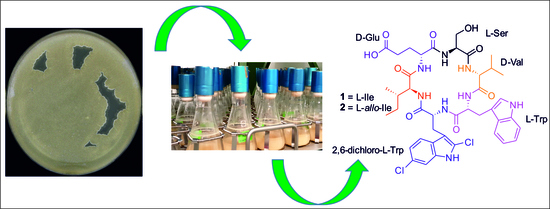
Amycolatomycins A and B, Cyclic Hexapeptides Isolated from an Amycolatopsis sp. 195334CR
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of Amycolatomycins
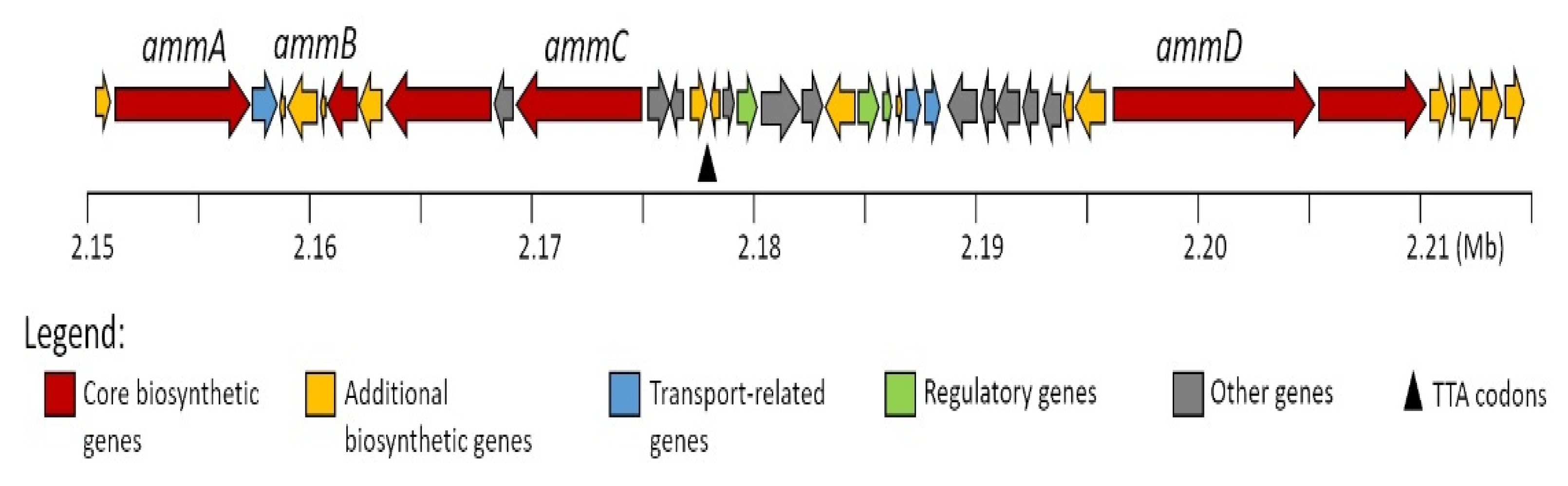
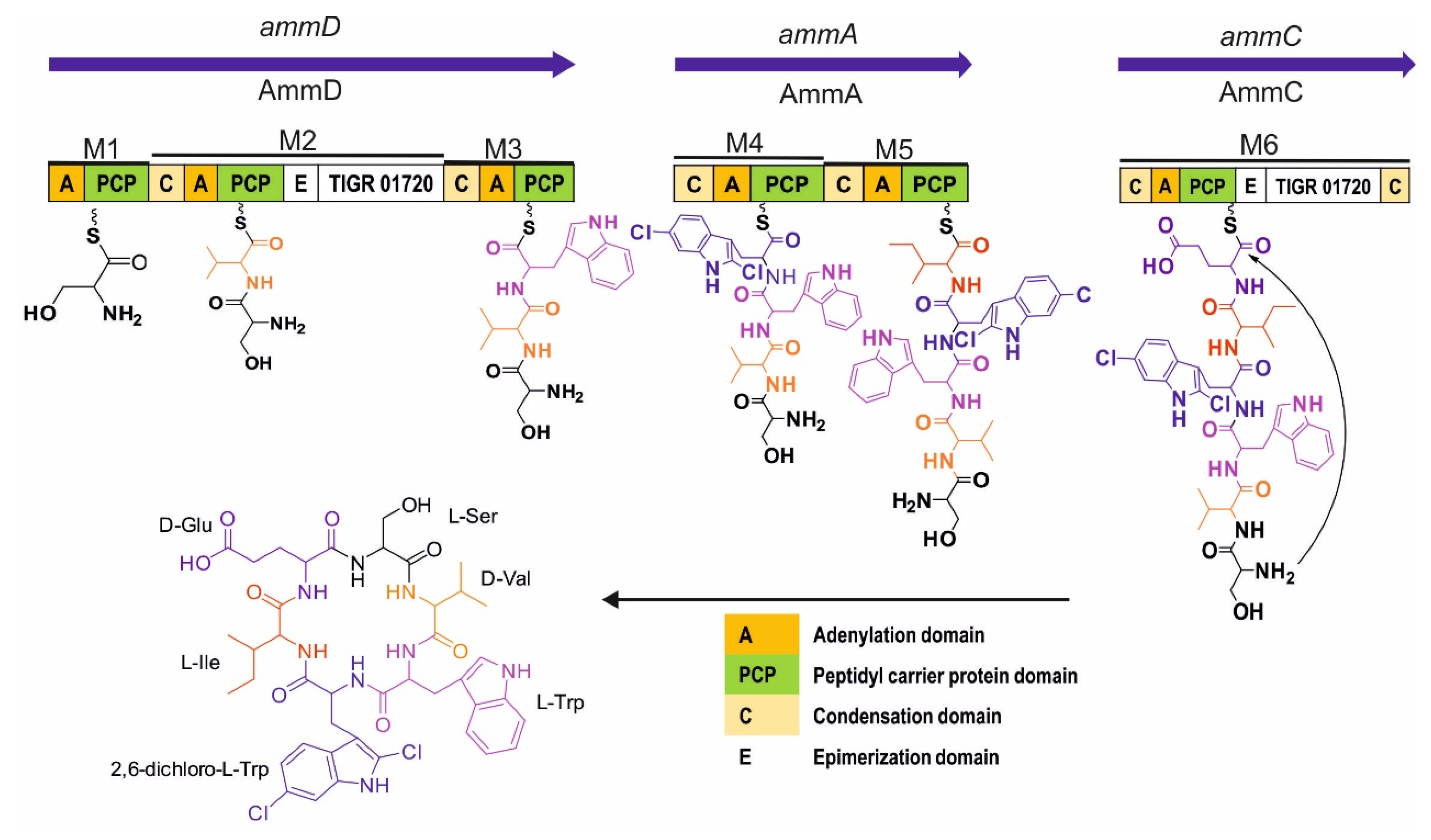
2.2. In Silico Analysis of the Amycolatomycin Biosynthetic Gene Clusters (BGCsBCGs)
2.3. Biological Activity of Amycolatomycin A
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Origin of the Strain
3.3. Analysis of 16S rRNA Sequences
3.4. Scale-Up Production, Extraction, and Isolation of Compounds
3.5. Ring-Opening and Partial Hydrolysis of Amycolatomycin A
3.6. Determination of Absolute Amino Acid Stereochemistry
3.7. Determination of Tryptophan Absolute Amino Acid Stereochemistry
3.8. Determination of Isoleucine Stereoisomer Absolute Stereochemistry with C4 HPLC–DAD/MS Marfey’s Analysis
3.9. Genomic DNA Isolation, Sequencing, and Bioinformatic Analysis
3.10. Antimicrobial and Cytotoxic Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stadler, M.; Dersch, P. How to overcome the antibiotic crisis – Facts, challenges, technologies & future perspectives. Curr Top. Microbiol Immunol. 2017, 398, 496. [Google Scholar]
- Genilloud, O. Mining actinomycetes for novel antibiotics in the omics era: Are we ready to exploit this new paradigm? Antibiotics 2018, 7, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef]
- Ding, T.; Yang, L.-J.; Zhang, W.-D.; Shen, Y.-H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Huang, H.; Wei, W.; Zhong, Y.; Tang, B.; Yuan, H.; Zhu, L.; Huang, W.; Ge, M.; Yang, S.; et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genom. 2014, 15, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Zhong, Y.; Yuan, H.; Wang, J.; Zheng, H.; Wang, Y.; Cen, X.; Xu, F.; Bai, J.; Han, X.; et al. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res. 2010, 20, 1096–1108. [Google Scholar] [CrossRef] [Green Version]
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org. Lett. 2017, 19, 1000–1003. [Google Scholar] [CrossRef]
- Xiao, Y.S.; Zhang, B.; Zhang, M.; Guo, Z.K.; Deng, X.Z.; Shi, J.; Li, W.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Rifamorpholines A-E, potential antibiotics from locust-associated actinobacteria: Amycolatopsis sp. Hca4. Org. Biomol. Chem. 2017, 15, 3909–3916. [Google Scholar] [CrossRef]
- Izuta, S.; Kosaka, S.; Kawai, M.; Miyano, R.; Matsuo, H.; Matsumoto, A.; Nonaka, K.; Takahashi, Y.; Omura, S.; Nakashima, T. Dipyrimicin A and B, microbial compounds isolated from Amycolatopsis sp. K16-0194. J. Antibiot. 2018, 71, 535–537. [Google Scholar] [CrossRef]
- Sheng, Y.; Fotso, S.; Serrill, J.D.; Shahab, S.; Santosa, D.A.; Ishmael, J.E.; Proteau, P.J.; Zabriskie, T.M.; Mahmud, T. Succinylated apoptolidins from Amycolatopsis sp. ICBB 8242. Org. Lett. 2015, 17, 2526–2529. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, K.K.; Pitcher, A.A.; Smart, B.P.; Spiciarich, D.R.; Iavarone, A.T.; Bertozzi, C.R. Isotopic signature transfer and mass pattern prediction (IsoStamp): An enabling technique for chemically-directed proteomics. ACS Chem. Biol. 2011, 6, 829–836. [Google Scholar] [CrossRef]
- Watts, K.R.; Morinaka, B.I.; Amagata, T.; Robinson, S.J.; Tenney, K.; Bray, W.M.; Gassner, N.C.; Lokey, R.S.; Media, J.; Valeriote, F.A.; et al. Biostructural features of additional jasplakinolide (jaspamide) analogues. J. Nat. Prod. 2011, 74, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s methods for amino acid analysis in natural products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramoto, K.; Kamiya, H. Recovery of tryptophan in peptides and proteins by high-temperature and short-term acid hydrolysis in the presence of phenol. Anal. Biochem. 1990, 189, 223–230. [Google Scholar] [CrossRef]
- Chan, C.-O.; Crich, D.; Natarajan, S. Enantiospecific Synthesis of Amino Acids: Preparation of (R)- and (S)-α- methylaspartic acid from (S)-tryptophan. Tetrahedron Lett. 1992, 33, 3405–3408. [Google Scholar] [CrossRef]
- Ranganathan, S.; Ranganathan, D.; Bhattacharyya, D. The Transformation of Tryptophan to Aspartic Acid in Peptides. J. Chem. Soc. Chem. Commun. 1987, 279, 1085–1086. [Google Scholar] [CrossRef]
- Anderson, Z.J.; Hobson, C.; Needley, R.; Song, L.; Perryman, M.S.; Kerby, P.; Fox, D.J. NMR-based assignment of isoleucine: Vs. allo -isoleucine stereochemistry. Org. Biomol. Chem. 2017, 15, 9372–9378. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challis, G.L.; Naismith, J.H. Structural aspects of non-ribosomal peptide biosynthesis. Curr. Opin. Struct. Biol. 2004, 14, 748–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amoutzias, G.D.; Van de Peer, Y.; Mossialos, D. Evolution and taxonomic distribution of non-ribosomal peptide and polyketide synthases. Future Microbiol. 2008, 3, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Haynes, S.W.; Ames, B.D.; Peng, W.; Vien, L.P.; Walsh, C.T.; Tang, Y. Cyclization of Fungal Nonribosomal Peptides by a Terminal Condensation-Like Domain. Nat. Chem. Biol. 2012, 8, 823–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romans-Fuertes, P.; Sondergaard, T.E.; Sandmann, M.I.H.; Wollenberg, R.D.; Nielsen, K.F.; Hansen, F.T.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. Identification of the non-ribosomal peptide synthetase responsible for biosynthesis of the potential anti-cancer drug sansalvamide in Fusarium solani. Curr. Genet. 2016, 62, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Sandargo, B.; Michehl, M.; Stadler, M.; Surup, F. Antifungal sesquiterpenoids, rhodocoranes F-L from submerged cultures of the wrinkled peach mushroom, Rhodotus palmatus. J. Nat. Prod. 2020, 83, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Alkhalaf, L.M.; Ryan, K.S. Biosynthetic manipulation of tryptophan in bacteria: Pathways and mechanisms. Chem. Biol. 2015, 22, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Therien, A.G.; Huber, J.L.; Wilson, K.E.; Beaulieu, P.; Caron, A.; Claveau, D.; Deschamps, K.; Donald, R.G.K.; Galgoci, A.M.; Gallant, M.; et al. Broadening the spectrum of β-lactam antibiotics through inhibition of signal peptidase type I. Antimicrob. Agents Chemother. 2012, 56, 4662–4670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, P.; Jamison, M.; La, S.; MacMillan, J.B. Inducamides A-C, chlorinated alkaloids from an RNA polymerase mutant strain of Streptomyces sp. Org. Lett. 2014, 16, 5656–5659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, J.; Satoa, M.; Ishibashia, M.; Shigemoria, H.; Nakamurab, T.; Ohizumic, Y. Keramamide A, a Novel Peptide from the Okinawan Marine Sponge Theonella sp. J. Chem. Soc. Perkin Trans. 1 1991, 2609–2611. [Google Scholar] [CrossRef]
- Jansen, R.; Kunze, B.; Reichenbach, H.; Hofle, G. Chondramides A-D, new cytostatic and antifungal cyclodepsipeptides from Chondromyces crocatus (Myxobacteria): Isolation and structure elucidation. Liebigs Ann. 1996, 2, 285–290. [Google Scholar]
- Primahana, G.; Risdian, C.; Mozef, T.; Sudarman, E.; Köck, M.; Wink, J.; Stadler, M. Nonocarbolines A–E, β-carboline antibiotics produced by the rare actinobacterium Nonomuraea sp. from Indonesia. Antibiotics 2020, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Mohr, K.I.; Stechling, M.; Wink, J.; Wilharm, E.; Stadler, M. Comparison of myxobacterial diversity and evaluation of isolation success in two niches: Kiritimati Island and German compost. Microbiol. Open 2016, 5, 268–278. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Pérez-Bonilla, M.; Oves-Costales, D.; González, I.; de la Cruz, M.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Krisynomycins, Imipenem Potentiators against Methicillin-Resistant Staphylococcus aureus, Produced by Streptomyces canus. J. Nat. Prod. 2020, 83, 2597–2606. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [Google Scholar] [CrossRef]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Wessel, A.; Luangsa-ard, J.J.; Stadler, M. Viridistratins A-C, antimicrobial and cytotoxic benzo[j]fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef] [PubMed]

| Unit | Pos | δH, Mult (J in Hz) | δC | Unit | Pos | δH, Mult (J in Hz) | δC |
|---|---|---|---|---|---|---|---|
| dcT | 1 | − | 170.8, C | Val | 1 | − | 170.3, C |
| 2 | 4.28, m | 54.4, CH | 2 | 4.04, d (br) (2.0) | 58.3, CH | ||
| 2NH | 8.66, d (8.4) | − | 2NH | 8.40, d (7.0) | |||
| 3 | 3.16, m; 2.92, dd (14.0, 10.4) | 27.0, CH2 | 3 | 2.27, m | 29.0, CH | ||
| 4 | − | 110.5, C | 4 | 0.85, d (6.8)ov | 19.3, CH3 | ||
| 5-Cl | − | 125.2, C | 5 | 0.84, d (6.8)ov | 16.9, CH3 | ||
| 6NH | 11.00, s | − | Ser | 1 | − | 170.3, C | |
| 7 | − | 134.4, C | 2 | 4.13, q (6.7 × 3) | 56.1, CH | ||
| 8 | 7.57, d (2.0) | 112.8, CH | 2NH | 8.14, s | − | ||
| 9-Cl | − | 123.1, C | 3 | 3.30a | 60.5, CH2 | ||
| 10 | 7.04, dd (8.5, 2.0) | 120.7, CH | Glu | 1 | − | 170.5, C | |
| 11 | 7.33, d (8.5, 2.0) | 117.4, CH | 2 | 4.42, q (6.7 × 3) | 51.1, CH | ||
| 12 | − | 128.3, C | 2NH | n.o | − | ||
| Trp | 1 | − | 170.3, C | 3 | 1.87, m | 28.2, CH2 | |
| 2 | 4.69, d (br) (6.0) | 53.5, CH | 4 | 2.19, t (8.0) | 30.2, CH2 | ||
| 2NH | n.o | 5 | − | 174.3, C | |||
| 3 | 3.11, m; 2.97, dd (14.0, 5.6) | 28.3, CH2 | Ile | 1 | − | 170.3, C | |
| 4 | − | 110.1, C | 2 | 4.09, s (br) | 57.8, CH | ||
| 5 | 7.13, s | 124.7, CH | 2NH | n.o | − | ||
| 6NH | 10.97, d (2.0) | − | 3 | 1.55, s (br) | 35.1, CH | ||
| 7 | − | 136.3, C | 4 | 1.22, m; 0.93, m | 24.8, CH2 | ||
| 8 | 7.35, d (2.0) | 110.7, CH | 5 | 0.75, t (7.4) | 10.8, CH3 | ||
| 9 | 6.99, dd (8.5, 2.0) | 118.5, CH | 6 | 0.51, d (6.2) | 14.6, CH3 | ||
| 10 | n.o | 125.5, C | |||||
| 11 | 7.56, s | 120.0, CH | |||||
| 12 | − | 126.4, C | |||||
| Gene (Nucleotide) | Protein (Amino Acid) | Proposed Function | Percent Identity and Similarity to Protein /Origin |
|---|---|---|---|
| ammA (6279) | AmmA (2092) | NRPS: C4 A4 (2,6-dichloro-Trp) PCP4 C5 A5 (Ile) PCP5 | 37%, 55%: AXN93581.1, PuwF-G [Cylindrospermum moravicum CCALA 993] |
| ammB (1290) | AmmB (429) | Halogenase | 77%, 88%: BAQ25509.1, FADH2-dependent_halogenase/Micromonospora sp. GMKU326 |
| ammC (5865) | AmmC (1954) | NRPS: C6 A6 (Glu) PCP6 E6 TIGR01720 CT | 40%, 52%: QBG38782.1, Atr217/Streptomyces atratus |
| ammD (9192) | AmmD (3063) | NRPS: A1(Ser) PCP1 C2 A2 (Val) PCP2 E2 TIGR01720 C3 A3 (Trp) PCP3 | 42%, 54%: ATU31794.1, NRPS/Streptomyces sp. KCB13F003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Primahana, G.; Risdian, C.; Mozef, T.; Wink, J.; Surup, F.; Stadler, M. Amycolatomycins A and B, Cyclic Hexapeptides Isolated from an Amycolatopsis sp. 195334CR. Antibiotics 2021, 10, 261. https://doi.org/10.3390/antibiotics10030261
Primahana G, Risdian C, Mozef T, Wink J, Surup F, Stadler M. Amycolatomycins A and B, Cyclic Hexapeptides Isolated from an Amycolatopsis sp. 195334CR. Antibiotics. 2021; 10(3):261. https://doi.org/10.3390/antibiotics10030261
Chicago/Turabian StylePrimahana, Gian, Chandra Risdian, Tjandrawati Mozef, Joachim Wink, Frank Surup, and Marc Stadler. 2021. "Amycolatomycins A and B, Cyclic Hexapeptides Isolated from an Amycolatopsis sp. 195334CR" Antibiotics 10, no. 3: 261. https://doi.org/10.3390/antibiotics10030261