Identification of Bioactive Compounds from Marine Natural Products and Exploration of Structure-Activity Relationships (SAR)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Active Marine Extracts
2.2. Isolation and Characterization of Bioactive Compounds
2.3. Structure—Activity Relationship
3. Materials and Methods
3.1. General
3.2. Natural Product Library
3.3. Bacterial Inhibition Assays
3.4. Evaluating
3.5. Purification of Natural Products from Extracts and Structure Elucidation
3.5.1. High Performance Liquid Chromatography (HPLC) Purification
3.5.2. Identification and Structure Elucidation
Identification and Elucidation of Purified Compounds from Sample Numbers 19033, 20608 and 26051
Identification and Elucidation of Fractions 44 from Sample Number 25663 (Compound 6)
Identification and Elucidation of Fractions 58 from Sample Number 25663 (Compound 7)
Identification and Elucidation of Purified Compounds from Sample Number 26104 (Compound 8)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bassetti, M.; Baguneid, M.; Bouza, E.; Dryden, M.; Nathwani, D.; Wilcox, M. European perspective and update on the management of complicated skin and soft tissue infections due to methicillin-resistant Staphylococcus aureus after more than 10 years of experience with linezolid. Clin. Microbiol. Infect. 2014, 20, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Richard, R.W.; Marisa, H.; Michael, Z.D. Antimicrobial Resistance in Methicillin-Resistant Staphylococcus aureus to Newer Antimicrobial Agents. Antimicrob. Agents Chemother. 2019, 63, 1216–1219. [Google Scholar]
- Dookie, N.; Rambaran, S.; Padayatchi, N.; Mahomed, S.; Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis: A review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 2018, 73, 1138–1151. [Google Scholar] [CrossRef] [Green Version]
- Pourakbari, B.; Mamishi, S.; Mohammadzadeh, M.; Mahmoudi, S. First-Line Anti-Tubercular Drug Resistance of Mycobacterium tuberculosis in IRAN: A Systematic Review. Front. Microbiol. 2016, 7, 1139. [Google Scholar] [CrossRef]
- Lopez-Avalos, G.; Gonzalez-Palomar, G.; Lopez-Rodriguez, M.; Vazquez-Chacon, C.A.; Mora-Aguilera, G.; Gonzalez-Barrios, J.A.; Villanueva-Arias, J.C.; Sandoval-Diaz, M.; Miranda-Hernández, U.; Alvarez-Maya, I. Genetic diversity of Mycobacterium tuberculosis and transmission associated with first-line drug resistance: A first analysis in Jalisco, Mexico. J. Glob. Antimicrob. Resist. 2017, 11, 90–97. [Google Scholar] [CrossRef]
- Indraningrat, A.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Perkins, M.V.; Zhang, W.; Franco, C.M.M. New marine natural products from sponges (Porifera) of the order Dictyoceratida (2001 to 2012); a promising source for drug discovery, exploration and future prospects. Biotechnol. Adv. 2016, 34, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Demerdash, A.; Atanasov, A.G.; Horbanczuk, O.K.; Tammam, M.A.; Abdel-Mogib, M.; Hooper, J.N.A.; Sekeroglu, N.; Al-Mourabit, A.; Kijjoa, A. Chemical Diversity and Biological Activities of Marine Sponges of the Genus Suberea: A Systematic Review. Mar. Drugs 2019, 17, 115. [Google Scholar] [CrossRef] [Green Version]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2008, 8, 69–85. [Google Scholar] [CrossRef]
- Hughes, C.C.; Fenical, W. Antibacterials from the Sea. Chem. A Eur. J. 2010, 16, 12512–12525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, G.; Bates, S.S.; La Barre, S. Bioactive Marine Molecules and Derivatives with Biopharmaceutical Potential. In Blue Biotechnology; Wiley: Hoboken, NJ, USA, 2018. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.M.S.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine Pharmacology in 2012–2013: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis, and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs. 2017, 15, 273. [Google Scholar]
- McGivern, J.G. Ziconotide: A review of its pharmacology and use in the treatment of pain. Neuropsychiatr. Dis. Treat. 2007, 3, 69–85. [Google Scholar] [CrossRef] [Green Version]
- Gordon, E.M.; Sankhala, K.K.; Chawla, N.; Chawla, S.P. Trabectedin for Soft Tissue Sarcoma: Current Status and Future Perspectives. Adv. Ther. 2016, 33, 1055–1071. [Google Scholar] [CrossRef] [Green Version]
- El-Damhougy, K.A.; El-Naggar, H.A.; Ibrahim, H.A.; Bashar, M.A.E.; Abou Senna, F.M. Biological activities of some marine sponge extracts from Aqaba Gulf, Red Sea, Egypt. Int. J. Fish. Aquat. Stud. 2017, 5. [Google Scholar] [CrossRef]
- Matobole, R.M.; van Zyl, L.J.; Parker-Nance, S.; Davies-Coleman, M.T.; Trindade, M. Antibacterial Activities of Bacteria Isolated from the Marine Sponges Isodictya compressa and Higginsia bidentifera Collected from Algoa Bay, South Africa. Mar. Drugs. 2017, 15, 47. [Google Scholar] [CrossRef] [Green Version]
- Van Soest, R.W.M.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; De Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N.A. Global diversity of sponges (Porifera). PLoS ONE 2012, 7, e35105. [Google Scholar] [CrossRef]
- Mohamed, M.; Radwan, S.P.M.; Samir, A. Ross, Two New Sulfated Sterols from the Marine Sponge Lendenfeldia dendyi NPC. Nat. Prod. Commun. 2007, 2, 1934578X0700200905. [Google Scholar]
- Liu, Y.; Liu, R.; Mao, S.-C.; Morgan, J.B.; Jekabsons, M.B.; Zhou, Y.-D.; Nagle, D.G. Molecular-Targeted Antitumor Agents. 19. Furospongolide from a Marine Lendenfeldia sp. Sponge Inhibits Hypoxia-Inducible Factor-1 Activation in Breast Tumor Cells. J. Nat. Prod. 2008, 71, 1854–1860. [Google Scholar] [CrossRef] [Green Version]
- Sera, Y.; Adachi, K.; Shizuri, Y. A New Epidioxy Sterol as an Antifouling Substance from a Palauan Marine Sponge, Lendenfeldia chondrodes. J. Nat. Prod. 1999, 62, 152–154. [Google Scholar] [CrossRef]
- Thakur, N.L.; Anil, A.C. Antibacterial Activity of the Sponge Ircinia Ramosa: Importance of its Surface-Associated Bacteria. J. Chem. Ecol. 2000, 26, 57–71. [Google Scholar] [CrossRef]
- Belma Konuklugġl, B.G. Antimicrobial activity of marine samples collected from the different coasts of turkey. Turk. J. Pharm. Sci. 2015, 12, 337–344. [Google Scholar]
- Mohamed, N.M.; Rao, V.; Hamann, M.T.; Kelly, M.; Hill, R.T. Monitoring Bacterial Diversity of the Marine Sponge Ircinia strobilina upon Transfer into Aquaculture. Appl. Environ. Microbiol. 2008, 74, 4133. [Google Scholar] [CrossRef] [Green Version]
- Evans-Illidge, E.A.; Logan, M.; Doyle, J.; Fromont, J.; Battershill, C.N.; Ericson, G.; Wolff, C.W.; Muirhead, A.; Kearns, P.; Abdo, D.; et al. Phylogeny drives large scale patterns in Australian marine bioactivity and provides a new chemical ecology rationale for future biodiscovery. PLoS ONE 2013, 8, e73800. [Google Scholar] [CrossRef]
- Dinarvand, M.; Spain, M.P.; Vafaee, F. Pharmacodynamic Functions of Synthetic Derivatives for Treatment of Methicillin-Resistant Staphylococcus aureus (MRSA) and Mycobacterium tuberculosis. Front. Microbiol. 2020, 11, 551189. [Google Scholar] [CrossRef]
- Simpson, M.; Poulsen, S.-A. An Overview of Australia′s Compound Management Facility: The Queensland Compound Library. ACS Chem. Biol. 2014, 9, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Compounds Australia. Available online: https://www.griffith.edu.au/griffith-sciences/compounds-australia (accessed on 20 September 2018).
- Bouslimani, A.; Sanchez, L.M.; Garg, N.; Dorrestein, P.C. Mass spectrometry of natural products: Current, emerging and future technologies. Nat. Prod. Rep. 2014, 31, 718–729. [Google Scholar] [CrossRef] [Green Version]
- Kind, T.; Fiehn, O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010, 2, 23–60. [Google Scholar] [CrossRef] [Green Version]
- Herbert Júnior, D.; Nathalya Isabel de, M.; Antônio Eduardo Miller, C. Electrospray Ionization Tandem Mass Spectrometry as a Tool for the Structural Elucidation and Dereplication of Natural Products: An Overview. In Tandem Mass Spectrometry—Applications and Principles; InTech: London, UK, 2012. [Google Scholar]
- McCracken, S.T. Synthetic Studies of Biologically Active Natural Products —Ascididemin and 6-Substituted 2-Pyranones. Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 2010. [Google Scholar]
- Lindsay, B.S. Studies in Marine Natural Product Synthesis, lsolation and Ecology. Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 1998. [Google Scholar]
- Da Silva Liberio, M. Chemical and Biological Investigations of Anticancer Compounds from Australian Ascidians. Ph.D. Thesis, Griffith University, Brisbane, Australia, 2014. [Google Scholar]
- Phuwapraisirisan, P.; Matsunaga, S.; van Soest, R.W.M.; Fusetani, N. Shinsonefuran, a cytotoxic furanosesterterpene with a novel carbon skeleton, from the deep-sea sponge Stoeba extensa1See Ref. 1.1. Tetrahedron Lett. 2004, 45, 2125–2128. [Google Scholar] [CrossRef]
- Carr, G. Bioactive Marine Natural Products: Isolation, Structure Elucidation and Synthesis of Pharmacophore Analogues. Ph.D. Thesis, The University of British Columbia, Vancouver, BC, Canada, 2010. [Google Scholar]
- Veltri, C.A. Identification of Cellular Targets of Marine Natural Products. Ph.D. Thesis, The University of Utah, Salt Lake City, UT, USA, 2009. [Google Scholar]
- Zhu, Q. A review of novel bacterial complex lipids: Implications for the pathogenesis of apical periodontitis. Iran. Endod. J. 2010, 5, 141–146. [Google Scholar]
- Bontemps, N.; Bry, D.; Lopez-Legentil, S.; Simon-Levert, A.; Long, C.; Banaigs, B. Structures and antimicrobial activities of pyridoacridine alkaloids isolated from different chromotypes of the ascidian Cystodytes dellechiajei. J. Nat. Prod. 2010, 73, 1044–1048. [Google Scholar] [CrossRef]
- Furuta, A.; Salam, K.A.; Hermawan, I.; Akimitsu, N.; Tanaka, J.; Tani, H.; Yamashita, A.; Moriishi, K.; Nakakoshi, M.; Tsubuki, M.; et al. Identifiction and biochemical characterization of halisulfate 3 and suvanine as novel inhibitors of hepatitis C virus NS3 helicase from a marine sponge. Mar. Drugs 2014, 12, 462–476. [Google Scholar] [CrossRef]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef] [Green Version]
- Ibarguren, M.; López, D.J.; Escribá, P.V. The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1518–1528. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, A.; Nara, M.; Sugiyama, Y.; Sakanaka, M.; Yachi, H.; Kitakata, A.; Nakagawa, A.; Minami, H.; Okuda, S.; Katoh, T.; et al. Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci. Biotechnol. Biochem. 2017, 81, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, J.H.H.L.; Seleghim, M.H.R.; Timm, C.; Grube, A.; Köck, M.; Nascimento, G.G.F.; Martins, A.C.T.; Silva, E.G.O.; de Souza, A.O.; Minarini, P.R.R.; et al. Antimicrobial and Antimycobacterial Activity of Cyclostellettamine Alkaloids from Sponge Pachychalina sp. Mar. Drugs 2006, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Febles, M.; Montalvao, S.; Crespin, G.D.; Norte, M.; Padron, J.M.; Tammela, P.; Fernandez, J.J.; Daranas, A.H. Synthesis and biological evaluation of crown ether acyl derivatives. Bioorganic Med. Chem. Lett. 2016, 26, 5591–5593. [Google Scholar] [CrossRef] [Green Version]
- Christopher, J.M.; Charles, W.R.; Thomas, R. Synthesis of marine alkaloid ascididemin. Tetrahedron Lett. 1990, 31, 4375–4376. [Google Scholar]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Gaush, C.R.; Hard, W.L.; Smith, T.F. Characterization of an established line of canine kidney cells (MDCK). Proceedings of the Society for Experimental Biology and Medicine. Soc. Exp. Biol. Med. 1966, 122, 931–935. [Google Scholar] [CrossRef]
- Tsuchiya, S.; Yamabe, M.; Yamaguchi, Y.; Kobayashi, Y.; Konno, T.; Tada, K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 1980, 26, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Poigny, S.; Nouri, S.; Chiaroni, A.; Guyot, M.; Samadi, M. Total Synthesis and Determination of the Absolute Configuration of Coscinosulfate. A New Selective Inhibitor of Cdc25 Protein Phosphatase. J. Org. Chem. 2001, 66, 7263–7269. [Google Scholar] [CrossRef]
- Bae, J.; Jeon, J.-E.; Lee, Y.-J.; Lee, H.-S.; Sim, C.J.; Oh, K.-B.; Shin, J. Sesterterpenes from the Tropical Sponge Coscinoderma sp. J. Nat. Prod. 2011, 74, 1805–1811. [Google Scholar] [CrossRef]
- Kernan, M.R.; Faulkner, D.J. Sesterterpene sulfates from a sponge of the family Halichondriidae. J. Org. Chem. 1988, 53, 4574–4578. [Google Scholar] [CrossRef]
- Loukaci, A.; Le Saout, I.; Samadi, M.; Leclerc, S.; Damiens, E.; Meijer, L.; Debitus, C.; Guyot, M. Coscinosulfate, a CDC25 phosphatase inhibitor from the sponge Coscinoderma mathewsi. Bioorganic Med. Chem. 2001, 9, 3049–3054. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nature reviews. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zidar, N.; Montalvão, S.; Hodnik, Ž.; Nawrot, D.; Žula, A.; Ilaš, J.; Kikelj, D.; Tammela, P.; Mašič, L. Antimicrobial Activity of the Marine Alkaloids, Clathrodin and Oroidin, and Their Synthetic Analogues. Mar. Drugs 2014, 12, 940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
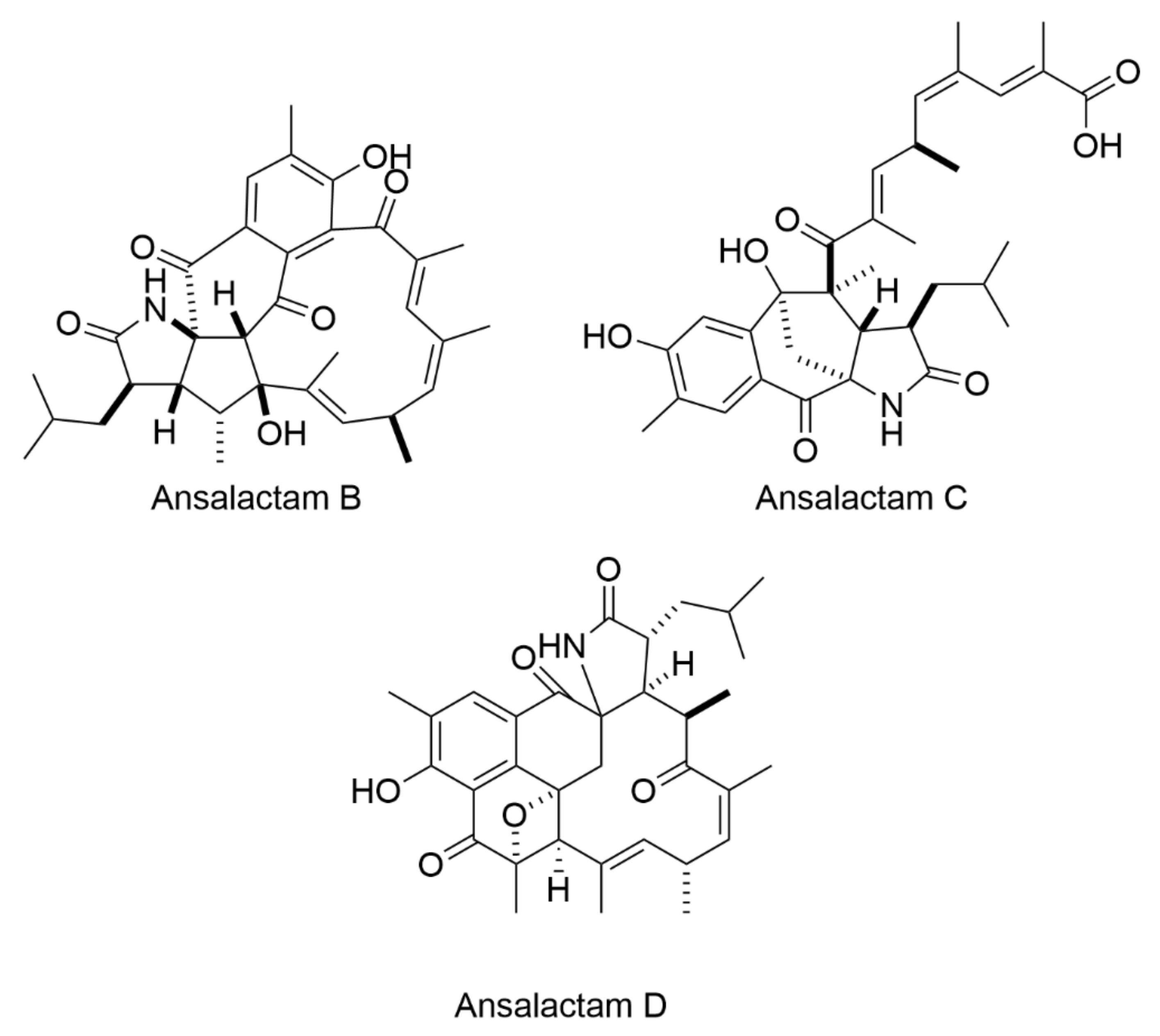
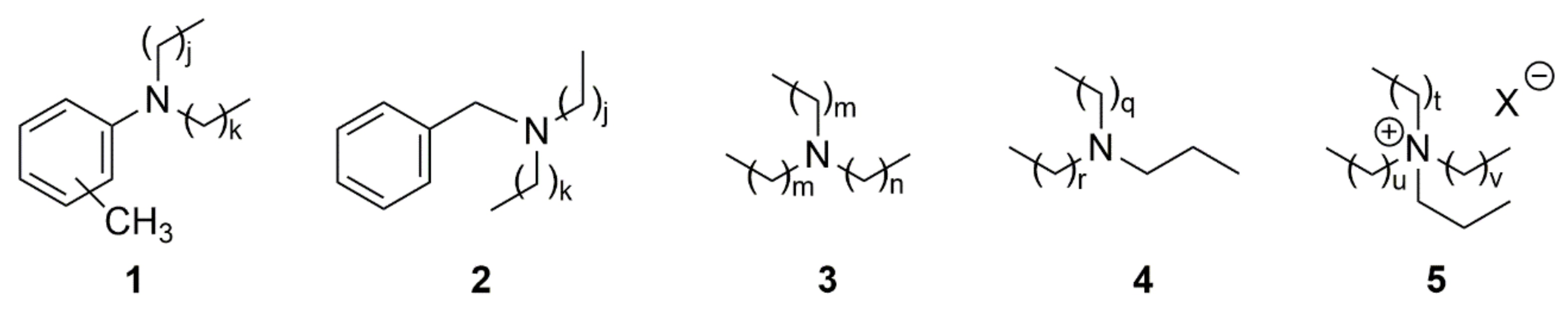
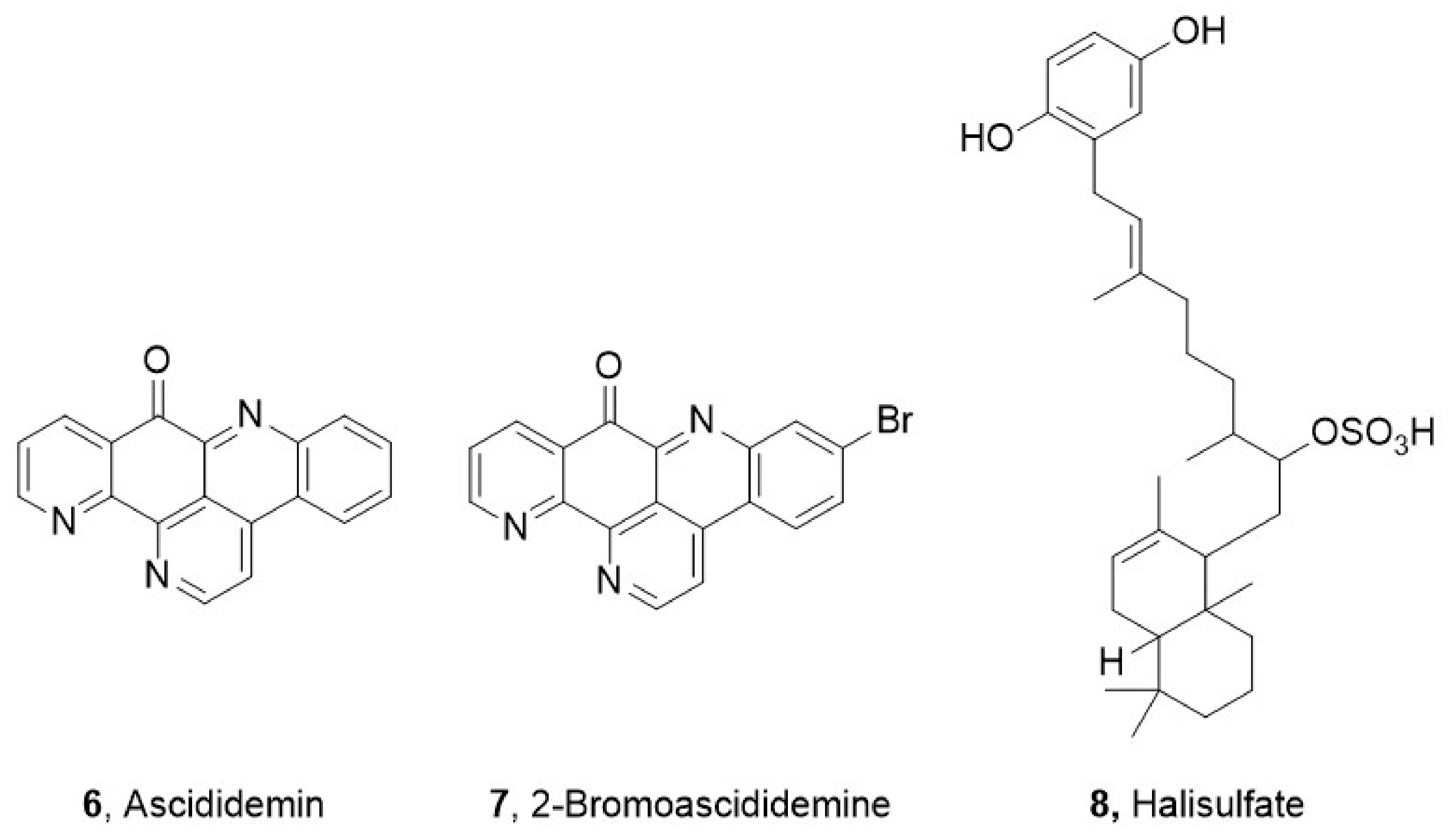

| Entry | AIMS Sample Code | QCL Sample Number | MIC (µg mL−1) | Cytotoxicity (% Cell Survival) | ||
|---|---|---|---|---|---|---|
| MRSA | Hep G2 | A549 | HEK | |||
| 1 | 19,033 | SN00733110 | 31.25 ± 0.9 | 91 ± 1.2 | 91 ± 0.3 | 98 ± 1.6 |
| 2 | 20,608 | SN00760947 | 31.25 ± 1.3 | 97 ± 1.0 | 101 ± 2.9 | 102 ± 0.7 |
| 3 | 20,608 | SN00760956 | 31.25 ± 0.4 | 100 ± 4.4 | 106 ± 1.8 | 95 ± 1.0 |
| 4 | 20,608 | SN00760958 | 62.5 ± 2.2 | 98 ± 0.6 | 108 ± 14 | 95 ± 2.5 |
| 5 | 26,051 | SN00731005 | 62.5 ± 2.8 | 101 ± 1.0 | 110 ± 2.7 | 98 ± 2.0 |
| 6 | 24,307 | SN00730755 | 31.25 ± 1.0 | 100 ± 1.0 | 106 ± 1.0 | 96 ± 4.0 |
| 7 | 25,663 | SN00732222 | 15.6 ± 1.5 | 100 ± 1.6 | 89 ± 2.0 | 97 ± 0.0 |
| 8 | 26,104 | SN00734298 | 62.5 ± 1.0 | 98 ± 0.5 | 100 ± 1.1 | 98 ± 1.1 |
| 9 | 22,565 | SN00739718 | 31.25 ± 0.4 | 100 ± 0.0 | 99 ± 0.4 | 97 ± 1.0 |
| Sample | Molecular Ion (m/z) | Molecular Formula | Proposed Structures | Spectra | MS/MS | NMR Analyses |
|---|---|---|---|---|---|---|
| 19,033 † | 326.37813 [M + H]+ | [C22H48N]+ Calc. = 326.37802 (∆m = 0.11 ppm) RDBE = 0 | 3 | Figure S1 | Table S2 Figure S11 | - |
| 20,608 | 332.33115 [M + H]+ | [C23H42N]+ Calc = 332.33118 (Δm = 0.03 ppm) RDBE = 4 ‡ | 1,2 | Figure S3 | Table S3 Figure S13 | - |
| 26,051 | 368.42508 [M + H]+ | [C25H54N]+ Calc. = 368.42495 (∆m = 0.13 ppm) RDBE = 0 | 4,5 | Figure S5 | Table S4 Figure S15 | - |
| 25,663 | 306.0635 [M+Na]+ | [C18H9ON3] Calc. = 306.0637 (∆m = 0.8 ppm) RDBE = 16 | 6 | Figure S7 | - | Table S5 Figure S20 |
| 25,663 | 361.9925 [M + H]+ | [C18H8BrN3O] Calc. = 361.9923 (∆m = 0.5 ppm) RDBE = 16 | 7 | Figure S7 | - | Table S6 Figure S21 |
| 26,104 | 467.117 [M + H]+ | [C30H17N2O4] Calc. = 467.118 (∆m = 1.9 ppm) RDBE = 0 | 8 | Figure S9 | - | Table S7 Figures S22–S25 |
| Name and CAS no. | Structures | MRSA | E. coli | P. aeruginosa | M. tuberculosis | HepG2 | HEK 293 | A549 | THP-1 |
|---|---|---|---|---|---|---|---|---|---|
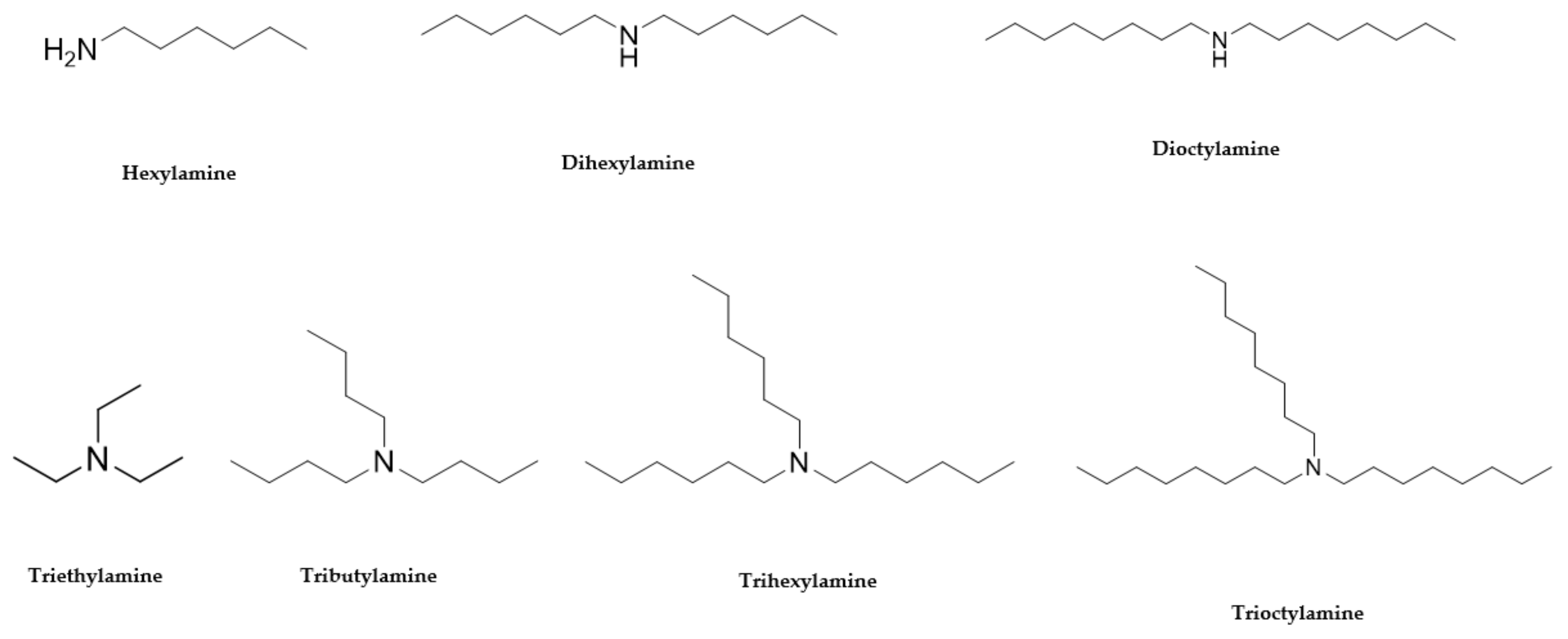
| Hexylamine 111-26-2 |  | >100 | >100 | 50 | >100 | >100 | >100 | >100 | >100 |
| Dihexylamine 143-16-8 |  | 12.5 | 12.5 | 6.2 | >100 | >100 | >100 | >100 | >100 |
| Dioctylamine 1120-48-5 | 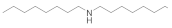 | 6.2 | 6.2 | 3 | >100 | >100 | >100 | >100 | >100 |
| Triethylamine 121-44-8 | 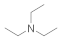 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| Tributylamine 102-82-9 | 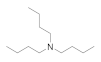 | >100 | >100 | 50 | >100 | >100 | >100 | >100 | >100 |
| Trihexylamine 102-86-3 | 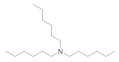 | 3 | 3 | 6.5 | >100 | >100 | 50 | >100 | >100 |
| Trioctylamine 1116-76-3 | 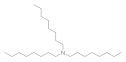 | 12.5 | 25 | 25 | 0.02 | >100 | 6 | 50 | 3.1 |
| Name and CAS no. | Structures | MRSA | E. coli | P. aeruginosa | M. tuberculosis | HepG2 | HEK 293 | A549 | THP-1 |
|---|---|---|---|---|---|---|---|---|---|
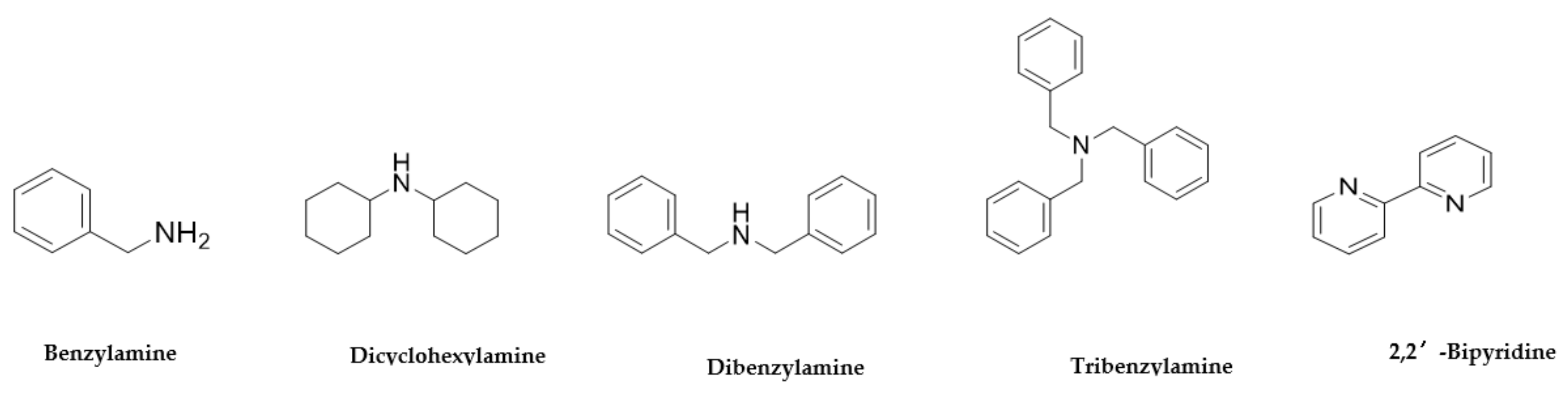
| Benzylamine 100-46-9 |  | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| Dicyclohexylamine 101-83-7 |  | >100 | >100 | 12.5 | >100 | >100 | >100 | >100 | >100 |
| Dibenzylamine 103-49-1 |  | >100 | >100 | 12.5 | >100 | >100 | >100 | >100 | >100 |
| Tribenzylamine 620-40-6 |  | >100 | >100 | 6.5 | >100 | >100 | >100 | >100 | >100 |
| 2,2′-Bipyridine 366-18-7 |  | >100 | >100 | >100 | 0.1 | 3 | 3 | >100 | 50 |
| Name and CAS no. | Structures | MRSA | E. coli | P. aeruginosa | M. tuberculosis | HepG2 | HEK 293 | A549 | THP-1 |
|---|---|---|---|---|---|---|---|---|---|
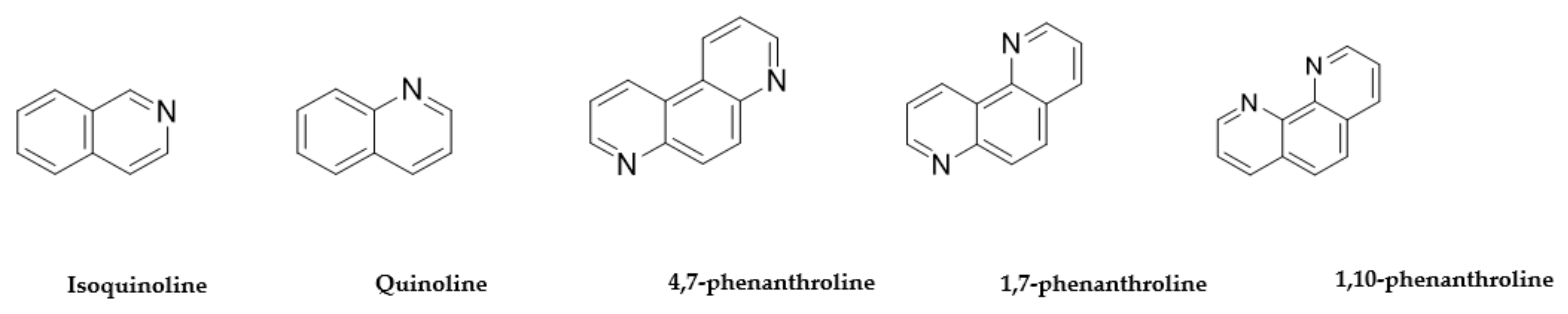
| Isoquinoline 119-65-3 |  | >100 | >100 | >100 | 0.1 | 3 | >100 | 50 | 3 |
| Quinoline 91-22-5 | 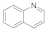 | >100 | >100 | >100 | 0.1 | 3 | 50 | 50 | 3 |
| 4,7-phenanthroline 230-07-9 |  | 1.5 | 1.5 | 2.5 | 2.5 | >100 | >100 | >100 | >100 |
| 1,7-phenanthroline 230-46-6 | 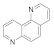 | 25 | 25 | 2.5 | >100 | >100 | >100 | >100 | >100 |
| 1,10-phenanthroline 66-71-7 | 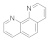 | >100 | 50 | 0.3 | 0.1 | 0.1 | 50 | 3 | 0.1 |
| Name and CAS no. | Structures | MRSA | E. coli | P. aeruginosa | TB. H37Rv | HepG2 | HEK 293 | A549 | THP-1 |
|---|---|---|---|---|---|---|---|---|---|
| 4,7-Dimethyl-1,10-Phenanthrolin 3248-05-3 |  | 0.3 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Neocuproine hemihydrate 34302-69-7 |  | 0.09 | 0.3 | 0.04 | 0.1 | 0.1 | 0.1 | 6.2 | 12 |
| 5,6-Dimethyl-1,10-phenanthroline 3002-81-1 | 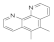 | 0.3 | 0.78 | >100 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3,4,7,8-Tetramethyl-1,10-phenanthroline 1660-93-1 | 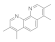 | 0.3 | 0.3 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1,10-Phenanthroline-5,6-dione 27318-90-7 | 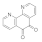 | 0.04 | 0.04 | 0.04 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Name and CAS no. | Structures | MRSA | E. coli | P. aeruginosa | TB. H37Rv | HepG2 | HEK 293 | A549 | THP-1 |
|---|---|---|---|---|---|---|---|---|---|
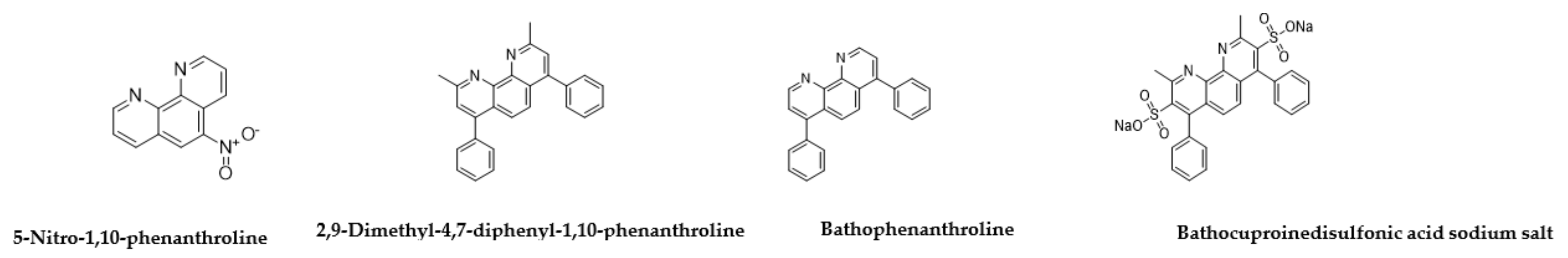
| 5-Nitro-1,10-phenanthroline 4199-88-6 |  | 0.09 | 0.78 | >100 | 0.1 | 0.1 | 0.1 | 12 | 0.1 |
| Bathophenanthroline 1662-01-7 | 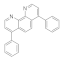 | 0.09 | 0.019 | 0.03 | 0.01 | 1.5 | 25 | 3 | 13 |
| 2,9-Dimethyl-4,7-diphenyl-1,10-phenanthroline 4733-39-5 | 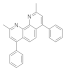 | 25 | 0.78 | 50 | 50 | 0.1 | 0.1 | >100 | >100 |
| Bathocuproinedisulfonic acid sodium salt 1257642-74-2 | 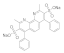 | >100 | >100 | 0.19 | 1.2 | 50 | 0.1 | >100 | >100 |
| Sample Name, structure and CAS no | Minimum Inhibitory/Toxicity Concentration of Drug (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| MRSA | E. coli | P. aeruginosa | M. tuberculosis H37Rv | HepG2 | HEK 293 | A549 | THP-1 | |
Hexylamine  111-26-2 111-26-2 | >100 | >100 | 50 | >100 | >100 | >100 | >100 | >100 |
Dihexylamine 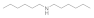 143-16-8 143-16-8 | 12.5 | 12.5 | 6.2 | >100 | >100 | 100 | >100 | >100 |
Dioctylamine 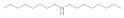 1120-48-5 1120-48-5 | 6.2 | 6.2 | 3 | >100 | >100 | >100 | >100 | >100 |
Triethylamine  121-44-8 121-44-8 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
Tributylamine 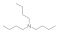 102-82-9 102-82-9 | >100 | >100 | 50 | >100 | >100 | >100 | >100 | >100 |
Trihexylamine  102-86-3 102-86-3 | 3 | 3 | 6.5 | >100 | >100 | 50 | 100 | >100 |
Trioctylamine  1116-76-3 1116-76-3 | 12.5 | 25 | 25 | 0.02 | >100 | 6 | 50 | 3.1 |
Benzylamine 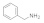 100-46-9 100-46-9 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
Dicyclohexylamine  101-83-7 101-83-7 | >100 | >100 | 12.5 | >100 | >100 | >100 | >100 | >100 |
Dibenzylamine 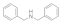 103-49-1 103-49-1 | >100 | >100 | 12.5 | >100 | >100 | >100 | >100 | >100 |
Tribenzylamine  620-40-6 620-40-6 | >100 | >100 | 6.5 | >100 | >100 | >100 | >100 | >100 |
2,2′-Bipyridyl 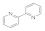 366-18-7 366-18-7 | >100 | >100 | >100 | 0.1 | 3 | 3 | >100 | 50 |
Isoquinoline 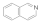 119-65-3 119-65-3 | >100 | >100 | >100 | 0.1 | 3 | 3 | >100 | 50 |
Quinoline 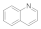 91-22-5 91-22-5 | >100 | >100 | >100 | 0.1 | 3 | 3 | 50 | 50 |
4,7-phenanthroline 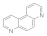 230-07-9 230-07-9 | 1.5 | 1.5 | 2.5 | 2.5 | >100 | >100 | >100 | >100 |
1,7-phenanthroline 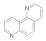 230-46-6 230-46-6 | 25 | 25 | 2.5 | >100 | >100 | >100 | >100 | >100 |
1,10 phenanthroline 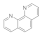 66-71-7 66-71-7 | >100 | 50 | 0.3 | 0.1 | 0.1 | 0.1 | 50 | 3 |
4,7-DImethyl-1,10-Phenanthroline 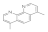 3248-05-3 3248-05-3 | 0.3 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
Neocuproine hemihydlyate 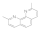 34302-69-7 34302-69-7 | 0.09 | 0.3 | 0.04 | 0.1 | 0.1 | 0.1 | 6.2 | 12 |
5,6-Dimethyl-1, 10-phenanthroline  3002-81-1 3002-81-1 | 0.3 | 0.78 | >100 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
3,4,7,8-Tetramethyl-1,10-phenanthroline 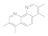 1660-93-1 1660-93-1 | 0.3 | 0.3 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
1,10-Phenanthroline-5,6-dione 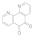 27318-90-7 27318-90-7 | 0.04 | 0.04 | 0.04 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
5-Nitro-1,10-phenanthroline 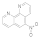 4199-88-6 4199-88-6 | 0.09 | 0.78 | >100 | 0.1 | 0.1 | 0.1 | 12 | 0.1 |
Bathophenanthroline 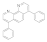 1662-01-7 1662-01-7 | 0.09 | 0.019 | 0.03 | 0.01 | 1.5 | 25 | 3 | 13 |
2,9-Dimethyl-1,7-diphenyl-1,10-phenatroline  4733-39-5 4733-39-5 | 25 | 0.78 | 50 | 50 | 0.1 | 0.1 | >100 | >100 |
Bathocuproinedisulfonic acid sodium salt  1257642-74-2 1257642-74-2 | >100 | >100 | 0.19 | 1.2 | 50 | 0.1 | >100 | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dinarvand, M.; Spain, M. Identification of Bioactive Compounds from Marine Natural Products and Exploration of Structure-Activity Relationships (SAR). Antibiotics 2021, 10, 337. https://doi.org/10.3390/antibiotics10030337
Dinarvand M, Spain M. Identification of Bioactive Compounds from Marine Natural Products and Exploration of Structure-Activity Relationships (SAR). Antibiotics. 2021; 10(3):337. https://doi.org/10.3390/antibiotics10030337
Chicago/Turabian StyleDinarvand, Mojdeh, and Malcolm Spain. 2021. "Identification of Bioactive Compounds from Marine Natural Products and Exploration of Structure-Activity Relationships (SAR)" Antibiotics 10, no. 3: 337. https://doi.org/10.3390/antibiotics10030337
APA StyleDinarvand, M., & Spain, M. (2021). Identification of Bioactive Compounds from Marine Natural Products and Exploration of Structure-Activity Relationships (SAR). Antibiotics, 10(3), 337. https://doi.org/10.3390/antibiotics10030337





