The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement
Abstract
:1. Introduction
2. Results
2.1. Antibiotic Consumption
2.2. Resistance Phenotype
2.3. Prevalence of the Resistance Genes
2.4. Whole Genome Sequencing (WGS)
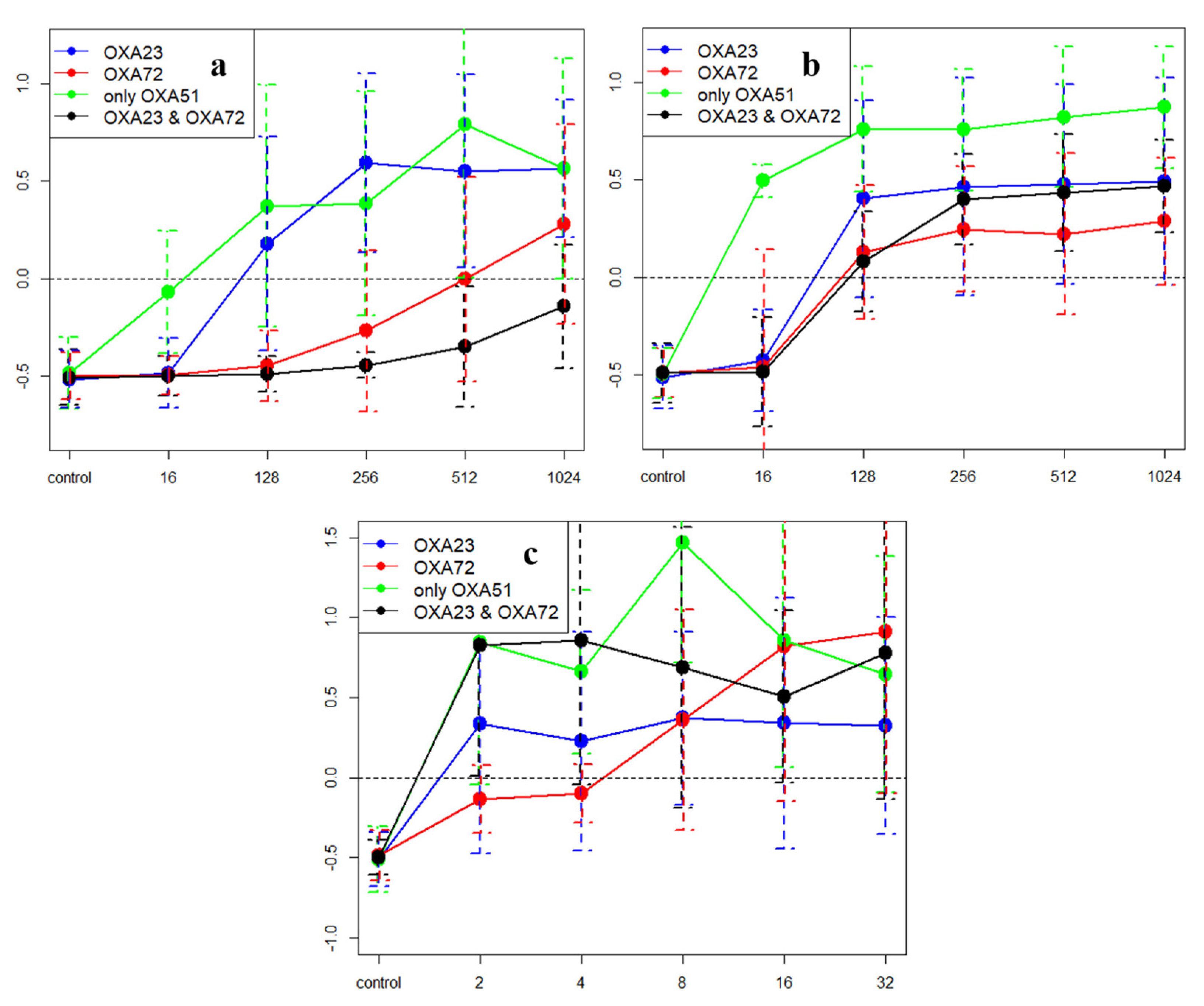
2.5. Time-Kill Assays
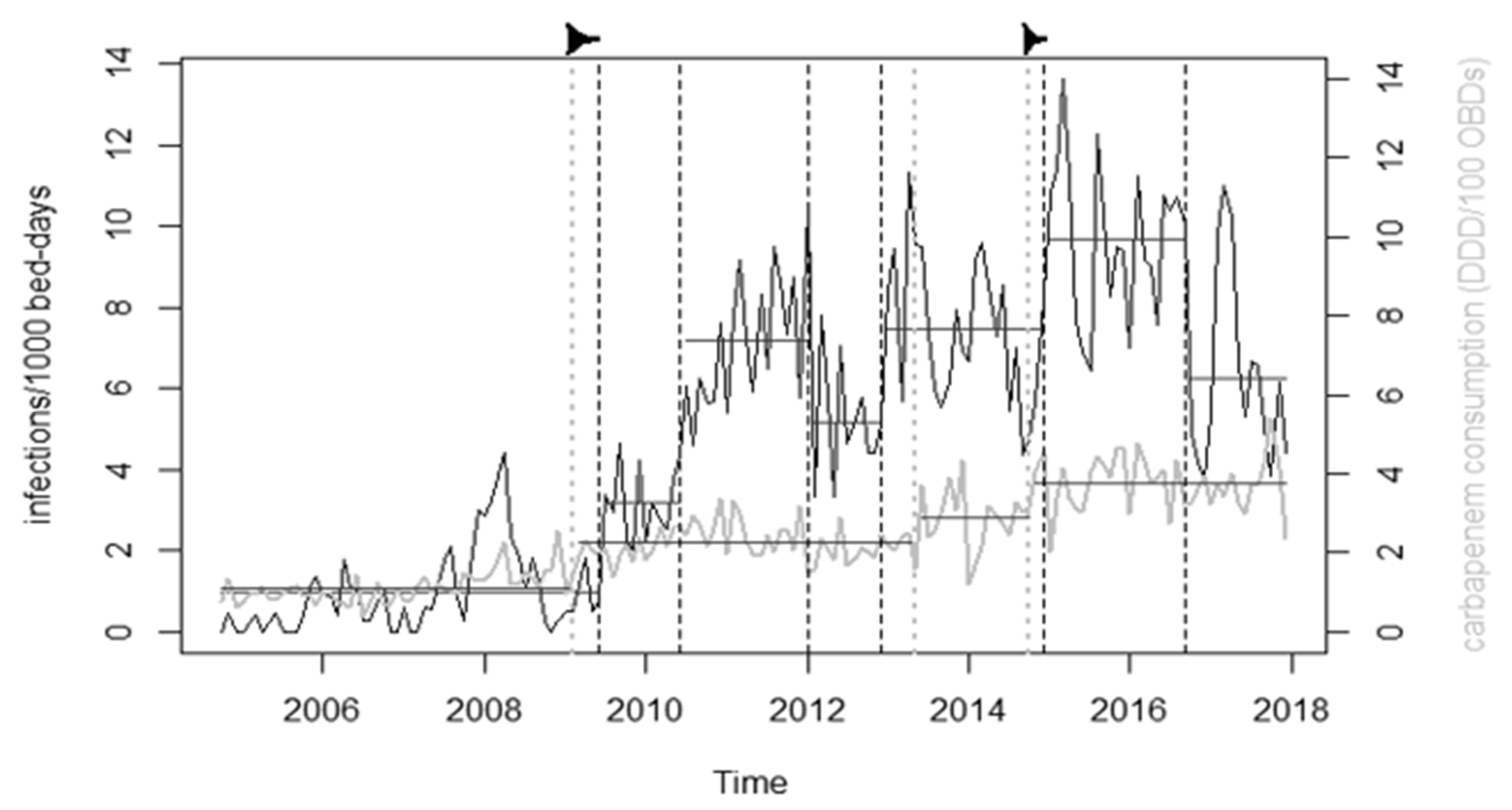
2.6. Change-Point Analysis
3. Discussion
4. Materials and Methods
4.1. Clinical Isolates
4.2. Antibiotic Consumption and Changepoint Analysis
4.3. Resistance Genes
4.4. Whole Genome Sequencing
4.5. Time-Kill Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergogne-Berezin, E.; Towner, K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996, 9, 148. [Google Scholar] [CrossRef]
- Lopez-Otsoa, F.; Gallego, L.; Towner, K.J.; Tysall, L.; Woodford, N.; Livermore, D.M. Endemic carbapenem resistance associated with OXA-40 carbapenemase among Acinetobacter baumannii isolates from a hospital in northern Spain. J. Clin. Microbiol. 2002, 40, 4741–4743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkshoorn, L.; Aucken, H.; Gerner-Smidt, P.; Janssen, P.; Kaufmann, M.E.; Garaizar, J.; Ursing, J.; Pitt, T.L. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 1996, 34, 1519–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paton, R.; Miles, R.S.; Hood, J.; Amyes, S.G.B. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 1993, 2, 81–87. [Google Scholar] [CrossRef]
- Bou, G.; Martínez-Beltrán, J. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2000, 44, 428–432. [Google Scholar] [PubMed] [Green Version]
- Héritier, C.; Poirel, L.; Fournier, P.E.; Claverie, J.M.; Raoult, D.; Nordmann, P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 4174–4179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.; Amyes, S.G.B. The sequences of seven class D β-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 2005, 11, 326–329. [Google Scholar]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.B.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Brown, S.; Amyes, S. OXA β-lactamases in Acinetobacter: The story so far. J. Antimicrob. Chemother. 2005, 57, 1–3. [Google Scholar] [CrossRef]
- Lolans, K.; Rice, T.W.; Munoz-Price, L.S.; Quinn, J.P. Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother. 2006, 50, 2941–2945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turton, J.F.; Kaufmann, M.E.; Glover, J.; Coelho, J.M.; Warner, M.; Pike, R.; Pitt, T.L. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 2005, 43, 3074–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marqué, S.; Poirel, L.; Héritier, C.; Brisse, S.; Blasco, M.D.; Filip, R.; Coman, G.; Naas, T.; Nordmann, P. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 2005, 43, 4885–4888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalla-Costa, L.M.; Coelho, J.M.; Souza, H.A.; Castro, M.E.; Stier, C.J.; Bragagnolo, K.L.; Rea-Neto, A.; Penteado-Filho, S.R.; Livermore, D.M.; Woodford, N. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J. Clin. Microbiol. 2003, 41, 3403–3406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, B.C.; Jeong, S.H.; Bae, I.K.; Kwon, S.B.; Lee, K.; Young, D.; Lee, J.H.; Song, J.S.; Lee, S.H. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 β-lactamase in Korea. J. Clin. Microbiol. 2005, 43, 2241–2245. [Google Scholar] [CrossRef] [Green Version]
- Naas, T.; Levy, M.; Hirschauer, C.; Marchandin, H.; Nordmann, P. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J. Clin. Microbiol. 2005, 43, 4826–4829. [Google Scholar] [CrossRef] [Green Version]
- Mózes, J.; Ebrahimi, F.; Gorácz, O.; Miszti, C.; Kardos, G. Effect of carbapenem consumption patterns on the molecular epidemiology and carbapenem resistance of Acinetobacter baumannii. J. Med. Microbiol. 2014, 63, 1654–1662. [Google Scholar] [CrossRef] [Green Version]
- Buisson, Y.; Van Nhieu, G.T.; Ginot, L.; Bouvet, P.; Schill, H.; Driot, L.; Meyran, M. Nosocomial outbreaks due to amikacin-resistant tobramycin-sensitive Acinetobacter species: Correlation with amikacin usage. J. Hosp. Infect. 2011, 15, 83–93. [Google Scholar]
- Villers, D.; Espaze, E.; Coste-Burel, M.; Giauffret, F.; Ninin, E.; Nicolas, F.; Richet, H. Nosocomial Acinetobacter baumannii infections: Microbiological and clinical epidemiology. Ann. Intern. Med. 1998, 129, 182–189. [Google Scholar] [CrossRef]
- Dortet, L.; Bonnin, R.A.; Bernabeu, S.; Escaut, L.; Vittecoq, D.; Girlich, D.; Imanci, D.; Fortineau, N.; Naas, T. First occurrence of OXA-72-producing Acinetobacter baumannii in Serbia. Antimicrob. Agents Chemother. 2016, 60, 5724–5730. [Google Scholar] [CrossRef] [Green Version]
- Povilonis, J.; Šeputienė, V.; Krasauskas, R.; Juškaitė, R.; Miškinytė, M.; Sužiedėlis, K.; Sužiedėlienė, E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013, 68, 1000–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnaud, G.; Zihoune, N.; Ricard, J.D.; Hippeaux, M.C.; Eveillard, M.; Dreyfuss, D.; Branger, C. Two sequential outbreaks caused by multidrug-resistant Acinetobacter baumannii isolates producing OXA-58 or OXA-72 oxacillinase in an intensive care unit in France. J. Hosp. Infect. 2010, 76, 358–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franolić-Kukina, I.; Bedenić, B.; Budimir, A.; Herljević, Z.; Vraneš, J.; Higgins, P.G. Clonal spread of carbapenem-resistant OXA-72-positive Acinetobacter baumannii in a Croatian university hospital. Int. J. Infect. Dis. 2011, 15, e706–e709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, S.C.; Yang, S.P.; Lee, Y.T.; Chuang, H.C.; Chen, C.P.; Chang, C.L.; Chen, T.-L.; Lu, P.L.; Hsueh, P.R.; Fung, C.P. Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne bla OXA-72 in Taiwan. BMC Infect. Dis. 2013, 13, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saavedra, S.Y.; Cayô, R.; Gales, A.C.; Leal, A.L.; Saavedra, C.H. Early dissemination of OXA-72-producing Acinetobacter baumannii strain in Colombia: A case report. Braz. J. Infect. Dis. 2014, 18, 678–680. [Google Scholar] [CrossRef] [Green Version]
- Vasconcelos, A.T.R.; Barth, A.L.; Zavascki, A.P.; Gales, A.C.; Levin, A.S.; Lucarevschi, B.R.; Cabral, B.G.; Brasiliense, D.M.; Rossi, F.; Furtado, G.H.C.; et al. The changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: A BrasNet report. Diagn. Microbiol. Infect. Dis. 2015, 83, 382–385. [Google Scholar] [CrossRef]
- Zarrilli, R.; Giannouli, M.; Rocco, F.; Loman, N.J.; Haines, A.S.; Constantinidou, C.; Pallen, M.J.; Triassi, M.; Di Nocera, P.P. Genome sequences of three Acinetobacter baumannii strains assigned to the multilocus sequence typing genotypes ST2, ST25, and ST78. J. Bacteriol. 2011, 193, 2359–2360. [Google Scholar] [CrossRef] [Green Version]
- Scerpella, E.G.; Wanger, A.R.; Armitige, L.; Anderlini, P.; Ericsson, C.D. Nosocomial outbreak caused by a multiresistant clone of Acinetobacter baumannii: Results of the case-control and molecular epidemiologic investigations. Infect. Control. Hosp. Epidemiol. 1995, 16, 92–97. [Google Scholar]
- Mulin, B.; Talon, D.; Viel, J.F.; Vincent, C.; Leprat, R.; Thouverez, M.; Michel-Briand, Y. Risk factors for nosocomial colonization with multiresistant Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 1995, 14, 569–576. [Google Scholar] [CrossRef]
- Manikal, V.M.; Landman, D.; Saurina, G.; Oydna, E.; Lal, H.; Quale, J. Endemic carbapenem-resistant Acinetobacter species in Brooklyn, New York: Citywide prevalence, interinstitutional spread, and relation to antibiotic usage. Clin. Infect. Dis. 2000, 31, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Go, E.S.; Urban, C.; Burns, J.; Mariano, N.; Mosinka-Snipas, K.; Rahal, J.J.; Kreiswirth, B.; Eisner, W. Clinical and molecular epidemiology of Acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet 1994, 344, 1329–1332. [Google Scholar] [CrossRef]
- Tóth, H.; Fésűs, A.; Kungler-Gorácz, O.; Balázs, B.; Majoros, L.; Szarka, K.; Kardos, G. Utilization of vector autoregressive and linear transfer models to follow up the antibiotic resistance spiral in gram-negative bacteria from cephalosporin consumption to colistin resistance. Clin. Infect. Dis. 2019, 69, 1410–1421. [Google Scholar]
- Villalobos, A.P.; Barrero, L.I.; Rivera, S.M.; Ovalle, M.V.; Valera, D. Vigilancia de infecciones asociadas a la atención en salud, resistencia bacteriana y consumo de antibióticos en hospitales de alta complejidad, Colombia. Biomédica 2014, 34, 67–80. [Google Scholar] [CrossRef] [Green Version]
- López-Lozano, J.M.; Lawes, T.; Nebot, C.; Beyaert, A.; Bertrand, X.; Hocquet, D.; Aldeyab, M.; Scott, M.; Conlon-Bingham, G.; Farren, D.; et al. A nonlinear time-series analysis approach to identify thresholds in associations between population antibiotic use and rates of resistance. Nat. Microbiol. 2019, 4, 1160–1172. [Google Scholar] [PubMed]
- Baym, M.; Lieberman, T.D.; Kelsic, E.D.; Chait, R.; Gross, R.; Yelin, I.; Kishony, R. Spatiotemporal microbial evolution on antibiotic landscapes. Science 2016, 353, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipsitch, M.; Samore, M.H. Antimicrobial use and antimicrobial resistance: A population perspective. Emerg. Infect. Dis. 2002, 8, 347. [Google Scholar] [CrossRef]
- Almagor, J.; Temkin, E.; Benenson, I.; Fallach, N.; Carmeli, Y.; DRIVE-AB Consortium. The impact of antibiotic use on transmission of resistant bacteria in hospitals: Insights from an agent-based model. PLoS ONE 2018, 13, e0197111. [Google Scholar]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Sokurenko, E.V.; Tchesnokova, V. Intensity and mechanisms of fluoroquinolone resistance within the H30 and H30Rx subclones of Escherichia coli sequence type 131 compared with other fluoroquinolone-resistant E. coli. Antimicrob. Agents Chemother. 2015, 59, 4471–4480. [Google Scholar] [CrossRef] [Green Version]
- Lawes, T.; López-Lozano, J.M.; Nebot, C.; Macartney, G.; Subbarao-Sharma, R.; Dare, C.R.; Edwards, G.F.S.; Gould, I.M. Turning the tide or riding the waves? Impacts of antibiotic stewardship and infection control on MRSA strain dynamics in a Scottish region over 16 years: Non-linear time series analysis. BMJ Open 2015, 5. [Google Scholar] [CrossRef]
- Frana, T.S.; Carlson, S.A.; Griffith, R.W. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 2001, 67, 445–448. [Google Scholar] [CrossRef] [Green Version]
- Vila, J.; Ruiz, J.; Navia, M.; Becerril, B.; Garcia, I.; Perea, S.; Lopez-Hernandez, I.; Alamo, I.; Ballester, F.; Planes, A.M.; et al. Spread of amikacin resistance in Acinetobacter baumannii strains isolated in Spain due to an epidemic strain. J. Clin. Microbiol. 1999, 37, 758–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaerts, P.; Galimand, M.; Bauraing, C.; Deplano, A.; Vanhoof, R.; De Mendonca, R.; Rodriguez-Villalobos, H.; Struelens, M.; Glupczynski, Y. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 2007, 59, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef] [PubMed]


| Isolate | Sample | Year | ST | CHDLs | Resistance Genes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bl | Qui | Agl | Sul | Mac | Phe | Tet | |||||
| 4151 Ac.No.: SRX9354489 | Blood | 2010 | 2 | blaOXA-23 blaOXA-69 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 8266 Ac.No.: SRX9354496 | Bronchial | 2010 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catb8 | tet(B) |
| 8419 Ac.No.: SRX9354497 | Bronchial | 2010 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 2738 Ac.No.: SRX9354478 | Throat | 2010 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 2037 Ac.No.: SRX9354477 | Abscess | 2010 | 1 | blaOXA-23 blaOXA-69 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-VIa | sul1 | ||||
| 8160 Ac.No.: SRX9354495 | Urine | 2010 | 1 | blaOXA-69 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-VIa | sul1 | ||||
| 8664 Ac. No.: SRX9354494 | Canule | 2010 | 45 | blaOXA-23 blaOXA-66 | blaADC-25 | aph(3′)-Ia aph(3′)-VIa | sul1 | catA1 | |||
| 2729 Ac.No.: SRX9354493 | Bronchial | 2010 | 45 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aph(3′)-Ia | sul1 | catA1 | tet(A) | |
| 13803 Ac.No.: SRX9354498 | Blood | 2010 | 79 | blaOXA-24 blaOXA-65 | blaADC-25 blaTEM-1B | aac(3)-IIa aac(6′)-Ian aph(3′’)-Ib aph(3′)-VIa aph(6)-Id armA | sul2 | cmlB1 | |||
| 5508 Ac.No.: SRX9354499 | Bronchial | 2017 | 2 | blaOXA-23 blaOXA-66 | blaOXA-23 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 6759 Ac.No.: SRX9354479 | Sputum | 2017 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 8363 Ac.No.: SRX9354482 | Bronchial | 2017 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 9032 Ac.No.: SRX9354481 | Sputum | 2017 | 2 | blaOXA-23 blaOXA-66 | blaADC-25 | aac(6′)-Ib-cr | aac(6′)-Ib3 aadA1 aph(3′’)-Ib aph(3′)-Ia aph(3′)-VIa aph(6)-Id armA | sul1 | mph(E) msr(E) | catB8 | tet(B) |
| 230 Ac.No.: SRX9354485 | Bronchial | 2017 | 636 | blaOXA-72 blaOXA-66 | aac(3)-Ia aadA1 aph(3′)-Ia | sul1 | catA1 | ||||
| 511 Ac.No.: SRX9354486 | Bronchial | 2017 | 636 | blaOXA-72 blaOXA-66 | aac(3)-Ia aadA1 aph(3′)-Ia | sul1 | catA1 | ||||
| 3603 Ac.No.: SRX9354487 | Bronchial | 2017 | 636 | blaOXA-72 blaOXA-23 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-Ia aph(3′)-VIa | sul1 | catA1 | |||
| 9930 Ac.No.: SRX9354492 | Bronchial | 2017 | 636 | blaOXA-72 blaOXA-23 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-Ia aph(3′)-VIa | sul1 | ||||
| 10135 Ac.No.: SRX9354488 | Nasal | 2017 | 636 | blaOXA-72 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 | sul1 | catA1 | |||
| 10177 Ac.No.: SRX9354490 | Throat | 2017 | 636 | blaOXA-72 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 | sul1 | catA1 | |||
| 10992 Ac.No.: SRX9354483 | Bronchial | 2017 | 636 | blaOXA-72 blaOXA-23 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-Ia aph(3′)-VIa | sul1 | ||||
| 8733 Ac.No.: SRX9354491 | Drain | 2017 | 636 | blaOXA-72 blaOXA-66 | blaADC-25 | aac(3)-Ia aadA1 aph(3′)-Ia | sul1 | ||||
| 3593 Ac.No.: SRX9354480 | Bronchial | 2017 | 492 | blaOXA-72 blaOXA-66 | blaADC-25 | aadA2 aph(3′’)-Ib aph(6)-IdarmA | sul1 sul2 | mph(E) msr(E) | tet(B) | ||
| 8429 Ac.No.: SRX9354484 | Trachea | 2017 | 492 | blaOXA-72 blaOXA-66 | blaADC-25 | aadA2 aph(3′’)-Ib aph(6)-IdarmA | sul1 sul2 | mph(E) msr(E) | tet(B) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balázs, B.; Tóth, Z.; Nagy, F.; Kovács, R.; Tóth, H.; Nagy, J.B.; Tóth, Á.; Szarka, K.; Majoros, L.; Kardos, G. The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement. Antibiotics 2021, 10, 127. https://doi.org/10.3390/antibiotics10020127
Balázs B, Tóth Z, Nagy F, Kovács R, Tóth H, Nagy JB, Tóth Á, Szarka K, Majoros L, Kardos G. The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement. Antibiotics. 2021; 10(2):127. https://doi.org/10.3390/antibiotics10020127
Chicago/Turabian StyleBalázs, Bence, Zoltán Tóth, Fruzsina Nagy, Renátó Kovács, Hajnalka Tóth, József Bálint Nagy, Ákos Tóth, Krisztina Szarka, László Majoros, and Gábor Kardos. 2021. "The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement" Antibiotics 10, no. 2: 127. https://doi.org/10.3390/antibiotics10020127








