Gentamicin Adsorption onto Soil Particles Prevents Overall Short-Term Effects on the Soil Microbiome and Resistome
Abstract
1. Introduction
2. Results
2.1. Gentamicin Effect on Soil Bacterial Communities
2.2. Gentamicin Effect on the Soil Resistome
3. Discussion
4. Materials and Methods
4.1. Soil Sampling
4.2. Microcosm Experiment
4.3. Determination of Gentamicin Effect on Soil Enrichments
4.4. Enrichment of Soil Bacteria in 1:10 TSB Medium Polluted with Gentamicin
4.5. DNA Extraction from Soil Microcosms
4.6. DNA Quantification and Quantitative PCR (qPCR) Assays
4.7. 16S rRNA Gene and cDNA Sequencing and Analysis
4.8. Metagenomic Sequencing and Analysis
4.9. Gentamicin Bioavailability and Adsorption in Soil
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delmont, T.O.; Robe, P.; Cecillon, S.; Clark, I.M.; Constancias, F.; Simonet, P.; Hirsch, P.R.; Vogel, T.M. Accessing the Soil Metagenome for Studies of Microbial Diversity. Appl. Environ. Microbiol. 2010, 77, 1315–1324. [Google Scholar] [CrossRef]
- Nesme, J.; Simonet, P. The soil resistome: A critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 2015, 17, 913–930. [Google Scholar] [CrossRef]
- Monier, J.-M.; Demanèche, S.; O Delmont, T.; Mathieu, A.; Vogel, T.M.; Simonet, P. Metagenomic exploration of antibiotic resistance in soil. Curr. Opin. Microbiol. 2011, 14, 229–235. [Google Scholar] [CrossRef]
- Naidoo, Y.; Valverde, A.; Cason, E.D.; Pierneef, R.E.; Cowan, D.A. A clinically important, plasmid-borne antibi-otic resistance gene (β-lactamase TEM-116) present in desert soils. Sci. Total Environ. 2020, 719, doi. [Google Scholar] [CrossRef]
- Perron, G.G.; Whyte, L.; Turnbaugh, P.J.; Goordial, J.; Hanage, W.P.; Dantas, G.; Desai, M.M. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse re-sistance mechanisms to modern antibiotics. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Goodman, R.M.; Handelsman, J. Uncultured soil bacteria are a reservoir of new antibiotic re-sistance genes. Environ. Microbiol. 2004, 6, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Willms, I.M.; Kamran, A.; Aßmann, N.F.; Krone, D.; Bolz, S.H.; Fiedler, F.; Nacke, H. Discovery of Novel Antibiotic Resistance Determinants in Forest and Grassland Soil Meta-genomes. Front. Microbiol. 2019, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xu, Z.; Riber, L.; Hansen, L.H.; Sørensen, S.J. Diverse gene functions in a soil mobilome. Soil Biol. Biochem. 2016, 101, 175–183. [Google Scholar] [CrossRef]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef]
- Hoelzer, K.; Wong, N.; Thomas, J.; Talkington, K.; Jungman, E.; Coukell, A. Antimicrobial drug use in food-producing animals and associated human health risks: What, and how strong, is the evidence? BMC Vet. Res. 2017, 13, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014, 22, 536–545. [Google Scholar] [CrossRef]
- Blau, K.; Jacquiod, S.; Sørensen, S.J.; Su, J.-Q.; Zhu, Y.-G.; Smalla, K.; Jechalke, S. Manure and Doxycycline Affect the Bacterial Community and Its Resistome in Lettuce Rhizosphere and Bulk Soil. Front. Microbiol. 2019, 10, 725. [Google Scholar] [CrossRef] [PubMed]
- Wolters, B.; Jacquiod, S.J.A.; Sørensen, S.J.; Widyasari-Mehta, A.; Bech, T.B.; Kreuzig, R.; Smalla, K. Bulk soil and maize rhizosphere resistance genes, mobile genetic elements and microbial com-munities are differently impacted by organic and inorganic fertilization. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Kang, Y.; Li, Q.; Yin, Z.; Shen, M.; Zhao, H.; Bai, Y.; Mei, L.; Hu, J. High diversity and abundance of cultivable tetracycline-resistant bacteria in soil following pig manure application. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Kopmann, C.; Rosendahl, I.; Groeneweg, J.; Weichelt, V.; Krögerrecklenfort, E.; Brandes, N.; Nordwig, M.; Ding, G.-C.; Siemens, J.; et al. Increased Abundance and Transferability of Resistance Genes after Field Application of Manure from Sulfadiazine-Treated Pigs. Appl. Environ. Microbiol. 2013, 79, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Binh, C.T.T.; Jechalke, S.; Kopmann, C.; Zimmerling, U.; Krögerrecklenfort, E.; Ledger, T.; González, B.; Top, E.M.; Smalla, K. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: Di-versification driven by class 1 integron gene cassettes. Front. Microbiol. 2012, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Solehati, Q.; Zimmerling, U.; Kleineidam, K.; Schloter, M.; Müller, T.; Focks, A.; Thiele-Bruhn, S.; Smalla, K. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of ma-nure containing sulfadiazine. Appl. Environ. Microbiol. 2011, 77, 2527–2530. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.D.; Wright, G.D. Lessons from the Environmental Antibiotic Resistome. Annu. Rev. Microbiol. 2017, 71, 309–329. [Google Scholar] [CrossRef]
- Pan, M.; Chu, L. Adsorption and degradation of five selected antibiotics in agricultural soil. Sci. Total Environ. 2016, 48–56. [Google Scholar] [CrossRef]
- Albero, B.; Tadeo, J.L.; Escario, M.; Miguel, E.; Pérez, R.A. Persistence and availability of veterinary antibiotics in soil and soil-manure systems. Sci. Total Environ. 2018, 643, 1562–1570. [Google Scholar] [CrossRef]
- Thiele-Bruhn, S. Pharmaceutical antibiotic compounds in soils—A review. J. Plant Nutr. Soil Sci. 2003, 166, 145–167. [Google Scholar] [CrossRef]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Brandt, K.K.; Amézquita, A.; Backhaus, T.; Boxall, A.; Coors, A.; Heberer, T.; Lawrence, J.R.; Lazorchak, J.; Schoenfeld, J.; Snape, J.R.; et al. Ecotoxicological assessment of antibiotics: A call for improved consideration of microorgan-isms. Environ. Int. 2015, 85, 189–205. [Google Scholar] [CrossRef]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef]
- Durso, L.M.; Cook, K.L. Impacts of antibiotic use in agriculture: What are the benefits and risks? Curr. Opin. Microbiol. 2014, 19, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krögerrecklenfort, E.; Wellington, E.M.H.; Egan, S.; Van Elsas, J.D.; Van Overbeek, L.; Collard, J.M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol. Ecol. 2002, 42, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Hadelsman, J. Status of the Microbial Census. Microbiol. Mol. Biol. Rev. 2004, 68, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Larsson, D.G.J. Concentrations of antibiotics predicted to select for resistant bacteria: Pro-posed limits for environmental regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Evolutionary Trajectories to Antibiotic Resistance. Annu. Rev. Microbiol. 2017, 71, 579–596. [Google Scholar] [CrossRef]
- Cairns, J.; Ruokolainen, L.; Hultman, J.; Tamminem, M.; Virta, M.; Hiltunen, T. Ecology determines how low antibiotic concentration impacts community composition and hori-zontal transfer of resistance genes. Commun. Biol. 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Bergk Pinto, B.; Maccario, L.; Dommergue, A.; Vogel, T.M.; Larose, C. Do Organic Substrates Drive Microbial Community Interactions in Arctic Snow? Front. Microbiol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Yang, D.; Zheng, Y.; Yang, Y.; He, Y.; Luo, L.; Zhou, Y. Current progress in the adsorption, transport and biodegradation of antibiotics in soil. J. Environ. Manag. 2019, 251, 109598. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Genet. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.; Simonet, P.; Vogel, T.M. Describing microbial communities and performing global comparisons in the ‘omic era. ISME J. 2012, 6, 1625–1628. [Google Scholar] [CrossRef] [PubMed]
- Hamp, T.J.; Jones, W.J.; Fodor, A.A. Effects of Experimental Choices and Analysis Noise on Surveys of the “Rare Biosphere”. Appl. Environ. Microbiol. 2009, 75, 3263–3270. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Dohrmann, A.B.; Christensen, B.T. Bacterial Preferences for Specific Soil Particle Size Frac-tions Revealed by Community Analyses. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Neumann, D.; Heuer, A.; Hemkemeyer, M.; Martens, R.; Tebbe, C.C. Response of microbial communities to long-term fertilization depends on their microhabitat. FEMS Microbiol. Ecol. 2013, 86, 71–84. [Google Scholar] [CrossRef]
- Martens, R.; Tebbe, C.C. Artificial soil studies reveal domain-specific preferences of microorganisms for the colo-nisation of different soil minerals and particle size fractions. FEMS Microbiol. Ecol. 2014, 90, 770–782. [Google Scholar]
- Bernier, S.P.; Surette, M.G. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef]
- Josephson, L.; Houle, P.; Haggerty, M. Stability of dilute solutions of gentamicin and tobramycin. Clin. Chem. 1979, 25, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Hottentrager, S.; Teske, A.; Wawer, C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—A new molecular approach to analyse the genetic diversity of mixed microbial communities. In Molecular Microbial Ecology Manual; Akkermans, A., van Elsas, J., de Bruijn, F., Eds.; Kluwer Academic Publishers: Dordrecht, Netherlands, 1995; pp. 1–23. [Google Scholar]
- Watanabe, K.; Kodama, Y.; Harayama, S. Design and evaluation of PCR primers to amplify bacterial 16S riboso-mal DNA fragments used for community fingerprinting. J. Microbiol. Methods 2001, 44, 253–262. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA se-quences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-2. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 30 December 2020).
- Minoche, A.E.; Dohm, J.C.; Himmelbauer, H. Evaluation of genomic high-throughput sequencing data generat-ed on Illumina HiSeq and Genome Analyzer systems. Genome Biol. 2011, 12, 1–15. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platformfor ’omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Guironnet, A.; Wiest, L.; Sanchez-Cid, C.; Vogel, T.M.; Vulliet, E. Aminoglycosides analysis optimization using Ion pairing Liquid Chromatography coupled to tandem Mass Spectrometry and application on wastewater samples. J. Chromatogr. A 2021. under review. [Google Scholar]
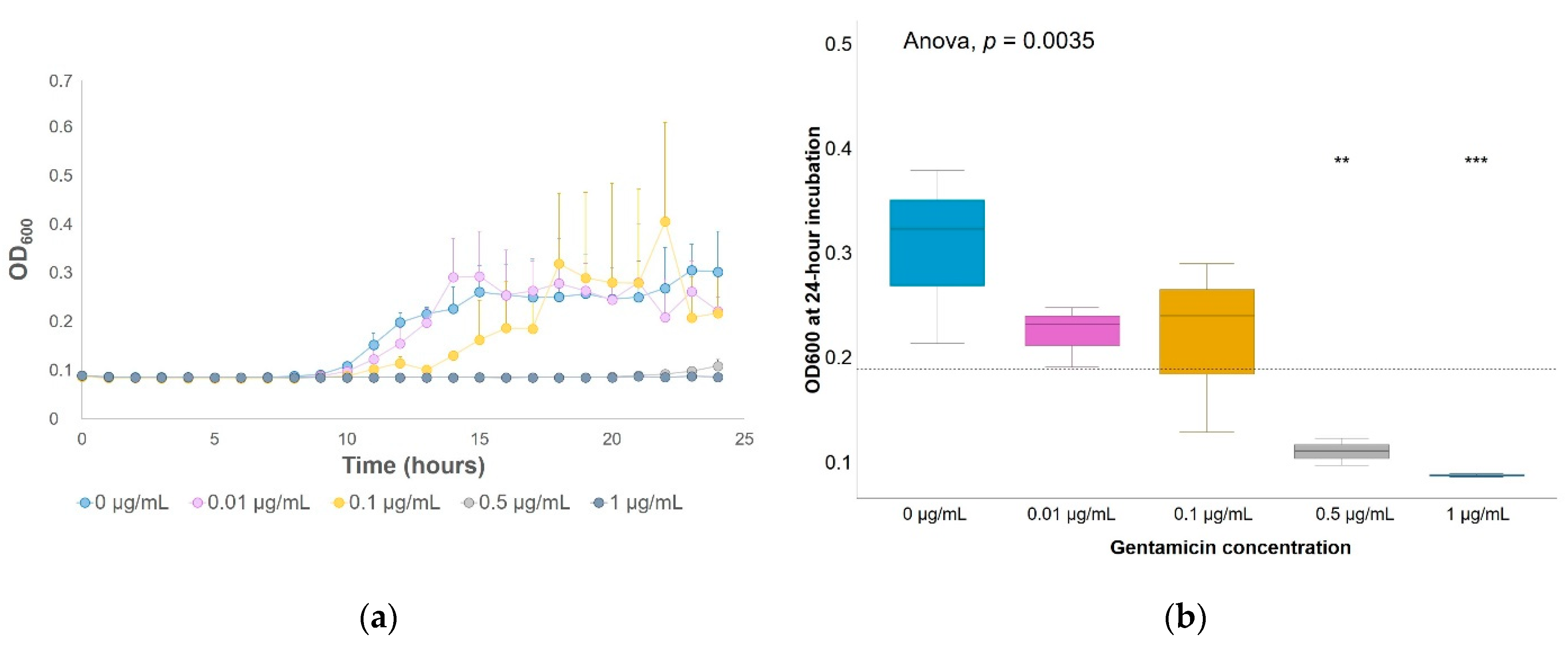
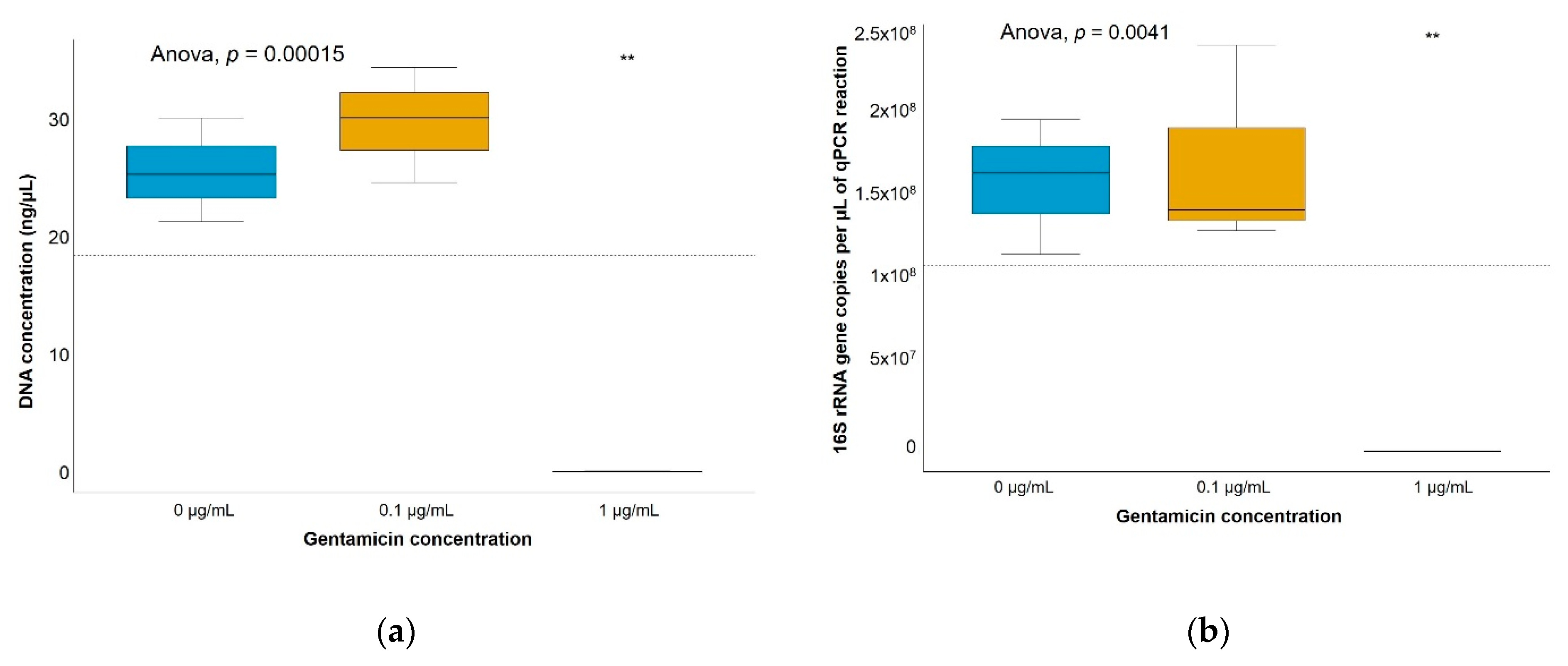

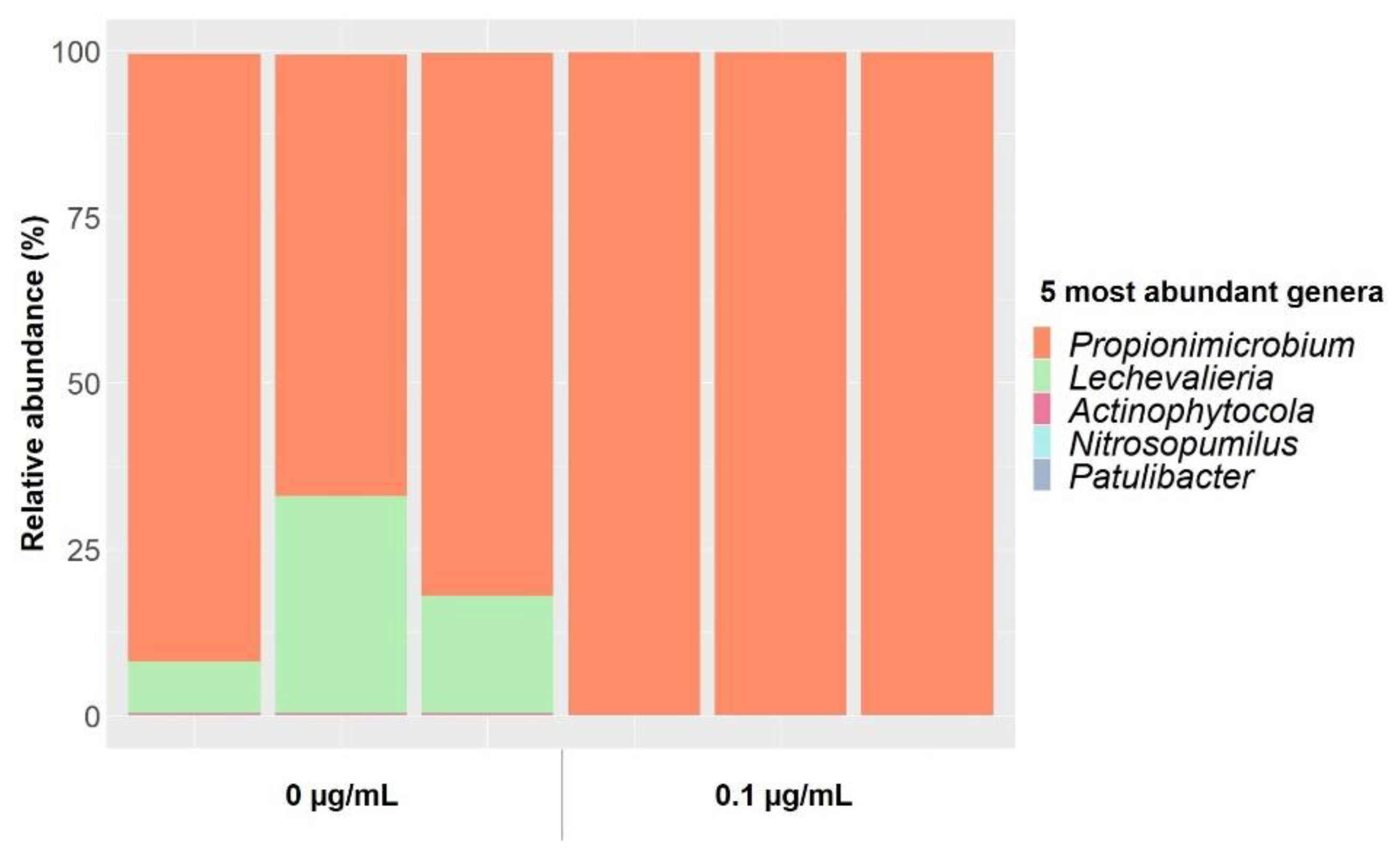
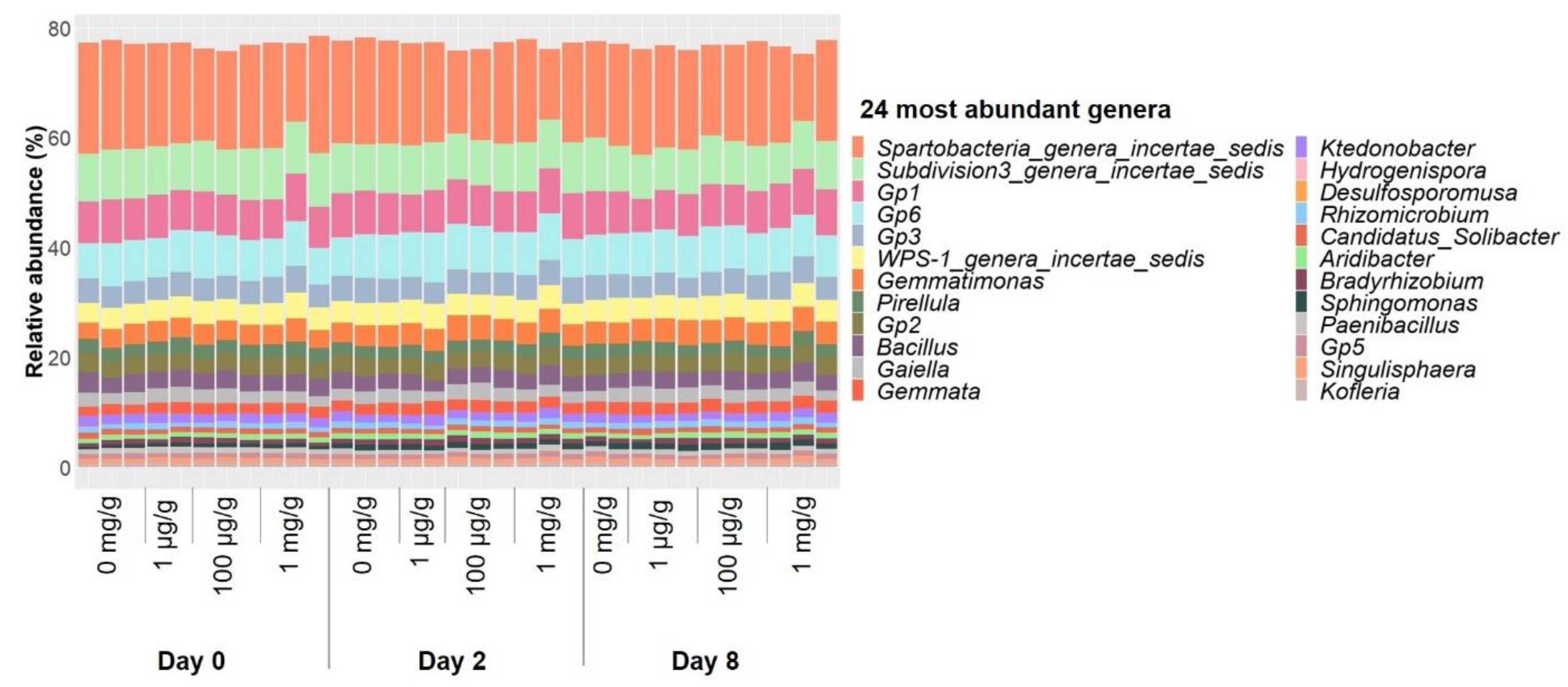
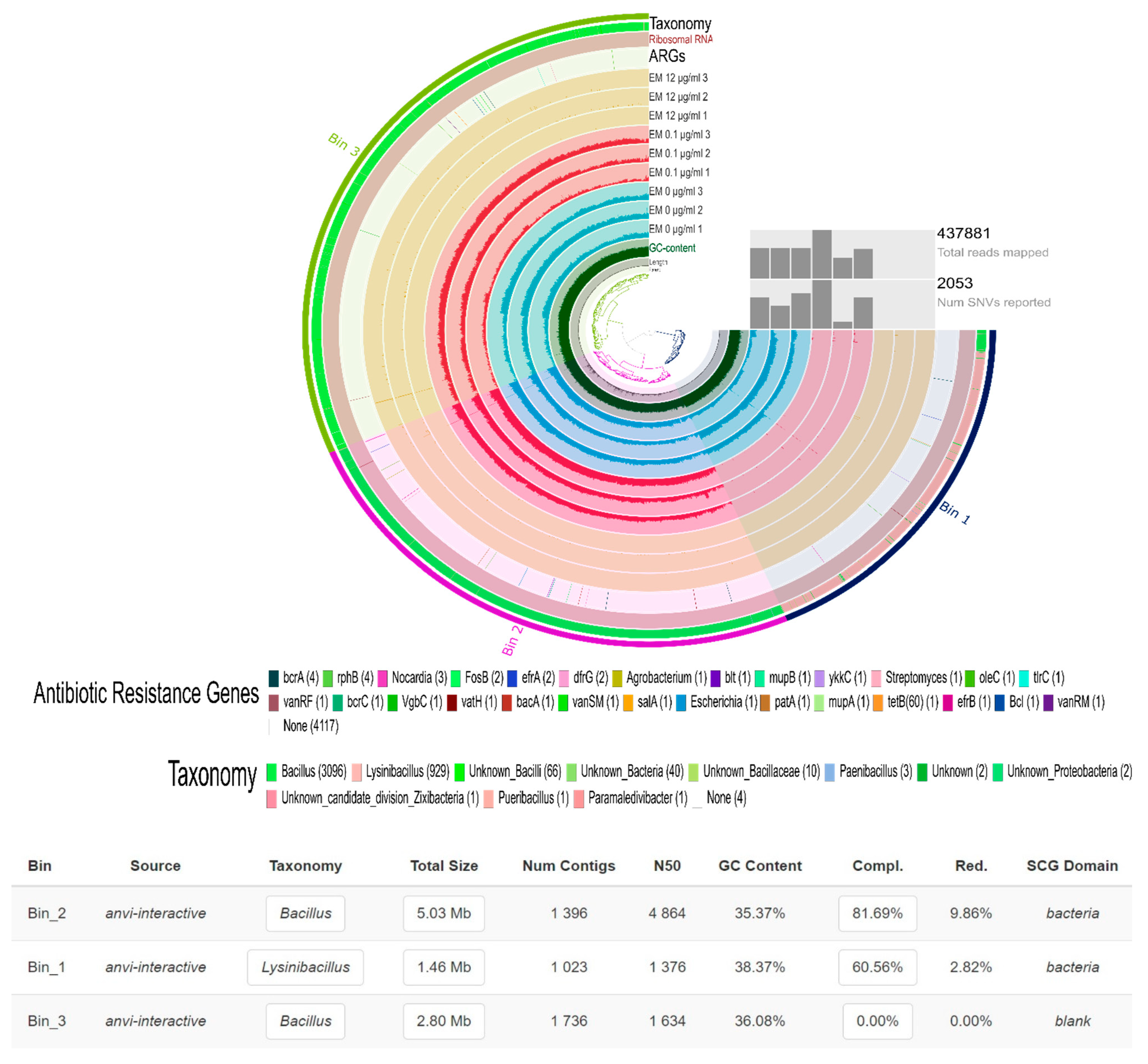
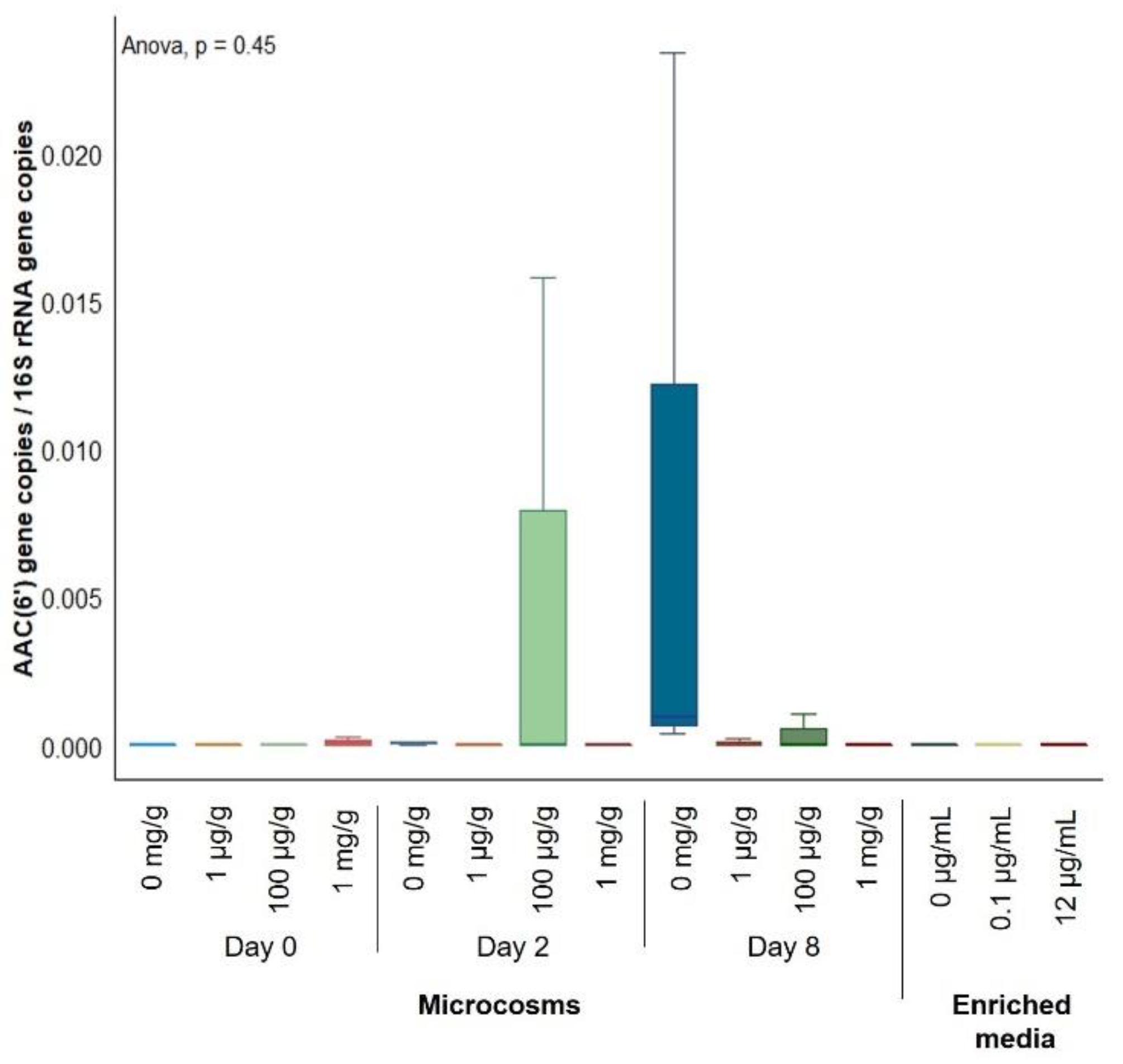
| [Gentamicin] | 0 µg/mL | 0.1 µg/mL | 12 µg/mL | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample/Gene | SC1 | SC2 | SC3 | SC4 | SC5 | SC6 | SC7 | SC8 | SC9 |
| muxB | 0 | 4.9 × 10−6 | 0 | 0 | 5.4 × 10−6 | 0 | 1.5 × 10−4 | 6.7 × 10−5 | 1.3 × 10−4 |
| adeF | 0 | 0 | 0 | 0 | 0 | 0 | 5.1 × 10−5 | 9.3 × 10−5 | 6.4 × 10−5 |
| Soil Parameter | La Côte de Saint André |
|---|---|
| Sand | 42.9% |
| Silt | 43.6% |
| Clay | 13.5% |
| pH | 7.24 |
| Organic matter | 2.92% |
| Organic C | 1.7% |
| Total N | 0.17% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Cid, C.; Guironnet, A.; Wiest, L.; Vulliet, E.; Vogel, T.M. Gentamicin Adsorption onto Soil Particles Prevents Overall Short-Term Effects on the Soil Microbiome and Resistome. Antibiotics 2021, 10, 191. https://doi.org/10.3390/antibiotics10020191
Sanchez-Cid C, Guironnet A, Wiest L, Vulliet E, Vogel TM. Gentamicin Adsorption onto Soil Particles Prevents Overall Short-Term Effects on the Soil Microbiome and Resistome. Antibiotics. 2021; 10(2):191. https://doi.org/10.3390/antibiotics10020191
Chicago/Turabian StyleSanchez-Cid, Concepcion, Alexandre Guironnet, Laure Wiest, Emmanuelle Vulliet, and Timothy M. Vogel. 2021. "Gentamicin Adsorption onto Soil Particles Prevents Overall Short-Term Effects on the Soil Microbiome and Resistome" Antibiotics 10, no. 2: 191. https://doi.org/10.3390/antibiotics10020191
APA StyleSanchez-Cid, C., Guironnet, A., Wiest, L., Vulliet, E., & Vogel, T. M. (2021). Gentamicin Adsorption onto Soil Particles Prevents Overall Short-Term Effects on the Soil Microbiome and Resistome. Antibiotics, 10(2), 191. https://doi.org/10.3390/antibiotics10020191






