A Closer Look at Dexamethasone and the SARS-CoV-2-Induced Cytokine Storm: In Silico Insights of the First Life-Saving COVID-19 Drug
Abstract
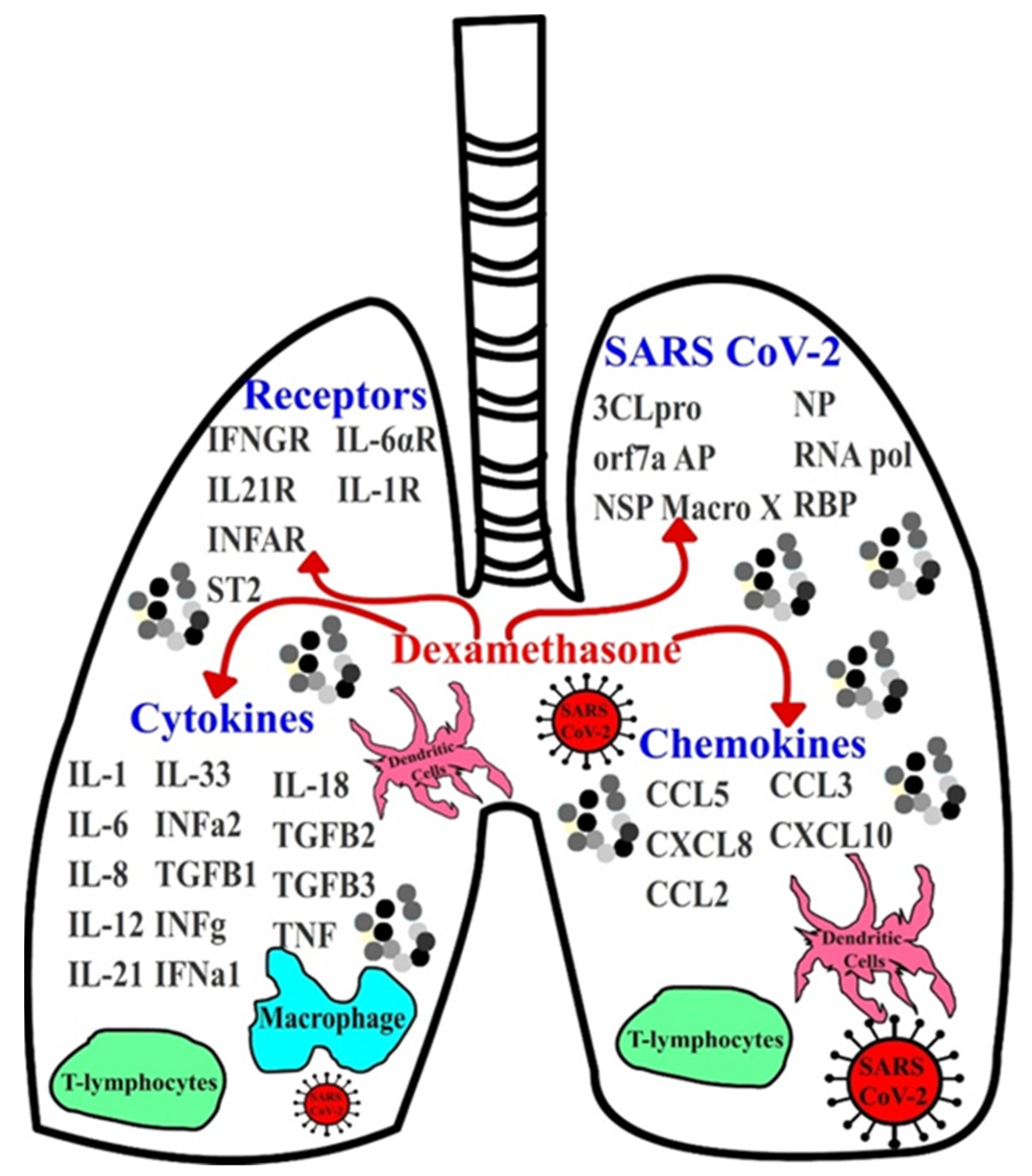
:1. Introduction
2. Materials and Methods
2.1. Cytokines, Chemokines, Receptors, SARS-CoV-2 Proteins, PDB Files
2.2. Docking of Dexamethasone
2.3. MD Simulations of Dexamethasone Complexes
3. Results and Discussion
3.1. Implications of Dexamethasone on Cytokine and Chemokine Suppression
3.2. Dexamethasone as a Potential SARS-CoV-2 Inhibitor
3.3. MD Simulations of Dexamethasone–Cytokine/Chemokine Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Group, R.C.; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with Covid-19—Preliminary Report. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Sharun, K.; Tiwari, R.; Dhama, J.; Dhama, K. Dexamethasone to combat cytokine storm in COVID-19: Clinical trials and preliminary evidence. Int. J. Surg. 2020, 82, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ahmet, A.; Ward, L.; Krishnamoorthy, P.; Mandelcorn, E.D.; Leigh, R.; Brown, J.P.; Cohen, A.; Kim, H. A practical guide to the monitoring and management of the complications of systemic corticosteroid therapy. Allergy Asthma Clin. Immunol. 2013, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammers, T.; Sofias, A.M.; van der Meel, R.; Schiffelers, R.; Storm, G.; Tacke, F.; Koschmieder, S.; Brümmendorf, T.H.; Kiessling, F.; Metselaar, J.M. Dexamethasone nanomedicines for COVID-19. Nat. Nanotechnol. 2020, 15, 622–624. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- BioSolveIT. SeeSAR Version 10.1. 2020. Available online: https://www.biosolveit.de (accessed on 9 March 2020).
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef] [Green Version]
- Warren, G.L.; Andrews, C.W.; Capelli, A.-M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Gastreich, M.; Lilienthal, M.; Briem, H.; Claussen, H. Ultrafast de novo docking combining pharmacophores and combinatorics. J. Comput.-Aided Mol. Des. 2006, 20, 717–734. [Google Scholar] [CrossRef]
- Schneider, N.; Lange, G.; Hindle, S.; Klein, R.; Rarey, M. A consistent description of HYdrogen bond and DEhydration energies in protein–ligand complexes: Methods behind the HYDE scoring function. J. Comput.-Aided Mol. Des. 2013, 27, 15–29. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrish-Novak, J.; Dillon, S.R.; Nelson, A.; Hammond, A.; Sprecher, C.; Gross, J.A.; Johnston, J.; Madden, K.; Xu, W.; West, J.; et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature 2000, 408, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003, 3, 133–146. [Google Scholar] [CrossRef]
- Schett, G.; Sticherling, M.; Neurath, M.F. COVID-19: Risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol. 2020, 20, 271–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contassot, E.; Beer, H.D.; French, L.E. Interleukin-1, inflammasomes, autoinflammation and the skin. Swiss Med. Wkly. 2012, 142, w13590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Veerdonk, F.L.; Netea, M.G. Blocking IL-1 to prevent respiratory failure in COVID-19. Crit. Care 2020, 24, 445. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Bickel, M. The role of interleukin-8 in inflammation and mechanisms of regulation. J. Periodontol. 1993, 64, 456–460. [Google Scholar] [PubMed]
- Mogensen, T.H.; Berg, R.S.; Paludan, S.R.; Østergaard, L. Mechanisms of dexamethasone-mediated inhibition of Toll-like receptor signaling induced by Neisseria meningitidis and Streptococcus pneumoniae. Infect. Immun. 2008, 76, 189–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monick, M.M.; Aksamit, T.R.; Geist, L.J.; Hunninghake, G.W. Dexamethasone inhibits IL-1 and TNF activity in human lung fibroblasts without affecting IL-1 or TNF receptors. Am. J. Physiol. 1994, 267, L33–L38. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.C.L.; Lam, C.W.K.; Tam, L.-S.; Wong, C.K. IL33: Roles in Allergic Inflammation and Therapeutic Perspectives. Front. Immunol. 2019, 10, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carsana, L.; Sonzogni, A.; Nasr, A.; Rossi, R.S.; Pellegrinelli, A.; Zerbi, P.; Rech, R.; Colombo, R.; Antinori, S.; Corbellino, M.; et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infect. Dis. 2020, 20, 1135–1140. [Google Scholar] [CrossRef]
- Sokol, C.L.; Luster, A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016303. [Google Scholar] [CrossRef] [Green Version]
- Maghazachi, A.A.; Al-Aoukaty, A.; Schall, T.J. CC chemokines induce the generation of killer cells from CD56+ cells. Eur. J. Immunol. 1996, 26, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Eldanasory, O.A.; Eljaaly, K.; Memish, Z.A.; Al-Tawfiq, J.A. Histamine release theory and roles of antihistamine in the treatment of cytokines storm of COVID-19. Travel. Med. Infect. Dis. 2020, 37, 101874. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, M.; Yamada, H.; Sato, K.; Shigemi, H.; Umeda, Y.; Morikawa, M.; Waseda, Y.; Anzai, M.; Kamide, Y.; Aoki-Saito, H.; et al. Extracellular acidification-induced CXCL8 production through a proton-sensing receptor OGR1 in human airway smooth muscle cells: A response inhibited by dexamethasone. J. Inflamm. 2019, 16, 4. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, S.; Wang, J.; Xue, Z.; Wang, C.; Wang, N. Dexamethasone inhibits SARS-CoV-2 spike pseudotyped virus viropexis by binding to ACE2. Virology 2021, 554, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Gresser, I. On intuition and the discovery of interferon. Cytokine Growth Factor Rev. 2015, 26, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Nüchel, J.; Ghatak, S.; Zuk, A.V.; Illerhaus, A.; Mörgelin, M.; Schönborn, K.; Blumbach, K.; Wickström, S.A.; Krieg, T.; Sengle, G.; et al. TGFB1 is secreted through an unconventional pathway dependent on the autophagic machinery and cytoskeletal regulators. Autophagy 2018, 14, 465–486. [Google Scholar] [CrossRef] [Green Version]
- Letterio, J.J.; Roberts, A.B. Regulation of immune responses by TGF-beta. Annu. Rev. Immunol. 1998, 16, 137–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grillo, F.; Barisione, E.; Ball, L.; Mastracci, L.; Fiocca, R. Lung fibrosis: An undervalued finding in COVID-19 pathological series. Lancet Infect. Dis. 2020, 21, e72. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, T.; Tukiyama, T.; Tokiwa, T. Effect of dexamethasone on binding activity of transcription factors nuclear factor-κB and activator protein-1 in SW982 human synovial sarcoma cells. Vitr. Cell. Dev. Biol. Anim. 2005, 41, 80–82. [Google Scholar] [CrossRef]
- Zehra, Z.; Luthra, M.; Siddiqui, S.M.; Shamsi, A.; Gaur, N.A.; Islam, A. Corona virus versus existence of human on the earth: A computational and biophysical approach. Int. J. Biol. Macromol. 2020, 161, 271–281. [Google Scholar] [CrossRef]
- Shamsi, A.; Mohammad, T.; Anwar, S.; Amani, S.; Khan, M.S.; Husain, F.M.; Rehman, M.T.; Islam, A.; Hassan, M.I. Potential drug targets of SARS-CoV-2: From genomics to therapeutics. Int. J. Biol. Macromol. 2021, 177, 1–9. [Google Scholar] [CrossRef]
- Shamsi, A.; Mohammad, T.; Anwar, S.; AlAjmi, M.F.; Hussain, A.; Rehman, M.T.; Islam, A.; Hassan, M.I. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: Possible implication in COVID-19 therapy. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, T.; Shamsi, A.; Anwar, S.; Umair, M.; Hussain, A.; Rehman, M.T.; AlAjmi, M.F.; Islam, A.; Hassan, M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: Towards the development of effective COVID-19 therapy. Virus Res. 2020, 288, 198102. [Google Scholar] [CrossRef]
- Frick, D.N.; Virdi, R.S.; Vuksanovic, N.; Dahal, N.; Silvaggi, N.R. Variable Macro X Domain of SARS-CoV-2 Retains the Ability to Bind ADP-ribose. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef] [PubMed]
- AlAjmi, M.F.; Azhar, A.; Owais, M.; Rashid, S.; Hasan, S.; Hussain, A.; Rehman, M.T. Antiviral potential of some novel structural analogs of standard drugs repurposed for the treatment of COVID-19. J. Biomol. Struct. Dyn. 2020, 39, 6676–6688. [Google Scholar] [CrossRef] [PubMed]




| Category | Target | ΔG [kcal/mol] | Ki [M] | IC50 [M] |
|---|---|---|---|---|
| Receptor | IL-33 receptor (IL-1RAcP) | −11.095 | 7.400 × 10−9 | 1.472 × 10−8 |
| Receptor | IFNG receptor (IFNGR) | −11.040 | 8.100 × 10−9 | 1.616 × 10−8 |
| Receptor | IL-21 receptor (IL21R) | −10.722 | 1.400 × 10−8 | 2.762 × 10−8 |
| Cytokine | IL-21 | −8.935 | 2.820 × 10−7 | 5.640 × 10−7 |
| Receptor | INFA2 receptor (INFAR) | −8.917 | 2.900 × 10−7 | 5.813 × 10−7 |
| Receptor | IL-33 receptor (ST2) | −8.890 | 3.000 × 10−7 | 6.080 × 10−7 |
| Cytokine | IL-12 | −8.793 | 3.590 × 10−7 | 7.170 × 10−7 |
| Chemokine | CCL5 | −8.514 | 5.740 × 10−7 | 1.150 × 10−6 |
| Cytokine | IL-1 | −8.429 | 6.620 × 10−7 | 1.320 × 10−6 |
| TGFβ-1 | −8.304 | 8.180 × 10−7 | 1.640 × 10−6 | |
| Cytokine | INF-γ | −8.272 | 8.630 × 10−7 | 1.730 × 10−6 |
| Cytokine | INFα2 | −8.205 | 9.670 × 10−7 | 1.930 × 10−6 |
| Receptor | IL-6αR-gp130 | −8.043 | 1.300 × 10−6 | 2.543 × 10−6 |
| NSP macro X | −7.929 | 1.540 × 10−6 | 3.083 × 10−6 | |
| Cytokine | IL-33 | −7.419 | 3.640 × 10−6 | 7.280 × 10−6 |
| Cytokine | IL-8 | −7.318 | 4.320 × 10−6 | 8.640 × 10−6 |
| Chemokine | CXCL8 | −7.318 | 4.320 × 10−6 | 8.640 × 10−6 |
| Cytokine | IL-6 | −6.820 | 1.000 × 10−5 | 2.000 × 10−5 |
| 3CLpro | −5.762 | 5.970 × 10−5 | 1.190 × 10−4 | |
| Receptor | IL-1RA receptor (IL−1R) | −5.597 | 7.900 × 10−5 | 1.570 × 10−4 |
| Chemokine | CCL2 | NA | ||
| Chemokine | CCL3 | N/A | ||
| Chemokine | CXCL10 | N/A | ||
| Cytokine | INFα-1 | N/A | ||
| NP | N/A | |||
| Cytokine | TGFβ-2 | N/A | ||
| Cytokine | TGFβ-3 | N/A | ||
| Cytokine | TNF-α | N/A | ||
| RNA Pol | N/A | |||
| RBP | N/A | |||
| Orf7a AP | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan, P.; Arnold, S.J.; Hsiao, N.-W.; Shu, C.-W. A Closer Look at Dexamethasone and the SARS-CoV-2-Induced Cytokine Storm: In Silico Insights of the First Life-Saving COVID-19 Drug. Antibiotics 2021, 10, 1507. https://doi.org/10.3390/antibiotics10121507
Morgan P, Arnold SJ, Hsiao N-W, Shu C-W. A Closer Look at Dexamethasone and the SARS-CoV-2-Induced Cytokine Storm: In Silico Insights of the First Life-Saving COVID-19 Drug. Antibiotics. 2021; 10(12):1507. https://doi.org/10.3390/antibiotics10121507
Chicago/Turabian StyleMorgan, Paul, Shareen J. Arnold, Nai-Wan Hsiao, and Chih-Wen Shu. 2021. "A Closer Look at Dexamethasone and the SARS-CoV-2-Induced Cytokine Storm: In Silico Insights of the First Life-Saving COVID-19 Drug" Antibiotics 10, no. 12: 1507. https://doi.org/10.3390/antibiotics10121507
APA StyleMorgan, P., Arnold, S. J., Hsiao, N. -W., & Shu, C. -W. (2021). A Closer Look at Dexamethasone and the SARS-CoV-2-Induced Cytokine Storm: In Silico Insights of the First Life-Saving COVID-19 Drug. Antibiotics, 10(12), 1507. https://doi.org/10.3390/antibiotics10121507








