First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Phenotypic Tests of K. pneumoniae MS3802
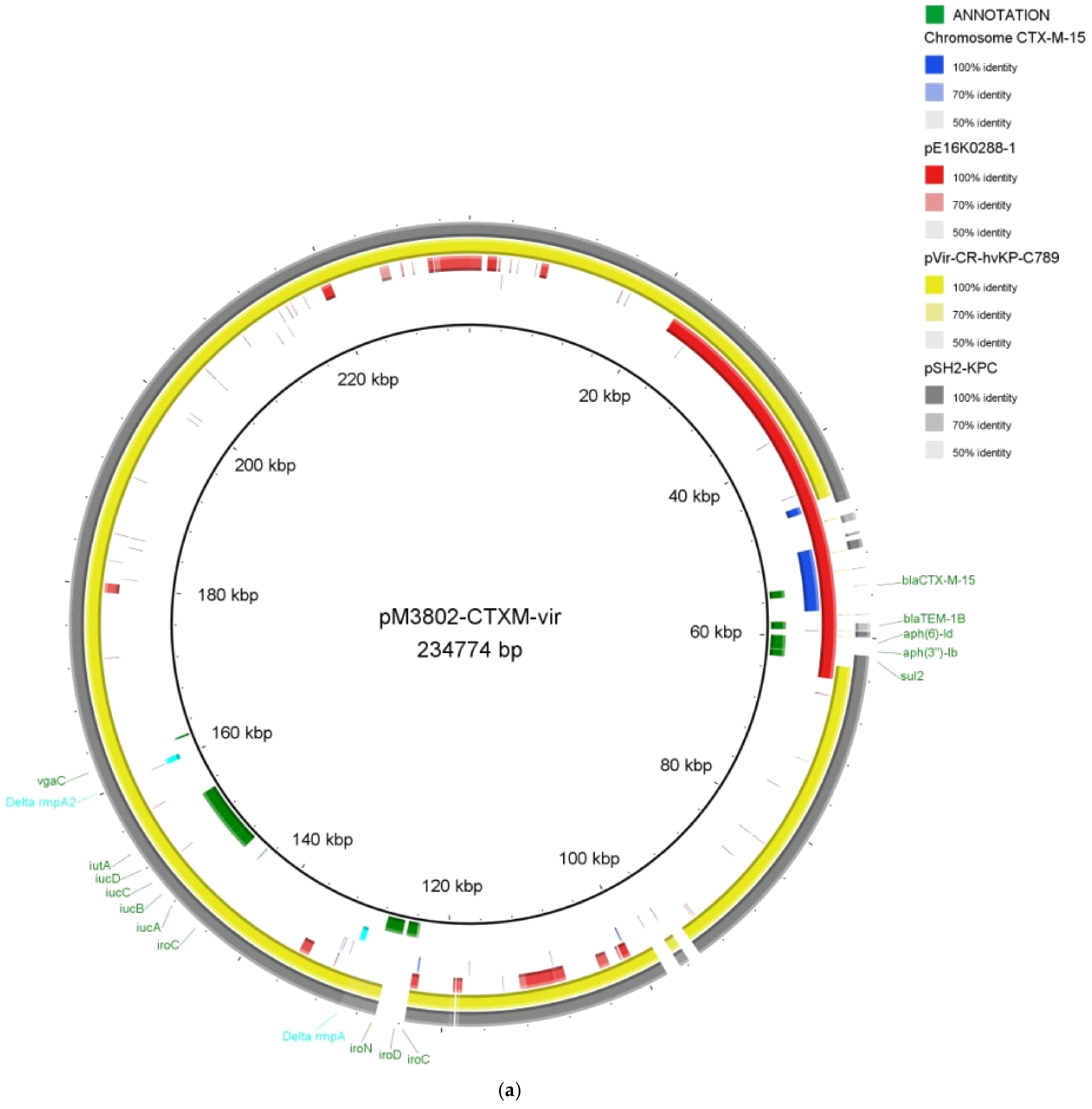
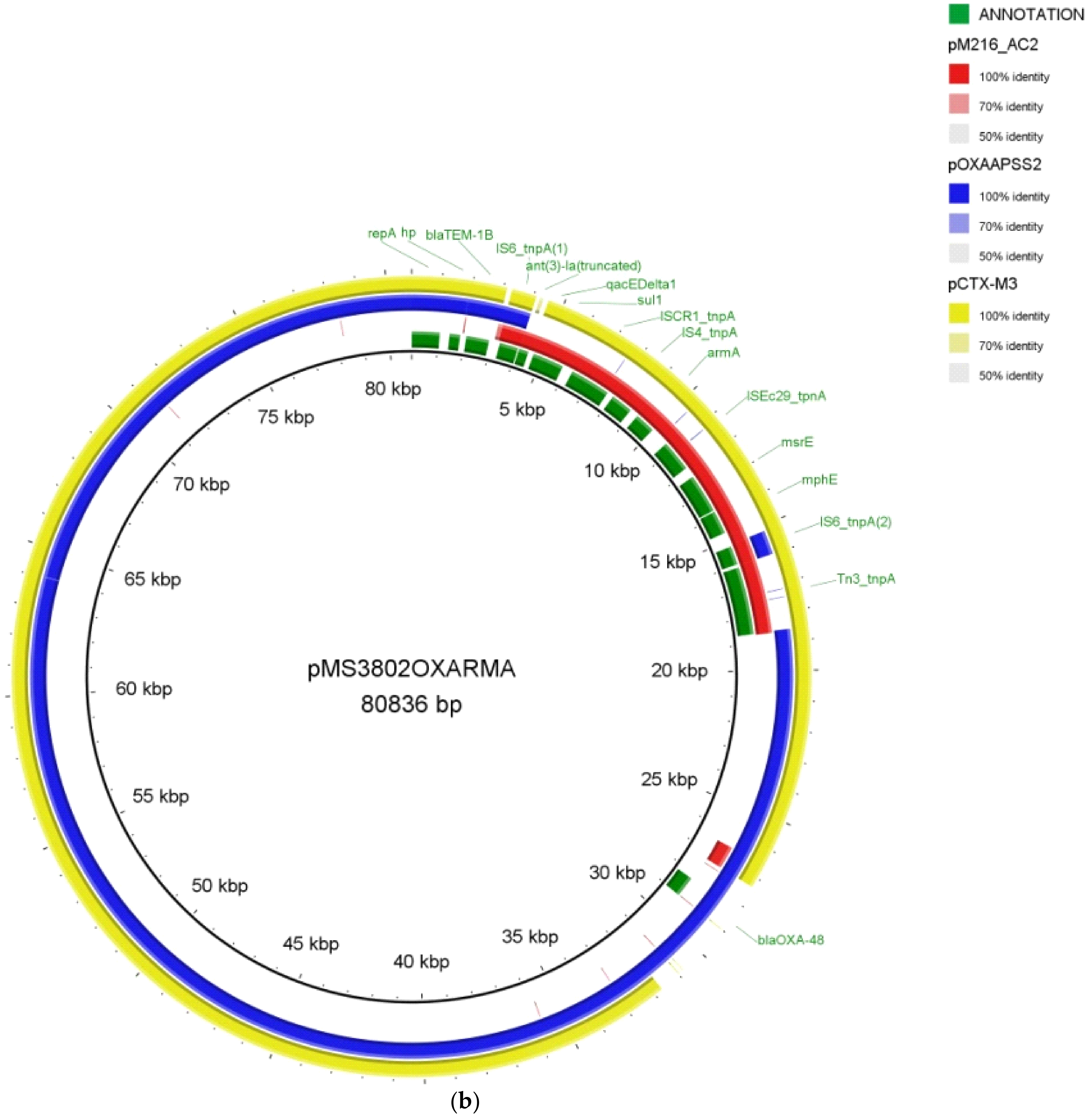
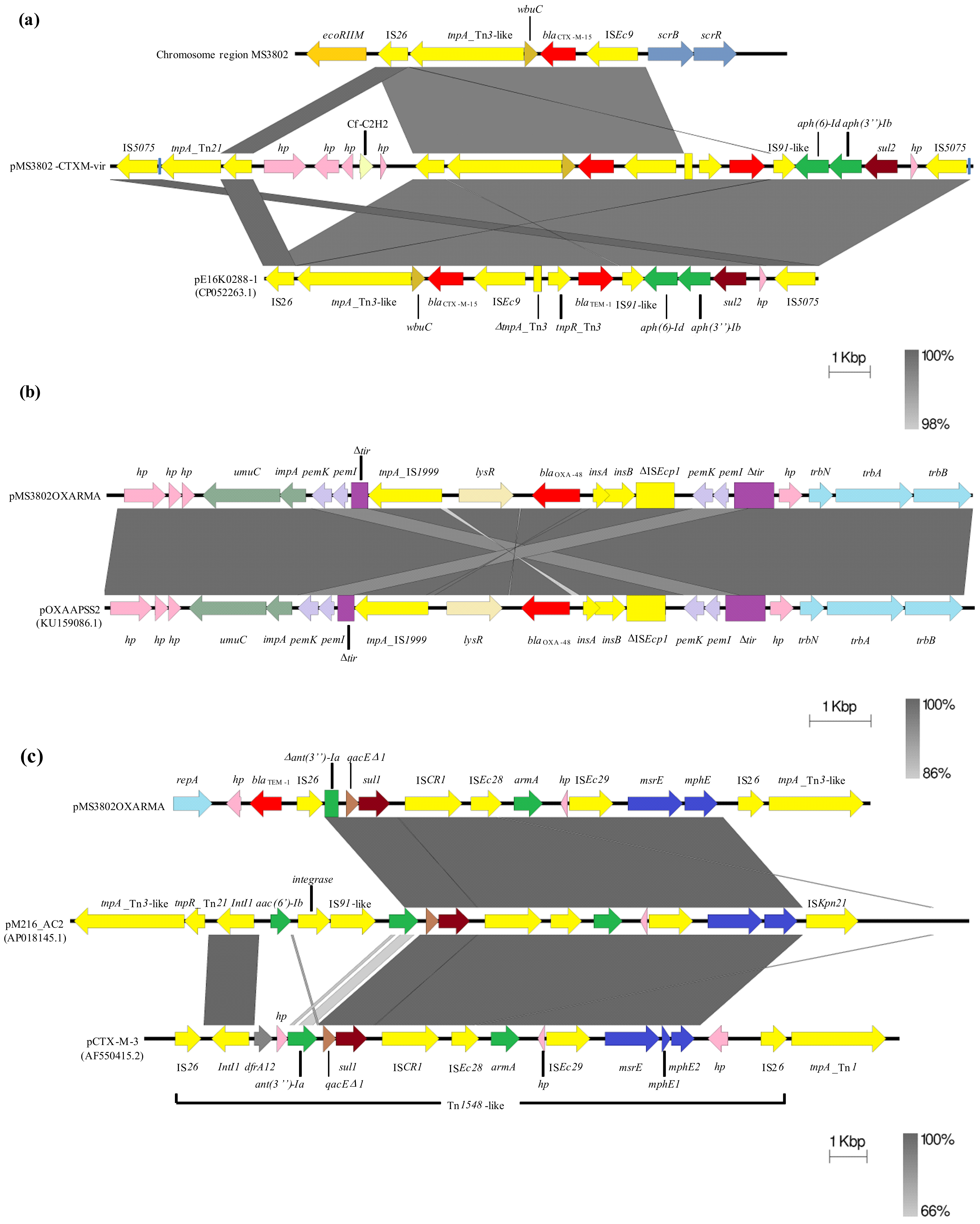
2.2. Whole-Genome Sequencing of the XDR K. pneumoniae MS3802
2.3. Demonstration of Transferability of MS3802 Plasmids
3. Conclusions
4. Materials and Methods
4.1. Strain Identification and Antimicrobial Susceptibility Testing
4.2. String Test
4.3. Whole-Genome Sequencing and Bioinformatics Analysis
4.4. Conjugation Experiments
4.5. Nucleotide Accession Numbers
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hamzaoui, Z.; Ocampo-Sosa, A.; Fernandez Martinez, M.; Landolsi, S.; Ferjani, S.; Maamar, E.; Saidani, M.; Slim, A.; Martinez-Martinez, L.; Boutiba-Ben Boubaker, I. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2018, 52, 898–905. [Google Scholar] [CrossRef]
- Liu, Y.C.; Cheng, D.L.; Lin, C.L. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 1986, 146, 1913–1916. [Google Scholar] [CrossRef]
- Lin, F.; Xu, Y.; Chang, Y.; Liu, C.; Jia, X.; Ling, B. Molecular Characterization of Reduced Susceptibility to Biocides in Clinical Isolates of Acinetobacter baumannii. Front. Microbiol. 2017, 8, 1836. [Google Scholar] [CrossRef]
- Cubero, M.; Grau, I.; Tubau, F.; Pallarés, R.; Dominguez, M.A.; Liñares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. 2016, 22, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-López, J.; García-Caballero, A.; Navarro-San Francisco, C.; Quereda, C.; Ruiz-Garbajosa, P.; Navas, E.; Dronda, F.; Morosini, M.I.; Cantón, R.; Diez-Aguilar, M. Hypermucoviscous Klebsiella pneumoniae: A challenge in community acquired infection. IDCases 2019, 17, e00547. [Google Scholar] [CrossRef]
- Cubero, M.; Marti, S.; Domínguez, M.Á.; González-Díaz, A.; Berbel, D.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae serotype K1 clinical isolates form robust biofilms at the air-liquid interface. PLoS ONE 2019, 14, e0222628. [Google Scholar] [CrossRef]
- Fang, C.-T.; Chuang, Y.-P.; Shun, C.-T.; Chang, S.-C.; Wang, J.-T. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 2004, 199, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Yeh, K.-M.; Chang, F.-Y.; Fung, C.-P.; Lin, J.-C.; Siu, L.K. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1. J. Med. Microbiol. 2006, 55, 803–804. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Chen, Y.S.; Wu, C.Y.; Chang, H.Y.; Lai, Y.C.; Peng, H.L. RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 2010, 192, 3144–3158. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.-R.; Lin, T.-L.; Chen, Y.-C.; Chou, H.-C.; Wang, J.-T. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011, 157, 3446–3457. [Google Scholar] [CrossRef]
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Catalán-Nájera, J.C.; Garza-Ramos, U.; Barrios-Camacho, H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence 2017, 8, 1111–1123. [Google Scholar] [CrossRef] [Green Version]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2019. [Google Scholar] [CrossRef] [Green Version]
- Brisse, S.; Fevre, C.; Passet, V.; Issenhuth-Jeanjean, S.; Tournebize, R.; Diancourt, L.; Grimont, P. Virulent clones of Klebsiella pneumoniae: Identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 2009, 4, e4982. [Google Scholar] [CrossRef] [Green Version]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.-S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.-H.; et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812–1820. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, C.; Wang, Q.; Wang, X.; Chen, H.; Li, H.; Zhang, F.; Li, S.; Wang, R.; Wang, H. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: Geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 2016, 60, 6115–6120. [Google Scholar] [CrossRef] [Green Version]
- Chew, K.L.; Lin, R.T.P.; Teo, J.W.P. Klebsiella pneumoniae in Singapore: Hypervirulent infections and the carbapenemase threat. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Dong, N.; Yang, X.; Zhang, R.; Chan, E.W.C.; Chen, S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg. Microbes Infect. 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.; Stanton, R.A.; Ansari, U.; McAllister, G.; Chan, M.Y.; Sula, E.; Grass, J.E.; Duffy, N.; Anacker, M.L.; Witwer, M.L.; et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Qu, T.; Zhou, J.; Jiang, Y.; Shi, K.; Li, B.; Shen, P.; Wei, Z.; Yu, Y. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 2015, 15, 161. [Google Scholar] [CrossRef] [Green Version]
- Turton, J.; Davies, F.; Turton, J.; Perry, C.; Payne, Z.; Pike, R. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blandm-5. Microorganisms 2019, 7, 326. [Google Scholar] [CrossRef] [Green Version]
- Mataseje, L.F.; Boyd, D.A.; Mulvey, M.R.; Longtin, Y. Two Hypervirulent Klebsiella pneumoniae isolates producing a blaKPC-2 carbapenemase from a canadian patient. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Shu, L.; Dong, N.; Lu, J.; Zheng, Z.; Hu, J.; Zeng, W.; Sun, Q.; Chan, E.W.C.; Zhou, H.; Hu, F.; et al. Emergence of OXA-232 Carbapenemase—Producing Klebsiella pneumoniae That Carries a pLVPK-Like Virulence Plasmid among Elderly Patients in China. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Long, D.; Xiang, T.-X.; Du, F.-L.; Wei, D.D.; Wan, L.-G.; Deng, Q.; Cao, X.-W.; Zhang, W. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 2019, 74, 1233–1240. [Google Scholar] [CrossRef]
- Chen, Y.T.; Chang, H.Y.; Lai, Y.C.; Pan, C.C.; Tsai, S.F.; Peng, H.L. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004, 337, 189–198. [Google Scholar] [CrossRef]
- Wu, K.-M.; Li, L.-H.; Yan, J.-J.; Tsao, N.; Liao, T.-L.; Tsai, H.-C.; Fung, C.-P.; Chen, H.-J.; Liu, Y.-M.; Wang, J.-T.; et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 2009, 191, 4492–4501. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.H.; Lu, P.L.; Liu, E.Y.M.; Chen, Y.Y.; Lin, F.M.; Lin, Y.T.; Chang, F.Y.; Lin, J.C. Rapid identification of capsular serotype K1/K2 Klebsiella pneumoniae in pus samples from liver abscess patients and positive blood culture samples from bacteremia cases via an immunochromatographic strip assay. Gut Pathog. 2019, 11. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Lin, D.; Zhang, R.; Chan, E.W.C.; Chen, S. Carriage of bla KPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 3317–3321. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Ma, G.; Li, C.; Jia, X.; Qin, C.; Yang, T.; Wang, L.; Jiang, X.; Ding, N.; Zhang, X.; et al. Emergence of a Multidrug-Resistant Hypervirulent Klebsiella pneumoniae Sequence Type 23 Strain with a Rare blaCTX-M-24-Harboring Virulence Plasmid. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jin, L.; Ouyang, P.; Wang, Q.; Wang, R.; Wang, J.; Gao, H.; Wang, X.; Wang, H. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: A multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 2020, 75, 327–336. [Google Scholar] [CrossRef]
- Xie, M.; Chen, K.; Ye, L.; Yang, X.; Xu, Q.; Yang, C.; Dong, N.; Chan, E.W.C.; Sun, Q.; Shu, L.; et al. Conjugation of Virulence Plasmid in Clinical Klebsiella pneumoniae Strains through Formation of a Fusion Plasmid. Adv. Biosyst. 2020, 4, 1900239. [Google Scholar] [CrossRef]
- Zhao, W.H.; Hu, Z.Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef]
- Partridge, S.R.; Hall, R.M. The IS1111 Family Members IS4321 and IS5075 Have Subterminal Inverted Repeats and Target the Terminal Inverted Repeats of Tn21 Family Transposons. J. Bacteriol. 2003, 185, 6371–6384. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.H.; Wu, C.C.; Kuo, J.T.; Chu, H.F.; Lee, D.Y.; Lin, C.T. Fnr-dependent rmpa and rmpa2 regulation of capsule polysaccharide biosynthesis in Klebsiella pneumoniae. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob. Agents Chemother. 2014, 58, 467–471. [Google Scholar] [CrossRef] [Green Version]
- Harmer, C.J.; Moran, R.A.; Hall, R.M. Movement of IS26-Associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Kalpoe, J.S.; Al Naiemi, N.; Poirel, L.; Nordmann, P. Detection of an ambler class d oxa-48-type β-lactamase in a Klebsiella pneumoniae strain in the netherlands. J. Med. Microbiol. 2011, 60, 677–678. [Google Scholar] [CrossRef] [Green Version]
- Galani, I.; Souli, M.; Panagea, T.; Poulakou, G.; Kanellakopoulou, K.; Giamarellou, H. Prevalence of 16S rRNA methylase genes in Enterobacteriaceae isolates from a Greek University Hospital. Clin. Microbiol. Infect. 2012, 18. [Google Scholar] [CrossRef] [Green Version]
- Pagani, L.; Dell’Amico, E.; Migliavacca, R.; D’Andrea, M.M.; Giacobone, E.; Amicosante, G.; Romero, E.; Rossolini, G.M. Multiple CTX-M-type extended-spectrum β-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in Northern Italy. J. Clin. Microbiol. 2003, 41, 4264–4269. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Dong, N.; Chen, K.; Yang, X.; Ye, L.; Chan, E.W.C.; Zhang, R.; Chen, S. A hybrid plasmid formed by recombination of a virulence plasmid and a resistance plasmid in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2020, 23, 466–470. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1-56238-839-8. [Google Scholar]
- Hernández, M.; Iglesias, M.R.; Rodríguez-Lázaro, D.; Gallardo, A.; Quijada, N.M.; Miguela-Villoldo, P.; Campos, M.J.; Píriz, S.; López-Orozco, G.; de Frutos, C.; et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinformatics 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive web: User-Friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [Green Version]
| Antimicrobial Agent | MIC (μg/mL) | |||
|---|---|---|---|---|
| K. pneumoniae MS3802 | E. coli J53-Pipr | E. coli J53-AKr | E. coli J53 (wt) | |
| Ampicillin | >128 (R) | >128 | >128 (R) | 64 (R) |
| Ampicillin/Sulbactam | >64 (R) | >64 | >64 (R) | 64 (R) |
| Amoxicillin/Clavulanic acid | >128 (R) | >128 (R) | >128 (R) | 8 (S) |
| Aztreonam | >128 (R) | <0.125 (S) | <0.125 (S) | 0.125 (S) |
| Piperacillin/tazobactam | >128 (R) | 128 (R) | 128 (R) | 2 (S) |
| Cefuroxime | >64 (R) | 8 (S) | 8 (S) | 8 (S) |
| Cefoxitin (*) | 16 (I) | 8 (S) | 8 (S) | 2 (S) |
| Cefotaxime | >128 (R) | 0.5 (S) | 0.5 (S) | 0.125 (S) |
| Ceftazdime | 64 (R) | 2 (I) | 4 (I) | 0.5 (S) |
| Cefepime | 8 (R) | 0.25 (S) | 0.25 (S) | <0.06 (S) |
| Ertapenem | 4 (R) | 0.25 (S) | 0.25 (S) | <0.06 (S) |
| Imipenem | 2 (S) | 0.5 (S) | 0.5 (S) | 0.125 (S) |
| Meropenem | 1 (S) | 0.5 (S) | 0.5 (S) | <0.06 (S) |
| Doripenem (*) | 1 (S) | 0.5 (S) | 0.5 (S) | <0.06 (S) |
| Gentamicin | 128 (R) | 128 (R) | 128 (R) | 0.5 (S) |
| Tobramycin | 128 (R | 128 (R) | 128 (R) | 0.125 (S) |
| Amikacin | >128 (R) | >128 (R) | >128 (R) | 0.125 (S) |
| Netilmicin (*) | >128 (R) | 128 (R) | >128 (R) | 0.125 (S) |
| Ciprofloxacin | 4 (R) | <0.06 (S) | <0.06 (S) | <0.06 (S) |
| Levofloxacin | 8 (R) | <0.06 (S) | <0.06 (S) | <0.06 (S) |
| Tigecycline | 2 (R) | 0.25 (S) | 0.125 (S) | <0.06 (S) |
| Tetracycline (*) | 1 (S) | 0.5 (S) | 1 (S) | 0.5 (S) |
| Minocycline (*) | 8 (I) | 4 (S) | 2 (S) | 4 (S) |
| Colistin | 16 (R) | 0.125 (S) | <0.06 (S) | <0.06 (S) |
| Trimethoprim/sulfamethoxazole | >128 (R) | 0.125 (S) | 0.125 (S) | <0.06 (S) |
| Cloramphenicol | 32 (R) | 8 (S) | 8 (S) | 8 (S) |
| Fosfomycin | >256 (R) | 8 (S) | 8 (S) | 8 (S) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández, M.; López-Urrutia, L.; Abad, D.; De Frutos Serna, M.; Ocampo-Sosa, A.A.; Eiros, J.M. First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics 2021, 10, 157. https://doi.org/10.3390/antibiotics10020157
Hernández M, López-Urrutia L, Abad D, De Frutos Serna M, Ocampo-Sosa AA, Eiros JM. First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics. 2021; 10(2):157. https://doi.org/10.3390/antibiotics10020157
Chicago/Turabian StyleHernández, Marta, Luis López-Urrutia, David Abad, Mónica De Frutos Serna, Alain A. Ocampo-Sosa, and José María Eiros. 2021. "First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain" Antibiotics 10, no. 2: 157. https://doi.org/10.3390/antibiotics10020157
APA StyleHernández, M., López-Urrutia, L., Abad, D., De Frutos Serna, M., Ocampo-Sosa, A. A., & Eiros, J. M. (2021). First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics, 10(2), 157. https://doi.org/10.3390/antibiotics10020157






