostafa, M.; Arab, R.M.; Rezk, M. Prevalence of Aeromonas hydrophila Infection in Wild and Cultured Tilapia Nilotica (O. niloticus). In Proceedings of the 8th International Symposium on Tilapia Aquaculture, Cairo, Egypt, 12–14 October 2008; pp. 1257–1271. [Google Scholar]
- Soto, E.; Griffin, M.; Arauz, M.; Riofrio, A.; Martinez, A.; Cabrejos, M.E. Edwardsiella ictalurias the Causative Agent of Mortality in Cultured Nile Tilapia. J. Aquat. Anim. Health 2012, 24, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Jantrakajorn, S.; Maisak, H.; Wongtavatchai, J. Comprehensive Investigation of Streptococcosis Outbreaks in Cultured Nile Tilapia, Oreochromis niloticus, and Red Tilapia, Oreochromis sp., of Thailand. J. World Aquac. Soc. 2014, 45, 392–402. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, W.; Hu, G.; Qiu, L.; Meng, S.; Song, C.; Fan, L.; Zhao, Z.; Bing, X.; Chen, J. Gut microbiota analysis of juvenile genetically improved farmed tilapia (Oreochromis niloticus) by dietary supplementation of different resveratrol concentrations. Fish Shellfish. Immunol. 2018, 77, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, S.H.; Francisco, J.; Campion, T.; Pimpinato, R.; Andrade, G.; Garcia, F.; Tornisielo, V. Multiresidue antimicrobial determination in Nile tilapia (Oreochromis Niloticus) cage farming by liquid chromatography tandem mass spectrometry. Aquaculture 2015, 447, 37–43. [Google Scholar] [CrossRef]
- Eissa, F.; Ghanem, K.; Al-Sisi, M. Occurrence and human health risks of pesticides and antibiotics in Nile tilapia along the Rosetta Nile branch, Egypt. Toxicol. Rep. 2020, 7, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Ström, G.H.; Björklund, H.; Barnes, A.C.; Da, C.T.; Nhi, N.H.Y.; Lan, T.T.; Magnusson, U.; Haldén, A.N.; Boqvist, S. Antibiotic Use by Small-Scale Farmers for Freshwater Aquaculture in the Upper Mekong Delta, Vietnam. J. Aquat. Anim. Heal. 2019, 31, 290–298. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Yang, K.; Chen, Y.; Fan, K. Characterization of a Bi-directional Promoter OtrRp Involved in Oxytetracycline Biosynthesis. Curr. Microbiol. 2019, 76, 1264–1269. [Google Scholar] [CrossRef]
- Chopra, I.; Robers, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarrete, P.; Mardones, P.; Opazo, R.; Espejo, R.; Romero, J. Oxytetracycline Treatment Reduces Bacterial Diversity of Intestinal Microbiota of Atlantic Salmon. J. Aquat. Anim. Health 2008, 20, 177–183. [Google Scholar] [CrossRef]
- Favero, L.M.; Facimoto, C.T.; Chideroli, R.T.; da Costa, A.R.; Umezu, D.F.; Honda, B.T.B.; de Oliveira, A.G.; Flaiban, K.K.M.D.C.; Di Santis, G.W.; Pereira, U.D.P. Administration of dehydrated oxytetracycline effectively reduces francisellosis mortality in Nile tilapia. Aquac. Res. 2021, 52, 4116–4126. [Google Scholar] [CrossRef]
- Roy, A.; Abraham, T.J.; Namdeo, M.S.; Singha, J.; Julinta, R.B.; Boda, S. Effects of Oral Oxytetracycline-Therapy on Wound Progression and Healing Following Aeromonas caviae Infection in Nile Tilapia (Oreochromis niloticus L.). Braz. Arch. Biol. Technol. 2019, 62. [Google Scholar] [CrossRef]
- Oviedo-Bolaños, K.; Rodríguez-Rodríguez, J.A.; Sancho-Blanco, C.; Barquero-Chanto, J.E.; Peña-Navarro, N.; Escobedo-Bonilla, C.M.; Umaña-Castro, R. Molecular identification of Streptococcus sp. and antibiotic resistance genes present in Tilapia farms (Oreochromis niloticus) from the Northern Pacific region, Costa Rica. Aquac. Int. 2021, 29, 2337–2355. [Google Scholar] [CrossRef]
- Limbu, S.; Zhou, L.; Sun, S.-X.; Zhang, M.-L.; Du, Z.-Y. Chronic exposure to low environmental concentrations and legal aquaculture doses of antibiotics cause systemic adverse effects in Nile tilapia and provoke differential human health risk. Environ. Int. 2018, 115, 205–219. [Google Scholar] [CrossRef] [PubMed]
- National Office of Animal Health NOAH Compendium. Available online: http://www.noahcompendium.co.uk (accessed on 29 August 2019).
- Bereded, N.; Abebe, G.; Fanta, S.; Curto, M.; Waidbacher, H.; Meimberg, H.; Domig, K. The Impact of Sampling Season and Catching Site (Wild and Aquaculture) on Gut Microbiota Composition and Diversity of Nile Tilapia (Oreochromis niloticus). Biology 2021, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Bereded, N.K.; Curto, M.; Domig, K.J.; Abebe, G.B.; Fanta, S.W.; Waidbacher, H.; Meimberg, H. Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia. Microorganisms 2020, 8, 1040. [Google Scholar] [CrossRef]
- Tan, H.Y.; Chen, S.-W.; Hu, S.-Y. Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish. Immunol. 2019, 92, 265–275. [Google Scholar] [CrossRef]
- Hallali, E.; Kokou, F.; Chourasia, T.K.; Nitzan, T.; Con, P.; Harpaz, S.; Mizrahi, I.; Cnaani, A. Dietary salt levels affect digestibility, intestinal gene expression, and the microbiome, in Nile tilapia (Oreochromis niloticus). PLoS ONE 2018, 13, e0202351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, Y.; Liu, Y.; Qiao, F.; Chen, L.; Liu, W.-T.; Du, Z.; Li, E. Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 2016, 454, 72–80. [Google Scholar] [CrossRef]
- Parata, L.; Mazumder, D.; Sammut, J.; Egan, S. Diet type influences the gut microbiome and nutrient assimilation of Genetically Improved Farmed Tilapia (Oreochromis niloticus). PLoS ONE 2020, 15, e0237775. [Google Scholar] [CrossRef]
- Sakyi, M.E.; Cai, J.; Ampofo-Yeboah, A.; Anokyewaa, M.A.; Wang, Z.; Jian, J. Starvation and re-feeding influence the growth, immune response, and intestinal microbiota of Nile tilapia (Oreochromis niloticus; Linnaeus 1758). Aquaculture 2021, 543, 736959. [Google Scholar] [CrossRef]
- Carlson, J.M.; Leonard, A.B.; Hyde, E.R.; Petrosino, J.F.; Primm, T.P. Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect. Drug Resist. 2017, 10, 143–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Limbu, S.M.; Shen, M.; Zhai, W.; Qiao, F.; He, A.; Du, Z.-Y.; Zhang, M. Environmental concentrations of antibiotics impair zebrafish gut health. Environ. Pollut. 2018, 235, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ma, L.; Wang, Y.; Fu, G.; Zhou, J.; Li, X.; Fang, W. Antimicrobial resistance and pulsed-field gel electrophoresis typing of Vibrio parahaemolyticus isolated from shrimp mariculture environment along the east coast of China. Mar. Pollut. Bull. 2018, 136, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Saharan, V.V.; Verma, P.; Singh, A.P. High prevalence of antimicrobial resistance in Escherichia coli, Salmonella spp. and Staphylococcus aureus isolated from fish samples in India. Aquac. Res. 2019, 51, 1200–1210. [Google Scholar] [CrossRef]
- Mok, J.S.; Cho, S.R.; Park, Y.J.; Jo, M.R.; Ha, K.S.; Kim, P.H.; Kim, M.J. Distribution and antimicrobial resistance of Vibrio parahaemolyticus isolated from fish and shrimp aquaculture farms along the Korean coast. Mar. Pollut. Bull. 2021, 171, 112785. [Google Scholar] [CrossRef] [PubMed]
- Delalay, G.; Berezowski, J.A.; Diserens, N.; Schmidt-Posthaus, H. An understated danger: Antimicrobial resistance in aquaculture and pet fish in Switzerland, a retrospective study from 2000 to 2017. J. Fish Dis. 2020, 43, 1299–1315. [Google Scholar] [CrossRef]
- Tyagi, A.; Singh, B.; ThammeGowda, N.K.B.; Singh, N.K. Shotgun metagenomics offers novel insights into taxonomic compositions, metabolic pathways and antibiotic resistance genes in fish gut microbiome. Arch. Microbiol. 2019, 201, 295–303. [Google Scholar] [CrossRef]
- Ye, L.; Liu, G.; Yao, T.; Lu, J. Monitoring of antimicrobial resistance genes in the spotted sea bass (Lateolabrax maculatus): Association with the microbiome and its environment in aquaculture ponds. Environ. Pollut. 2021, 276, 116714. [Google Scholar] [CrossRef]
- Saenz, J.S.; Marques, T.V.; Barone, R.S.C.; Cyrino, J.E.P.; Kublik, S.; Nesme, J.; Schloter, M.; Rath, S.; Vestergaard, G. Oral administration of antibiotics increased the potential mobility of bacterial resistance genes in the gut of the fish Piaractus mesopotamicus. Microbiome 2019, 8, 197. [Google Scholar] [CrossRef]
- Shefat, S.H.T. Vaccines for Use in Finfish Aquaculture. Acta Sci. Pharm. Sci. 2018, 2, 15–19. [Google Scholar]
- Holman, D.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ghanbari, M.; Klose, V.; Crispie, F.; Cotter, P.D. The dynamics of the antibiotic resistome in the feces of freshly weaned pigs following therapeutic administration of oxytetracycline. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, T.J.; Julinta, R.B.; Roy, A.; Singha, J.; Patil, P.K.; Kumar, K.A.; Paria, P.; Behera, B.K. Dietary therapeutic dose of oxytetracycline negatively influences the antioxidant capacity and immune-related genes expression in Nile tilapia Oreochromis niloticus (L.). Environ. Toxicol. Pharmacol. 2021, 87, 103685. [Google Scholar] [CrossRef] [PubMed]
- Limbu, S.M.; Zhang, H.; Luo, Y.; Chen, L.-Q.; Zhang, M.; Du, Z.-Y. High carbohydrate diet partially protects Nile tilapia (Oreochromis niloticus) from oxytetracycline-induced side effects. Environ. Pollut. 2019, 256, 113508. [Google Scholar] [CrossRef] [PubMed]
- Reda, R.; Ibrahim, R.; Ahmed, E.-N.G.; El-Bouhy, Z. Effect of oxytetracycline and florfenicol as growth promoters on the health status of cultured Oreochromis niloticus. Egypt. J. Aquat. Res. 2013, 39, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Robinson, K.; Becker, S.; Xiao, Y.; Lyu, W.; Yang, Q.; Zhu, H.; Yang, H.; Zhao, J.; Zhang, G. Differential Impact of Subtherapeutic Antibiotics and Ionophores on Intestinal Microbiota of Broilers. Microorganisms 2019, 7, 282. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, C.; Coronado, J.; Silva, A.; Romero, J. Cetobacterium Is a Major Component of the Microbiome of Giant Amazonian Fish (Arapaima gigas) in Ecuador. Animals 2018, 8, 189. [Google Scholar] [CrossRef] [Green Version]
- Suphoronski, S.A.; Chideroli, R.; Facimoto, C.T.; Mainardi, R.M.; de Souza, F.P.; Lopera-Barrero, N.M.; Jesus, G.F.A.; Martins, M.L.; Di Santis, G.W.; De Oliveira, A.; et al. Effects of a phytogenic, alone and associated with potassium diformate, on tilapia growth, immunity, gut microbiome and resistance against francisellosis. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.; Li, H.; Feng, J.; Chen, Y.; Nie, G.; Zhang, J. Effects of cadmium exposure on the composition and diversity of the intestinal microbial community of common carp (Cyprinus carpio L.). Ecotoxicol. Environ. Saf. 2018, 171, 92–98. [Google Scholar] [CrossRef]
- Zhou, L.; Limbu, S.; Qiao, F.; Du, Z.-Y.; Zhang, M. Influence of Long-Term Feeding Antibiotics on the Gut Health of Zebrafish. Zebrafish 2018, 15, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, T.; Keerthisinghe, T.P.; Chen, H.; Li, M.; Chu, W.; Yang, J.; Hu, Z.; Snyder, S.A.; Dong, W.; et al. Long-term oxytetracycline exposure potentially alters brain thyroid hormone and serotonin homeostasis in zebrafish. J. Hazard. Mater. 2020, 399, 123061. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Fernandes, J.; Kiron, V. Antibiotic-Induced Perturbations Are Manifested in the Dominant Intestinal Bacterial Phyla of Atlantic Salmon. Microorganisms 2019, 7, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.; Hiney, M.; Samuelsen, O.B. Bacterial resistance to antimicrobial agents used in fish farming: A critical evaluation of method and meaning. Annu. Rev. Fish Dis. 1994, 4, 273–313. [Google Scholar] [CrossRef]
- Kholil, I.; Hossain, M.M.; Neowajh, S.; Islam, S.; Kabir, M. Comparative efficiency of some commercial antibiotics against Pseudomonas infection in fish. Int. J. Fish. Aquat. Stud. 2015, 2, 114–117. [Google Scholar] [CrossRef]
- Serrano, P.H. Responsible use of antibiotics in aquaculture. In FAO Fisheries Technical Paper; FAO: Rome, Italy, 2005; Volume 469, p. 9. ISBN 9251054363. [Google Scholar]
- Zhang, Y.; Wen, B.; David, M.A.; Gao, J.-Z.; Chen, Z.-Z. Comparative analysis of intestinal microbiota of discus fish (Symphysodon haraldi) with different growth rates. Aquaculture 2021, 540, 736740. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, Q.; Wang, A.; Liu, Y.; Teame, T.; Ran, C.; Yang, Y.; He, S.; Zhou, W.; Olsen, R.E.; et al. Effects of dietary sodium acetate on food intake, weight gain, intestinal digestive enzyme activities, energy metabolism and gut microbiota in cultured fish: Zebrafish as a model. Aquaculture 2020, 523, 735188. [Google Scholar] [CrossRef]
- Wang, H.; Gu, Y.; Zhou, H.; Chen, J.; Wang, M.; Jiang, H.; Cao, H. Plesiomonas shigelloides, a potential pathogen of enteritis in Ictalurus punctatus. Isr. J. Aquac. Bamidgeh 2020, 72, 1–11. [Google Scholar] [CrossRef]
- Adesiyan, I.M.; Bisi-Johnson, M.A.; Ogunfowokan, A.O.; Okoh, A. Incidence and antimicrobial susceptibility fingerprints of Plesiomonas shigelliodes isolates in water samples collected from some freshwater resources in Southwest Nigeria. Sci. Total. Environ. 2019, 665, 632–640. [Google Scholar] [CrossRef]
- Jun, J.W.; Kim, J.H.; Jr, C.H.C.; Shin, S.P.; Han, J.E.; Jeong, D.S.; Park, S.C. Isolation and molecular detection of Plesiomonas shigelloides containing tetA gene from Asian arowana (Scleropages formosus) in a Korean aquarium. African J. Microbiol. Res. 2011, 5, 5019–5021. [Google Scholar] [CrossRef]
- Martins, A.; Pinheiro, T.; Imperatori, A.; Freire, S.; Sá-Freire, L.; Moreira, B.; Bonelli, R. Plesiomonas shigelloides: A notable carrier of acquired antimicrobial resistance in small aquaculture farms. Aquaculture 2018, 500, 514–520. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [Green Version]
- Fatahi-Bafghi, M. Antibiotic resistance genes in the Actinobacteria phylum. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1599–1624. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2017, 10, 626–640. [Google Scholar] [CrossRef] [Green Version]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- De Vries, L.E.; Vallès, Y.; Agersø, Y.; Vaishampayan, P.A.; García-Montaner, A.; Kuehl, J.V.; Christensen, H.; Barlow, M.; Francino, M.P. The Gut as Reservoir of Antibiotic Resistance: Microbial Diversity of Tetracycline Resistance in Mother and Infant. PLoS ONE 2011, 6, e21644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talwar, C.; Nagar, S.; Lal, R.; Negi, R.K. Fish Gut Microbiome: Current Approaches and Future Perspectives. Indian J. Microbiol. 2018, 58, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Narrowe, A.B.; Albuthi-Lantz, M.; Smith, E.P.; Bower, K.J.; Roane, T.M.; Vajda, A.M.; Miller, C.S. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome 2015, 3, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Fjellheim, A.J.; Playfoot, K.J.; Skjermo, J.; Vadstein, O. Inter-individual variation in the dominant intestinal microbiota of reared Atlantic cod (Gadus morhua L.) larvae. Aquac. Res. 2011, 43, 1499–1508. [Google Scholar] [CrossRef]
- Kim, H.B.; Borewicz, K.; White, B.A.; Singer, R.S.; Sreevatsan, S.; Tu, Z.J.; Isaacson, R.E. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Veter. Microbiol. 2011, 153, 124–133. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Hughes, R.J.; Denman, S.; Moore, R.J. Highly Variable Microbiota Development in the Chicken Gastrointestinal Tract. PLoS ONE 2013, 8, e84290. [Google Scholar] [CrossRef] [Green Version]
- Webster, T.M.U.; Consuegra, S.; Hitchings, M.; de Leaniz, C.G. Interpopulation Variation in the Atlantic Salmon Microbiome Reflects Environmental and Genetic Diversity. Appl. Environ. Microbiol. 2018, 84, e00691-18. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, G.S.; Desai, A.R.; Nilson, S.A.; Van Kessel, A.G.; Drew, M.D.; Hill, J.E. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture 2010, 307, 95–104. [Google Scholar] [CrossRef]
- Valdés, N.; Gonzalez, A.; Garcia, V.; Tello, M. Analysis of the Microbiome of Rainbow Trout (Oncorhynchus mykiss) Exposed to the Pathogen Flavobacterium psychrophilum 10094. Microbiol. Resour. Announc. 2020, 9, e01562-19. [Google Scholar] [CrossRef] [Green Version]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Van Braekel, J.; Fu, Q.; Roosens, N.H.; De Keersmaecker, S.C.; Vanneste, K. Targeting the 16S rRNA Gene for Bacterial Identification in Complex Mixed Samples: Comparative Evaluation of Second (Illumina) and Third (Oxford Nanopore Technologies) Generation Sequencing Technologies. Int. J. Mol. Sci. 2019, 21, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bara, M.; Joffe, A.R. The ethical dimension in published animal research in critical care: The public face of science. Crit. Care 2014, 18, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyons, P.P.; Turnbull, J.; Dawson, K.A.; Crumlish, M. Exploring the microbial diversity of the distal intestinal lumen and mucosa of farmed rainbow troutOncorhynchus mykiss(Walbaum) using next generation sequencing (NGS). Aquac. Res. 2015, 48, 77–91. [Google Scholar] [CrossRef]
- Knudsen, B.E.; Bergmark, L.; Munk, P.; Lukjancenko, O.; Priemé, A.; Aarestrup, F.; Pamp, S.J. Impact of Sample Type and DNA Isolation Procedure on Genomic Inference of Microbiome Composition. mSystems 2016, 1, e00095-16. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Cheng, Z.; Lei, S.; Liu, L.; Lv, X.; Chen, L.; Wu, M.; Wang, C.; Tian, B.; Song, Y. Community composition, diversity, and metabolism of intestinal microbiota in cultivated European eel (Anguilla anguilla). Appl. Microbiol. Biotechnol. 2018, 102, 4143–4157. [Google Scholar] [CrossRef]
- Kiruthiga, R.; Thanislass, J.; Antony, P.X.; Lydia, D.C.; Uma-Maheswari, D.; Venkatesa-Perumal, S. Analysis of V3 Region of 16S rRNA for the Characterization of Rumen Microbiome of Goat. J. Anim. Sci. Res. 2018, 2. [Google Scholar] [CrossRef]
- Ma, L.; Xie, Y.; Han, Z.; Giesy, J.P.; Zhang, X. Responses of earthworms and microbial communities in their guts to Triclosan. Chemosphere 2016, 168, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, Y.-B.; Xu, J.-X.; Ling, J.-Y.; Chen, J.-L.; Zhou, J.-L.; Zheng, L.; Du, Q.-P. Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 2017, 168, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- McMurdie, P.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Reshaping Data with thereshapePackage. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Oksanen, J. vegan: Community Ecology Package, R Package Version 1.8-5; 2007. Available online: https://CRAN.R-project.org/package=vegan (accessed on 20 May 2019).
- Yue, J.C.; Clayton, M.K. A Similarity Measure Based on Species Proportions. Commun. Stat. Theory Methods 2005, 34, 2123–2131. [Google Scholar] [CrossRef] [Green Version]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Fox, J.; Muenchen, R.; Putler, D. Package ‘RcmdrMisc’; 2018. Available online: https://CRAN.R-project.org/package=RcmdrMisc (accessed on 1 April 2020).
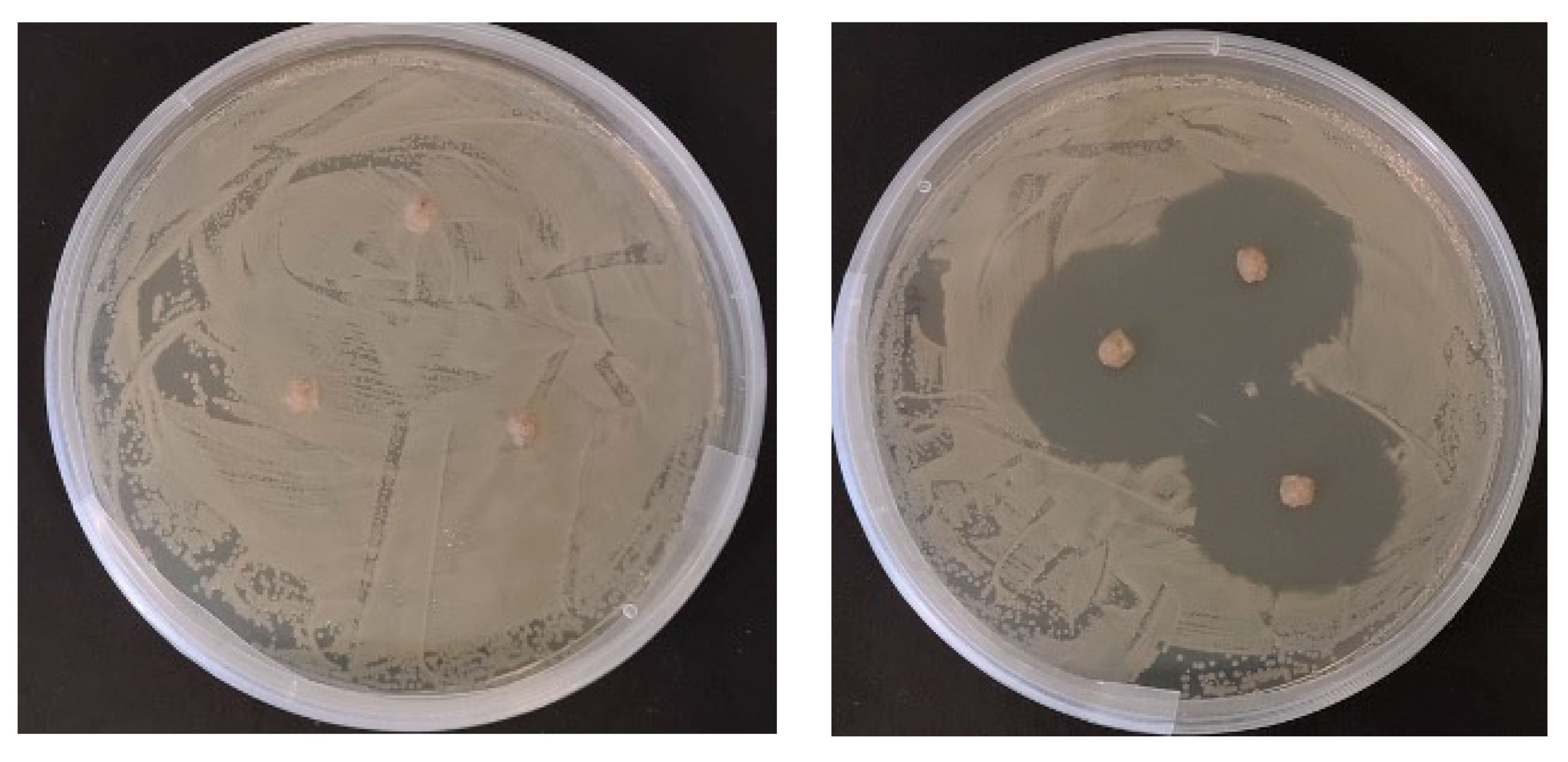

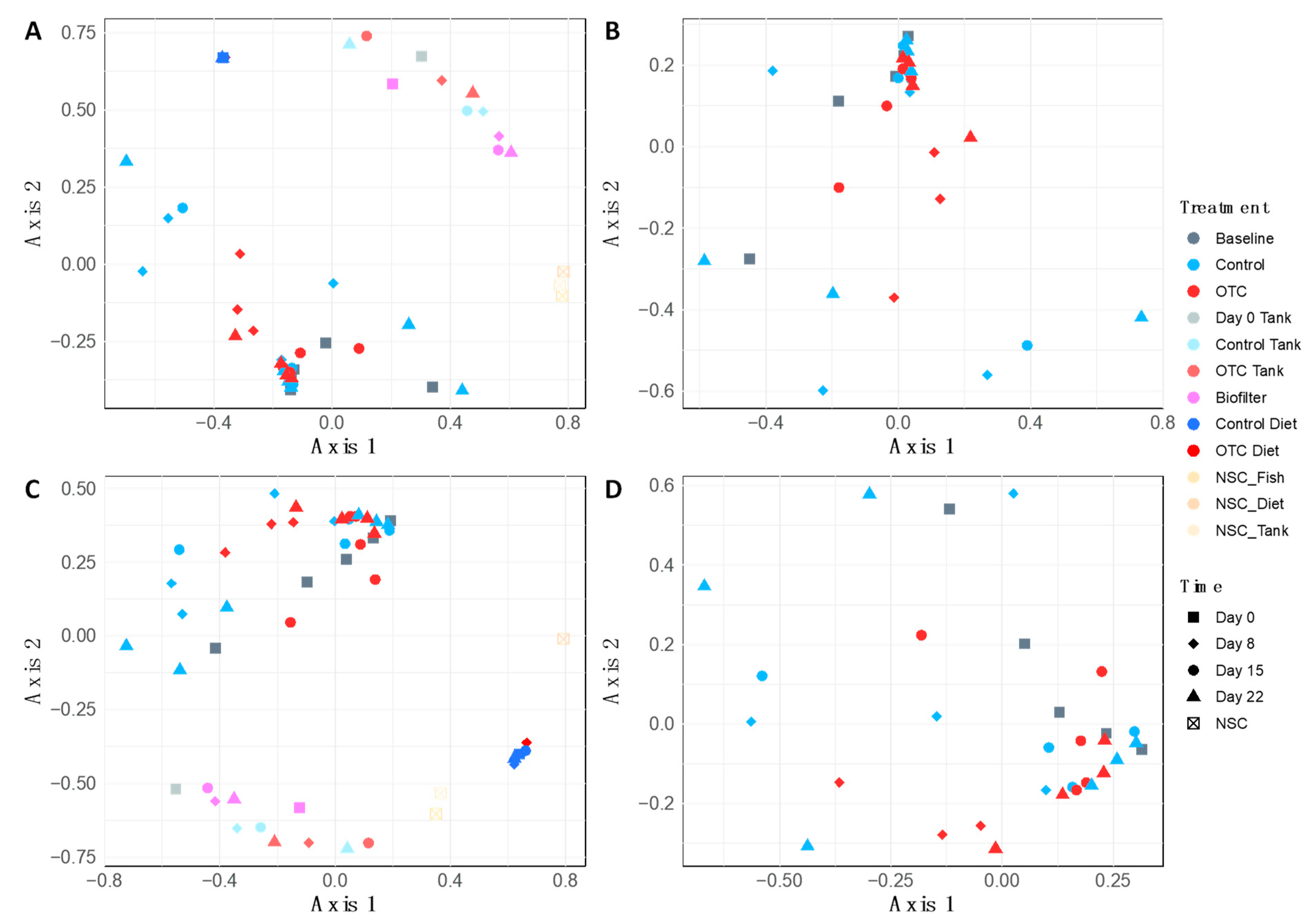
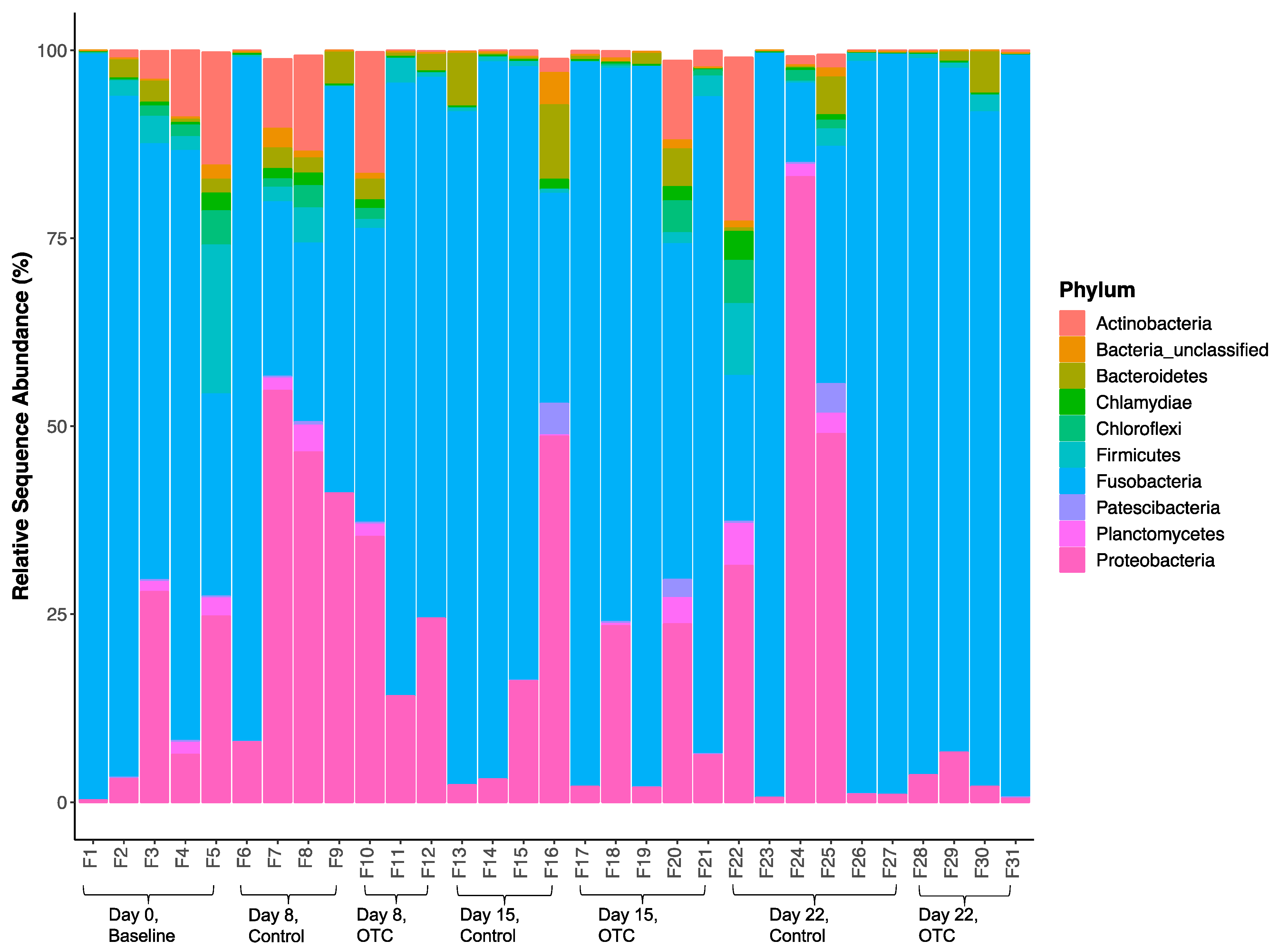
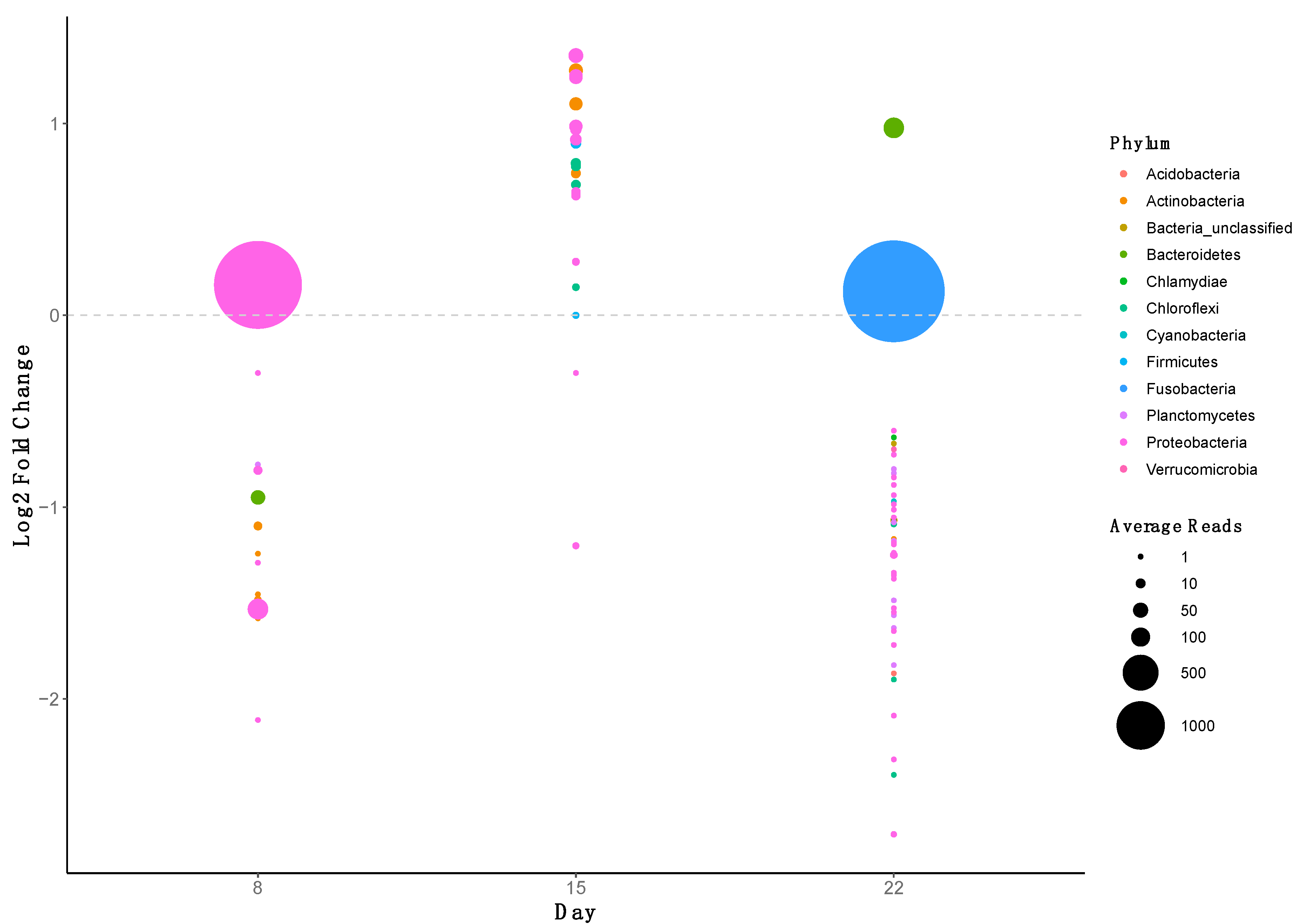
| Treatment | Day | Length (cm) | Weight (g) | Growth Rate (g/day) | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Baseline | 0 | 15.00 | 0.83 | 57.75 | 8.39 | 0.37 | 0.20 |
| Control | 8 | 14.63 | 0.95 | 51.88 | 8.29 | 0.33 | 0.37 |
| OTC | 8 | 14.95 | 1.41 | 54.22 | 14.12 | 0.31 | 0.25 |
| Control | 15 | 15.65 | 1.18 | 61.15 | 14.00 | 0.45 | 0.27 |
| OTC | 15 | 14.72 | 1.09 | 54.20 | 12.03 | 0.27 | 0.18 |
| Control | 22 | 15.65 | 1.04 | 62.65 | 12.90 | 0.32 | 0.22 |
| OTC | 22 | 15.17 | 0.71 | 56.37 | 8.42 | 0.28 | 0.18 |
| Phylum | Day 0 | Day 8 | Day 15 | Day 22 | |||
|---|---|---|---|---|---|---|---|
| Baseline | Control | OTC | Control | OTC | Control | OTC | |
| Actinobacteria | 5.78 ± 6.24 | 5.57 ± 6.48 | 5.55 ± 9.24 | 0.76 ± 0.83 | 2.89 ± 4.39 | 4.19 ± 8.66 | 0.14 ± 0.17 |
| Bacteria_unclassified | 0.48 ± 0.77 | 0.87 ± 1.24 | 0.26 ± 0.46 | 1.11 ± 2.11 | 0.34 ± 0.52 | 0.40 ± 0.53 | 0.02 ± 0.03 |
| Bacteroidetes | 1.58 ± 1.24 | 2.39 ± 1.74 | 1.95 ± 1.13 | 4.47 ± 4.86 | 1.51 ± 2.07 | 1.00 ± 1.96 | 1.84 ± 2.67 |
| Chlamydiae | 0.63 ± 0.97 | 0.81 ± 0.82 | 0.38 ± 0.66 | 0.33 ± 0.66 | 0.48 ± 0.79 | 0.84 ± 1.50 | 0.01 ± 0.01 |
| Chloroflexi | 1.62 ± 1.76 | 1.02 ± 1.40 | 0.49 ± 0.85 | 0.01 ± 0.01 | 1.06 ± 1.80 | 1.39 ± 2.22 | 0.00 ± 0.00 |
| Firmicutes | 5.53 ± 8.07 | 1.83 ± 20.02 | 1.88 ± 1.49 | 0.78 ± 0.18 | 1.00 ± 1.12 | 2.26 ± 3.69 | 1.07 ± 1.03 |
| Fusobacteria | 70.58 ± 28.91 | 47.98 ± 32.05 | 64.15 ± 22.24 | 73.58 ± 30.96 | 79.57 ± 21.57 | 59.44 ± 43.18 | 93.66 ± 4.19 |
| Patescibacteria | 0.10 ± 0.12 | 0.16 ± 0.20 | 0.08 ± 0.13 | 1.05 ± 2.10 | 0.53 ± 1.07 | 0.73 ± 1.58 | 0.00 ± 0.00 |
| Planctomycetes | 1.18 ± 1.04 | 1.32 ± 1.70 | 0.56 ± 0.96 | 0.06 ± 0.09 | 0.80 ± 1.51 | 1.67 ± 2.22 | 0.05 ± 0.09 |
| Proteobacteria | 12.46 ± 12.88 | 37.59 ± 20.50 | 24.61 ± 10.59 | 17.52 ± 21.68 | 11.47 ± 11.14 | 27.70 ± 33.76 | 3.21 ± 2.61 |
| ARG | Inverse Simpson | Shannon Diversity Index | ||
|---|---|---|---|---|
| R | p | R | p | |
| intI1 | 0.58 | 0.0008 | 0.57 | 0.0011 |
| tetA | 0.2 | 0.3000 | 0.24 | 0.2000 |
| tetM | 0.69 | 0.0005 | 0.55 | 0.0092 |
| tetX | 0.21 | 0.3700 | 0.45 | 0.0470 |
| Primer | Target | Sequence (5′–‘3) | Size (bp) | Ta°C | Eff. (%) | R2 | Application | Source |
|---|---|---|---|---|---|---|---|---|
| 341F | 16S rRNA (V3-4) | CCTACGGGNGGCWGCAG | 464 | 60 | 114.37 | 0.99 | 16S rRNA qPCR | [75,76] |
| 805R | GACTACHVGGGTATCTAATCC | |||||||
| Probe | FAM-ATTACCGCGGCTGCTGG-MGBEQ | |||||||
| 16S_V4F | 16S rRNA (V4) | [Illumina adapter]-AYTGGGYDTAAAGNG | 245 | 54 | N/A | N/A | Illumina Libraries | [77] |
| 16S_V4R Cocktail | [Illumina adapter] TACCRGGGTHTCTAATCC | |||||||
| [Illumina adapter]-TACCAGAGTATCTAATTC | ||||||||
| [Illumina adapter]-CTACDSRGGTMTCTAATC | ||||||||
| [Illumina adapter]-TACNVGGGTATCTAATC | ||||||||
| intI1_F | Class 1 integrase protein | CCTCCCGCACGATGATC | 280 | 63 | 101 | 0.99 | ARG qPCR | [78] |
| intI1_R | TCCACGCATCGTCAGGC | |||||||
| tetA_F | Tetracycline efflux pump | GCTACATCCTGCTTGCCTTC | 210 | 64 | 98 | 0.99 | ARG qPCR | [78] |
| tetA_R | CATAGATCGCCGTGAAGAGG | |||||||
| tetM_F | Tetracycline ribosomal protection | AGTGGAGAAATCCCTGCTCGGT | 149 | 66 | 107 | 0.99 | ARG qPCR | [78] |
| tetM_R | TGACTATTTGGACGACGGGGCT | |||||||
| tetX_F | Enzymatic modification of tetracycline | GAAAGAGACAACGACCGAGAG | 131 | 63 | 99 | 0.99 | ARG qPCR | [78] |
| tetX_R | ACACCCATTGGTAAGGCTAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Payne, C.J.; Turnbull, J.F.; MacKenzie, S.; Crumlish, M. Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus). Antibiotics 2021, 10, 1213. https://doi.org/10.3390/antibiotics10101213
Payne CJ, Turnbull JF, MacKenzie S, Crumlish M. Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus). Antibiotics. 2021; 10(10):1213. https://doi.org/10.3390/antibiotics10101213
Chicago/Turabian StylePayne, Christopher J., James F. Turnbull, Simon MacKenzie, and Margaret Crumlish. 2021. "Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus)" Antibiotics 10, no. 10: 1213. https://doi.org/10.3390/antibiotics10101213
APA StylePayne, C. J., Turnbull, J. F., MacKenzie, S., & Crumlish, M. (2021). Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus). Antibiotics, 10(10), 1213. https://doi.org/10.3390/antibiotics10101213






