Genome-Wide Identification, Characterization, and Expression Analysis of DDE_Tnp_4 Family Genes in Eriocheir sinensis
Abstract
:1. Introduction
2. Results
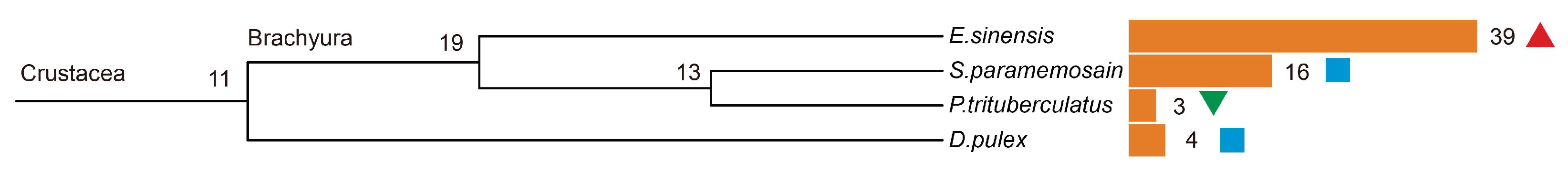
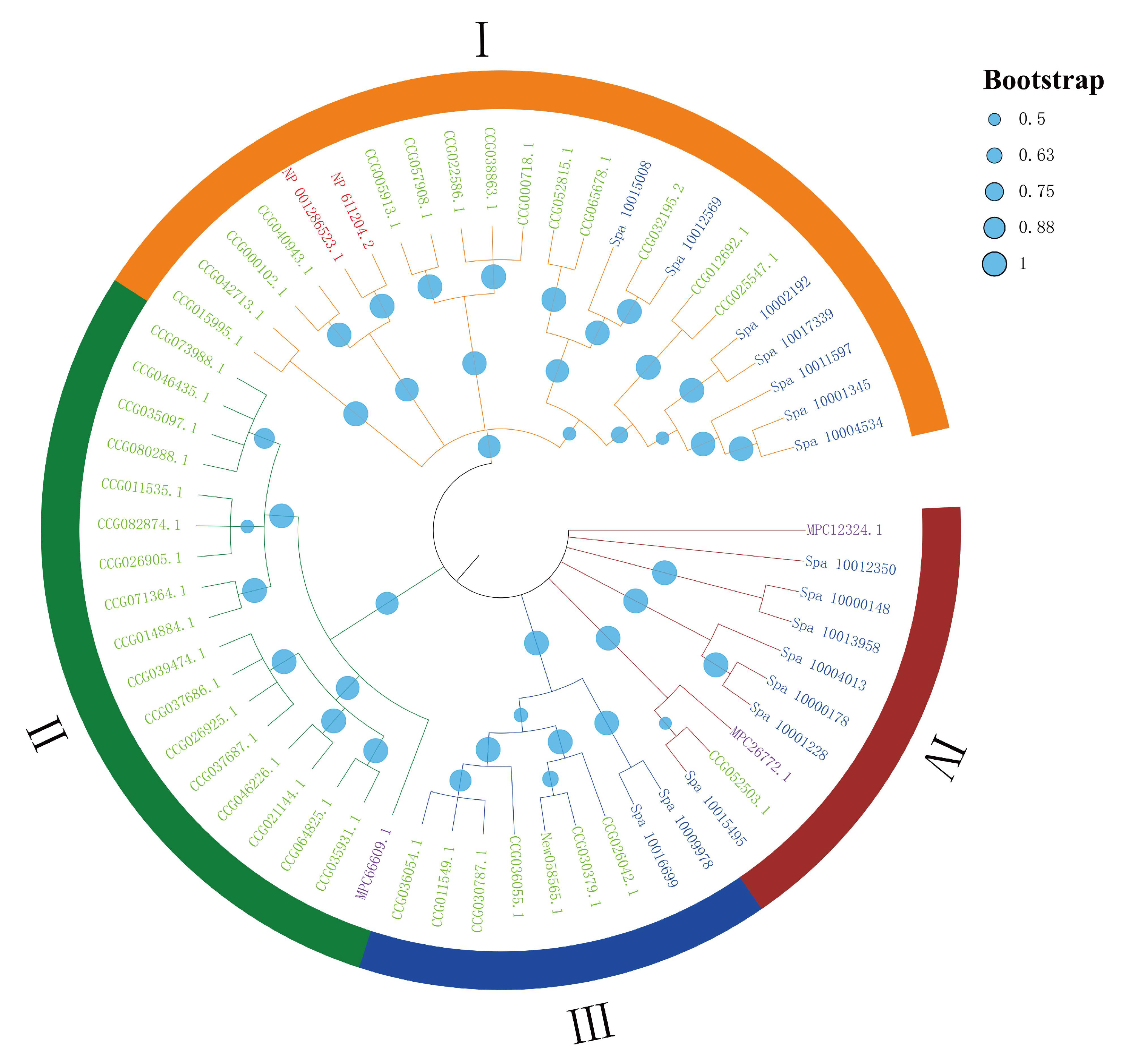
2.1. The Expansion of DDE_Tnp_4 Family
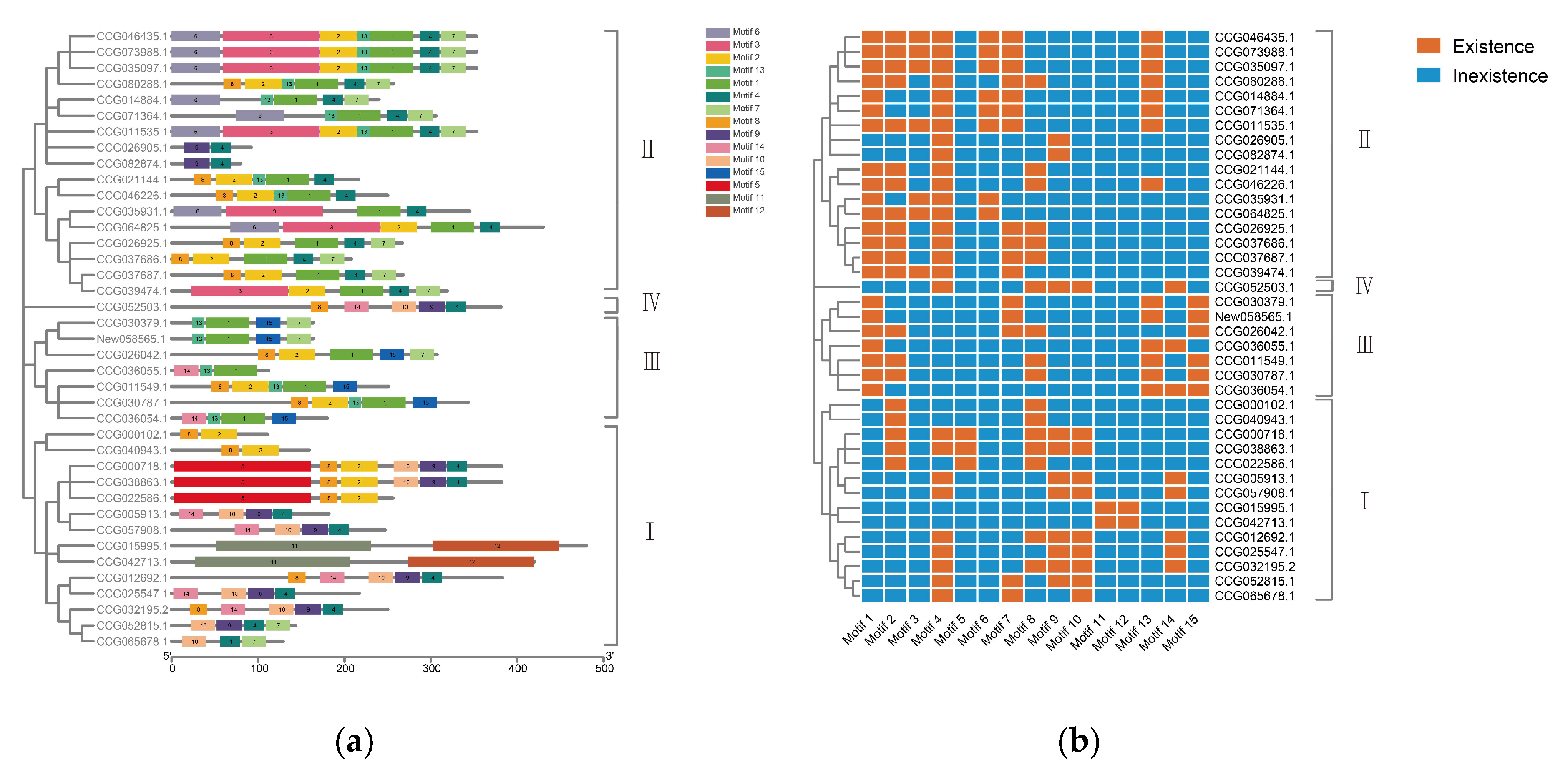
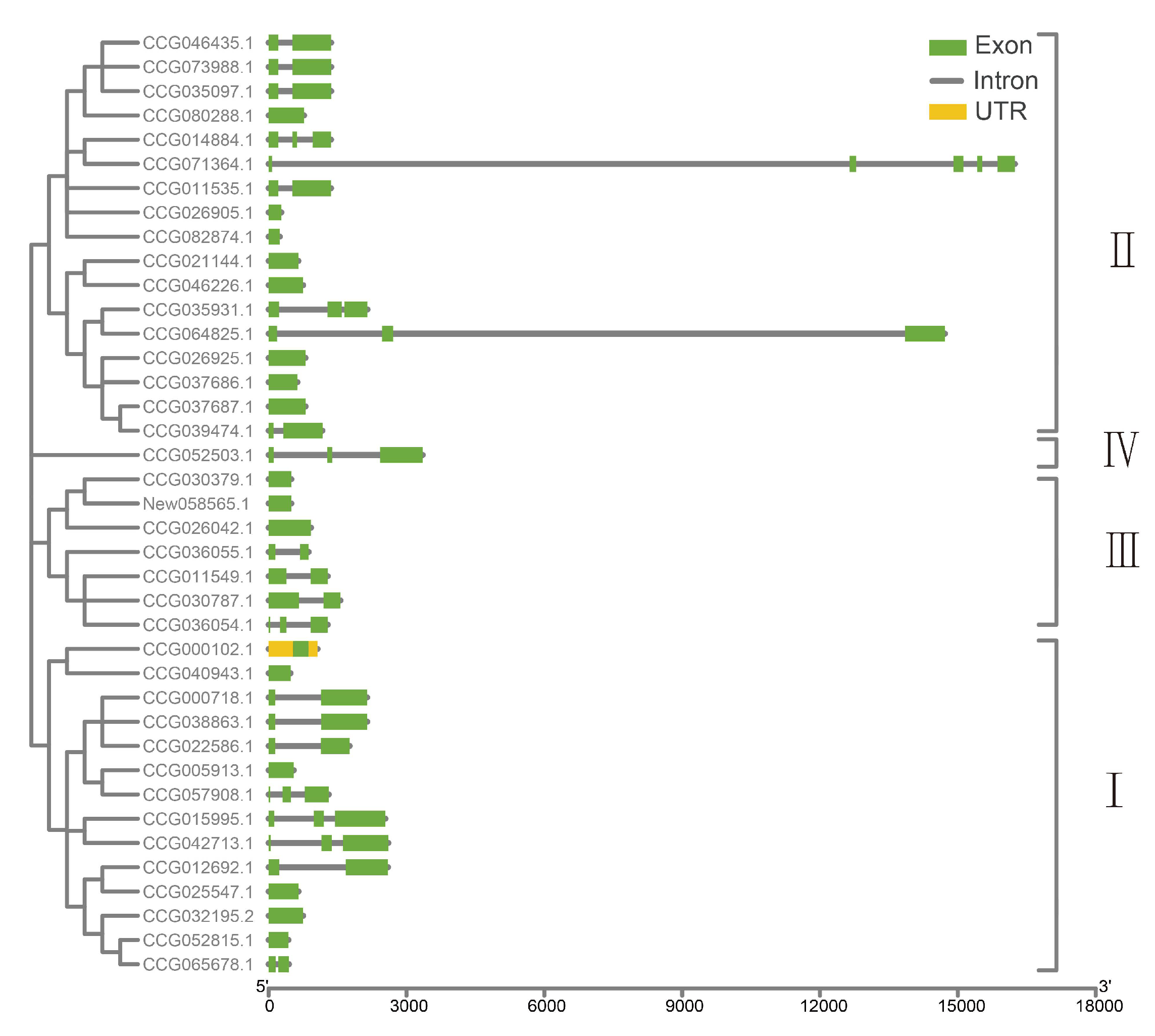
2.2. Conserved Motif and Gene Structure of DDE_Tnp_4 Family Genes in E. sinensis
2.3. Synteny and Duplication of DDE_Tnp_4 Family Genes in E. sinensis
2.4. Gene Expression of E. sinensis DDE_Tnp_4 Family Genes under Acute High Salinity Stress and Air Exposure Stress
3. Discussion
4. Materials and Methods
4.1. Identification of DDE_Tnp_4 Family Genes
4.2. Gene Family Expansion and Contraction Analysis
4.3. Gene Family Expansion and Contraction Analysis
4.4. Localization and Synteny Analysis of DDE_Tnp_4 Family Genes
4.5. Localization and Synteny Analysis of DDE_Tnp_4 Family Genes
4.6. Expression Profiling of DDE_Tnp_4 Family Genes under Stress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, L.; Cui, Z.; Song, C.; Liu, Y.; Hui, M.; Wang, C. Flow cytometric analysis of DNA content for four commercially important crabs in China. Acta Oceanol. Sinica 2016, 35, 7–11. [Google Scholar] [CrossRef]
- Herborg, L.M.; Rushton, S.P.; Clare, A.S.; Bentley, M.G. The invasion of the Chinese mitten crab (Eriocheir sinensis) in the United Kingdom and its comparison to continental Europe. Biol. Invasions 2005, 7, 959–968. [Google Scholar] [CrossRef]
- Dittel, A.I.; Epifanio, C.E. Invasion biology of the Chinese mitten crab Eriocheir sinensis: A brief review. J. Exp. Mar. Biol. Ecol. 2009, 374, 79–92. [Google Scholar] [CrossRef]
- Long, X.; Wu, X.; Zhao, L.; Ye, H.; Cheng, Y.; Zeng, C. Effects of salinity on gonadal development, osmoregulation and metabolism of adult male Chinese mitten crab, Eriocheir sinensis. PLoS ONE 2017, 12, e0179036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wang, J.; Hou, X.; Yue, W.; Li, Z.; Wang, C. Gene expression profiles of gill provide insights into the aerial respiration capacity of the Chinese mitten crab, Eriocheir sinensis. Aquaculture 2019, 506, 148–153. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, D.; Wang, F.; Dong, S. Hypothermal effects on survival, energy homeostasis and expression of energy-related genes of swimming crabs Portunus trituberculatus during air exposure. J. Therm. Biol. 2016, 60, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Liu, P.; Wang, Y.; Gao, B.; Chen, P.; Li, J. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS ONE 2013, 8, e82155. [Google Scholar]
- Long, X.; Wu, X.; Zhao, L.; Ye, H.; Cheng, Y.; Zeng, C. Physiological responses and ovarian development of female Chinese mitten crab Eriocheir sinensis subjected to different salinity conditions. Front. Physiol. 2018, 8, 1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.H.; Ma, H.L.; Deng, Y.Q.; Feng, J.; Chen, X.L.; Guo, Z.X. Transcriptome analysis and histopathology of the mud crab (Scylla paramamosain) after air exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2020, 228, 108652. [Google Scholar] [CrossRef]
- Li, E.; Wang, S.; Li, C.; Wang, X.; Chen, K.; Chen, L. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol. Genomics 2014, 46, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Jiang, K.; Zhang, F.; Zhang, D.; Shen, A.; Jiang, M.; Shen, X.; Ma, L. Effects of various salinities on Na(+)-K(+)-ATPase, Hsp70 and Hsp90 expression profiles in juvenile mitten crabs, Eriocheir sinensis. Genet. Mol. Res. 2012, 11, 978–986. [Google Scholar] [CrossRef]
- Bao, J.; Xing, Y.N.; Jiang, H.B.; Li, X.D. Identification of immune-related genes in gills of Chinese mitten crabs (Eriocheir sinensis) during adaptation to air exposure stress. Fish. Shellfish Immunol. 2019, 84, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, L.B.; Sun, S.F.; Liu, S.Y.; Liu, M.J.; Lin, J. Phylogenetic and ion-response analyses reveal a relationship between gene expansion and functional divergence in the Ca2+/cation antiporter family in Angiosperms. Plant. Mol. Biol. 2021, 105, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Zhao, L.; Xun, X.; Lou, J.; Li, M.; Li, X.; Wang, S.; Zhang, L.; Hu, X.; Bao, Z. Genome-Wide Identification and Characterization of SODs in Zhikong Scallop Reveals Gene Expansion and Regulation Divergence after Toxic Dinoflagellate Exposure. Mar. Drugs. 2019, 17, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, C.B.; Kim, H.S.; Kang, H.M.; Lee, J.S. ATP-binding cassette (ABC) proteins in aquatic invertebrates: Evolutionary significance and application in marine ecotoxicology. Aquat. Toxicol. 2017, 185, 29–39. [Google Scholar] [CrossRef]
- Ru, H.; Mi, W.; Zhang, P.; Alt, F.W.; Schatz, D.G.; Liao, M.; Wu, H. DNA melting initiates the RAG catalytic pathway. Nat. Struct. Mol. Biol. 2018, 25, 732–742. [Google Scholar] [CrossRef]
- Hickman, A.B.; Chandler, M.; Dyda, F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 50–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roussigne, M.; Kossida, S.; Lavigne, A.C.; Clouaire, T.; Ecochard, V.; Glories, A.; Amalric, F.; Girard, J.P. The THAP domain: A novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 2003, 28, 66–69. [Google Scholar] [CrossRef]
- Sabogal, A.; Lyubimov, A.Y.; Corn, J.E.; Berger, J.M.; Rio, D.C. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat. Struct. Mol. Biol. 2010, 17, 117–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Qian, X.; Zhu, F. Molecular characterization of shrimp harbinger transposase derived 1 (HARBI1)-like and its role in white spot syndrome virus and Vibrio alginolyticus infection. Fish Shellfish Immunol. 2018, 78, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Sumiyama, K.; Amemiya, C.T. A Living Fossil in the Genome of a Living Fossil: Harbinger Transposons in the Coelacanth Genome. Mol. Biol. Evol. 2012, 29, 985–993. [Google Scholar] [CrossRef] [Green Version]
- Vidal, N.M.; Grazziotin, A.L.; Iyer, L.M.; Aravind, L.; Venancio, T.M. Transcription factors, chromatin proteins and the diversification of Hemiptera. Insect Biochem. Mol. Biol. 2016, 69, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Malik, H.S.; Henikoff, S. Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster. PloS Genet. 2005, 1, 429–443. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.L.; Hu, T.H.; Wang, W.H.; Hu, H.J.; Wei, Q.Z.; Bao, C.L. Investigation of evolutionary and expressional relationships in the function of the leucine-rich repeat receptor-like protein kinase gene family (LRR-RLK) in the radish (Raphanus sativus L.). Sci. Rep. 2019, 9, 6937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honoo, S.; Toshio, S. Toll-Like Receptors of Deuterostome Invertebrates. Front. Immunol. 2012, 3, 34. [Google Scholar]
- Kapitonov, V.V.; Jurka, J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004, 23, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Alzohairy, A.M.; Gyulai, G.; Jansen, R.K.; Bahieldin, A. Transposable elements domesticated and neofunctionalized by eukaryotic genomes. Plasmid 2013, 69, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.C.; Lu, Q.H.; Xiong, F.; Hao, X.Y.; Wang, L.; Zheng, M.X.; Li, N.N.; Ding, C.Q.; Wang, X.C.; Yang, Y.J. Genome-wide identification, characterization, and expression analysis of nucleotide-binding leucine-rich repeats gene family under environmental stresses in tea (Camellia sinensis). Genomics 2020, 112, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ksepka, D.T.; Parham, J.F.; Allman, J.F.; Benton, M.J.; Carrano, M.T.; Cranston, K.A.; Donoghue, P.C.; Head, J.J.; Hermsen, E.J.; Irmis, R.B.; et al. The Fossil Calibration Database-A New Resource for Divergence Dating. Syst. Biol. 2015, 64, 853–859. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 8, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; Mccue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. A Stat. 2011, 174, 245. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zheng, J.; Yang, Y.; Cui, Z. Genome-Wide Identification, Characterization, and Expression Analysis of DDE_Tnp_4 Family Genes in Eriocheir sinensis. Antibiotics 2021, 10, 1430. https://doi.org/10.3390/antibiotics10121430
Xu Y, Zheng J, Yang Y, Cui Z. Genome-Wide Identification, Characterization, and Expression Analysis of DDE_Tnp_4 Family Genes in Eriocheir sinensis. Antibiotics. 2021; 10(12):1430. https://doi.org/10.3390/antibiotics10121430
Chicago/Turabian StyleXu, Yuanfeng, Jinbin Zheng, Yanan Yang, and Zhaoxia Cui. 2021. "Genome-Wide Identification, Characterization, and Expression Analysis of DDE_Tnp_4 Family Genes in Eriocheir sinensis" Antibiotics 10, no. 12: 1430. https://doi.org/10.3390/antibiotics10121430
APA StyleXu, Y., Zheng, J., Yang, Y., & Cui, Z. (2021). Genome-Wide Identification, Characterization, and Expression Analysis of DDE_Tnp_4 Family Genes in Eriocheir sinensis. Antibiotics, 10(12), 1430. https://doi.org/10.3390/antibiotics10121430





