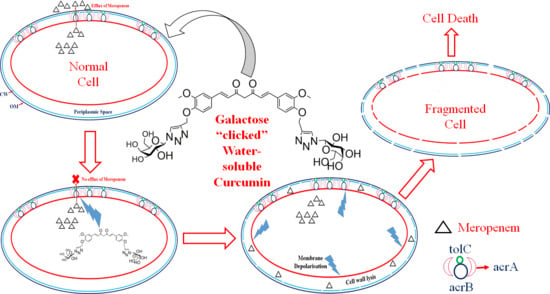
Galactose-Clicked Curcumin-Mediated Reversal of Meropenem Resistance among Klebsiella pneumoniae by Targeting Its Carbapenemases and the AcrAB-TolC Efflux System
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Susceptibility Profile
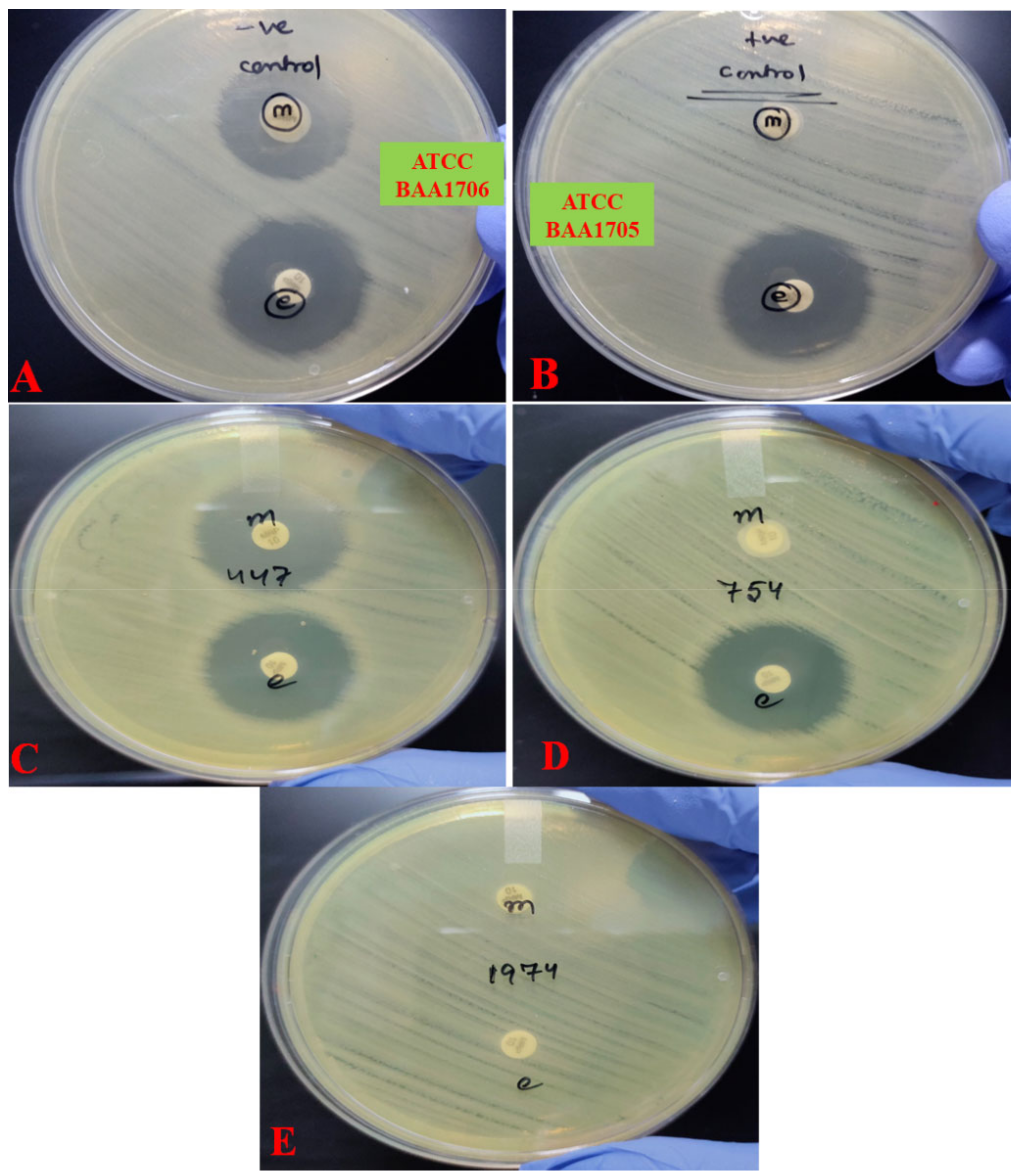
2.2. Detection of Carbapenemase Production
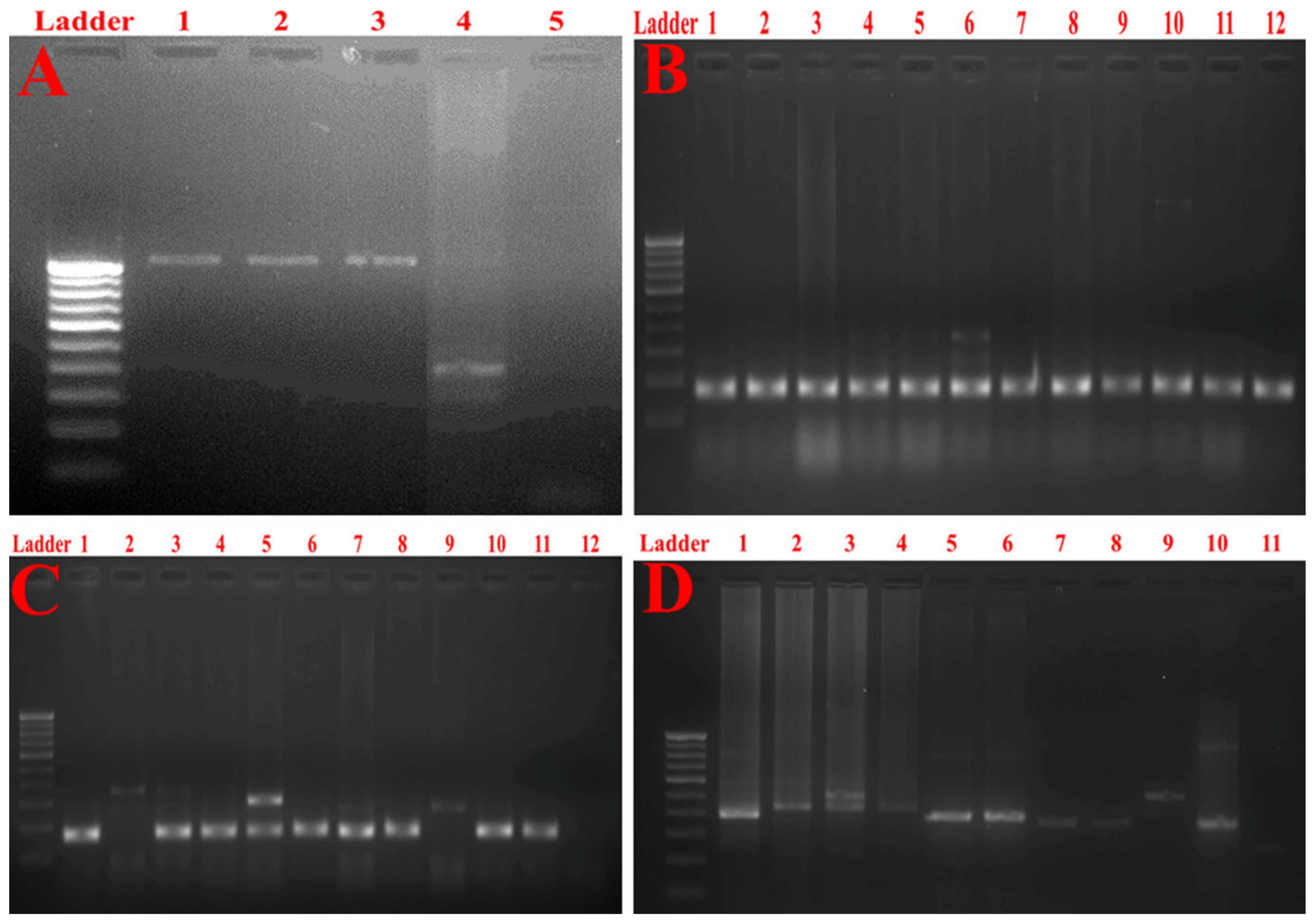
2.3. Molecular Typing of Klebsiella Pneumoniae Isolates Using Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR
2.4. Synthesis of Water-Soluble Curcumin (Curaq)
2.5. Photophysical Behaviour and Solubility
2.6. Minimum Inhibitory Concentration (MIC) Determination among Carbapenemase Producers
2.6.1. Minimum Inhibitory Concentration (MIC) Determination
2.6.2. Alteration in Minimum Inhibitory Concentration (MIC) after Adding CCCP, Verapamil, Valinomycin and Curaq
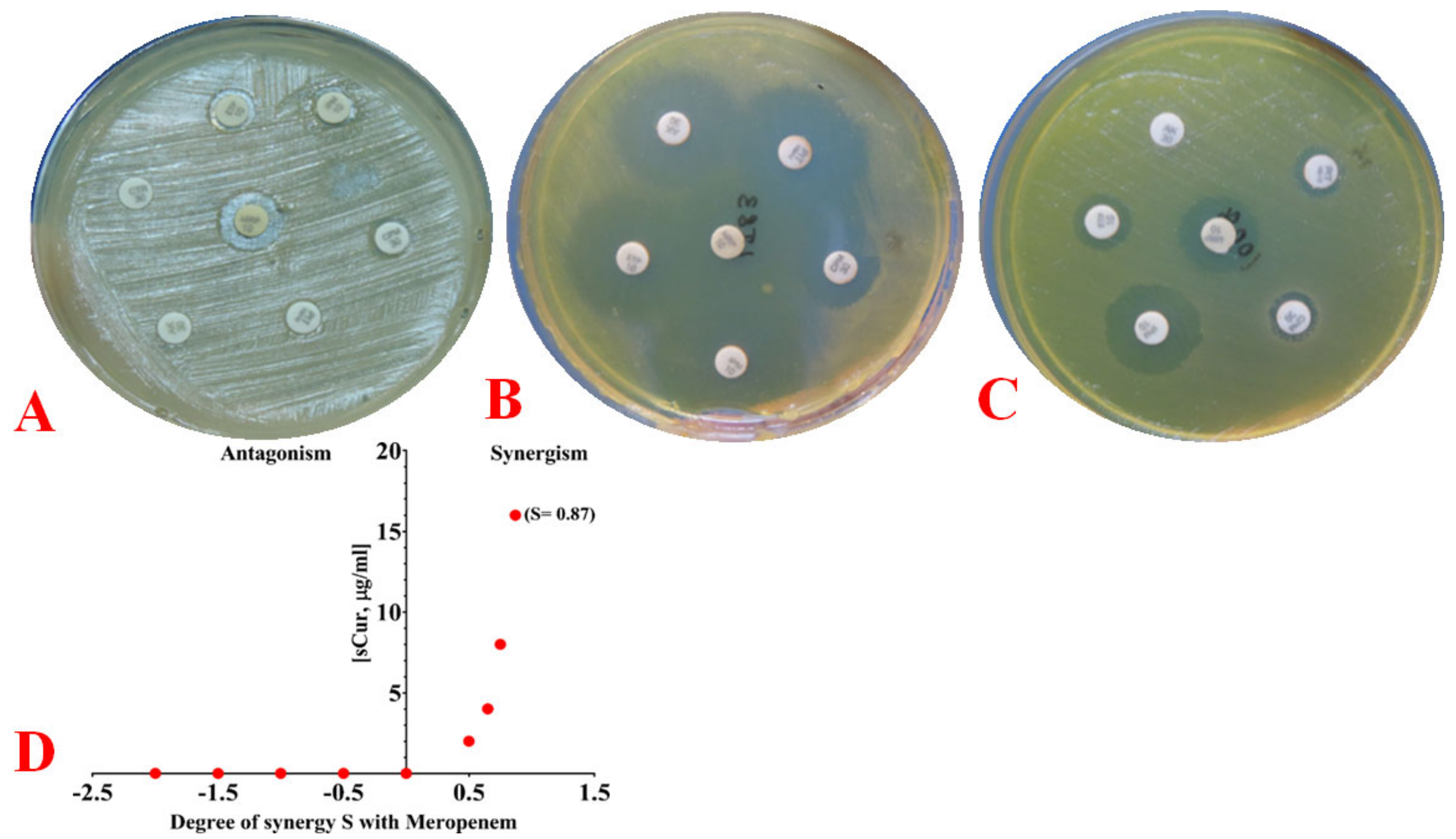
2.6.3. Phenotypic Observation of Synergy and Its Quantification Using the Bliss Model
2.7. Detection of Multidrug Resistance and Virulence Determinants Using PCR
2.8. Detection of Major Efflux Pumps System
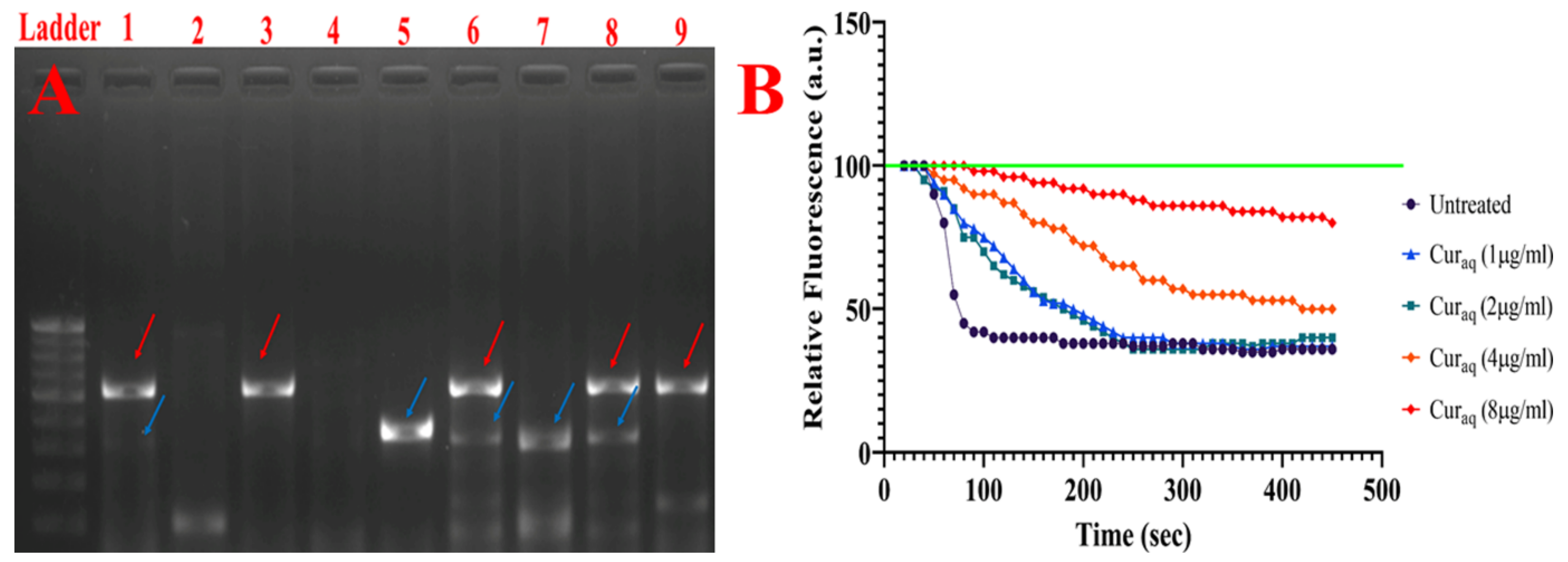
2.9. 1,2′-Dinaphthylamine Accumulation Assay
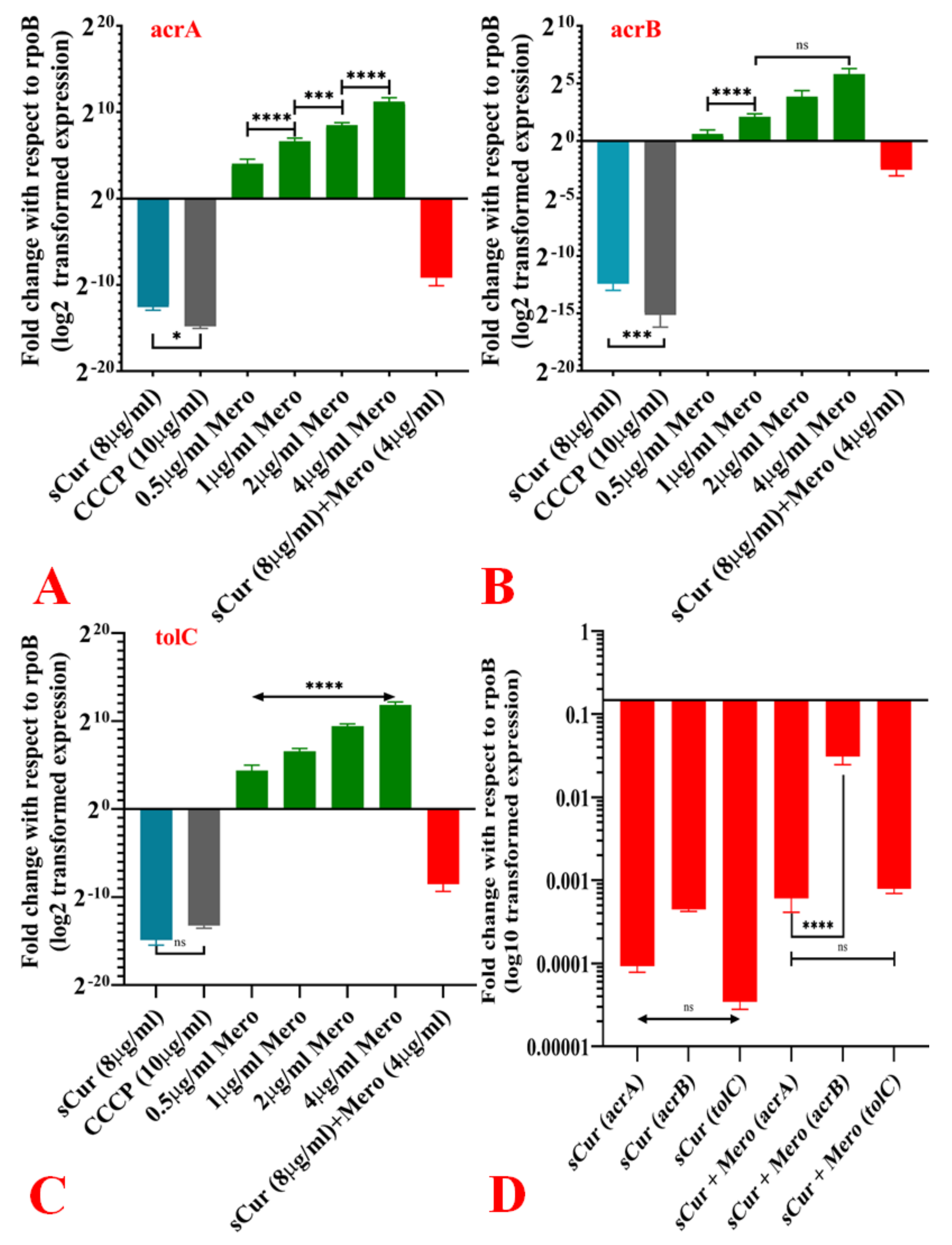
2.10. Expression Pattern of acrA, acrB and TolC in the Presence of Curaq
2.11. Effect over Carbapenemases
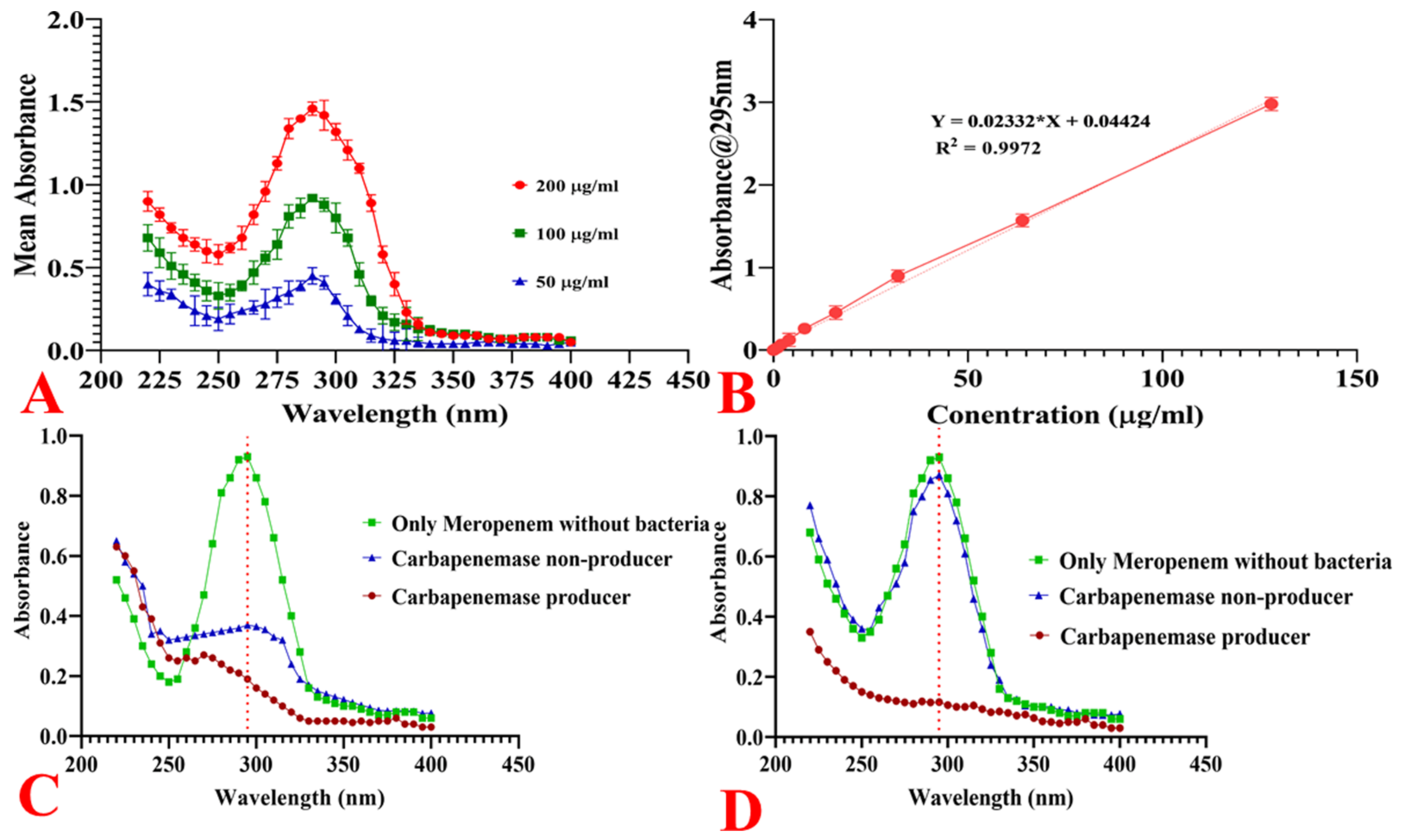
2.11.1. Spectrophotometric Analysis of Meropenem
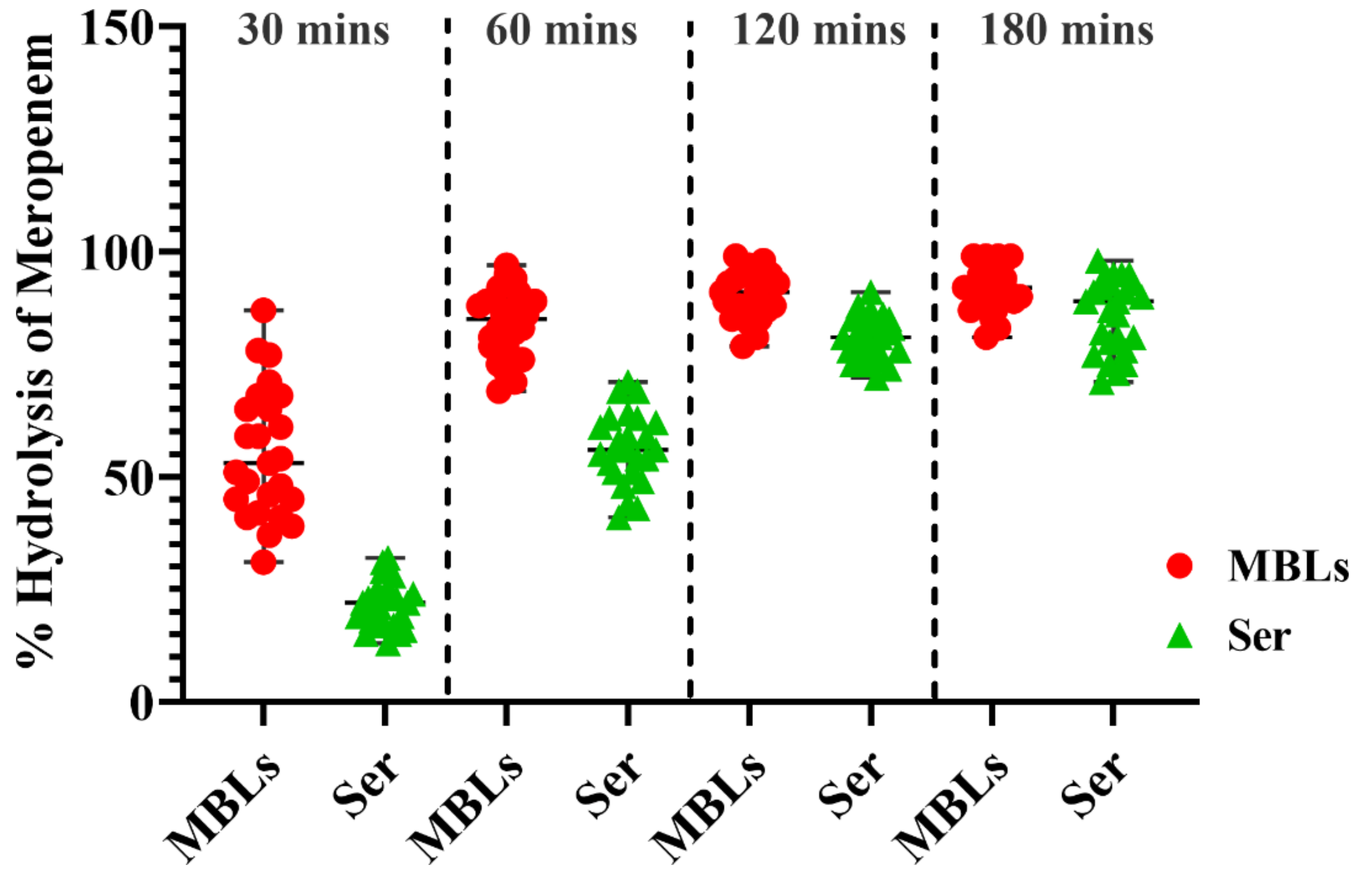
2.11.2. Hydrolysis of Meropenem by Bacterial Cells
2.11.3. Quantification of Bacterial Hydrolytic Activity to Meropenem
2.11.4. Carbapenemases Inhibition Kinetics by Curaq
2.12. Measurement of the Electrical Potential
2.13. Membrane Depolarisation Assay
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Bacterial Isolation and Culture Conditions
4.3. Antimicrobial Susceptibility Testing
4.4. Detection of Carbapenemases
4.5. Molecular Typing of Klebsiella Pneumoniae Isolates Using Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR
4.6. Synthesis of Water-Soluble Curcumin
4.6.1. Synthesis of Galactose Azide (2,3,4,6-Tetra-O-Acetyl-β-D-Galactopyranosyl Azide) (2)
4.6.2. Synthesis of Curcumin Di-Alkyne (4)
4.6.3. Synthesis of Curcumin Clicked Di-Acetylated Galactose (5)
4.6.4. Synthesis of De-O-Acetylated Curcumin Di-Galactose (6)
4.7. Photophysical Behavior and Solubility
4.8. Minimum Inhibitory Concentration (MIC) Determination
4.8.1. Antibiotics and Efflux Inhibitors
4.8.2. Phenotypic Evaluation of Drug Synergy
4.8.3. Quantifying Synergy Using the Bliss Independence Model and Calculating the Fractional Inhibitory Concentration Index
4.9. Determination of Various Virulence Factors
4.10. Detection of Major Efflux Pumps System
4.11. Mechanistic Underpinnings
4.11.1. 1,2′-Dinaphthylamine (1,2′-DNA) Efflux Assay
4.11.2. Quantitative Real-Time PCR (qRT-PCR) for Determination of Expression of acrA, acrB and TolC in Presence and Absence of Soluble Curcumin or CCCP, Along with Meropenem at Different Concentrations
4.11.3. Spectrophotometric Analysis of Meropenem
4.11.4. Meropenem Hydrolysis by Bacterial Cells
4.11.5. Estimation of Background Absorbance
4.11.6. Quantification of Bacterial Meropenem Hydrolytic Activities
4.11.7. Quantification of the Effect of Curaq on Bacterial Meropenem Hydrolytic Activities
4.11.8. Preparation of Cell-Free Extracts
4.11.9. Protein Measurement
4.11.10. Inhibition Assays on Carbapenemases
4.11.11. Measurement of the Electrical Potential
4.11.12. Membrane Depolarisation Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podschun, R.; Ullmann, U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.K.; Russo, T.A.; Karchmer, A.W. Hypervirulent Klebsiella pneumoniae. Open Forum Infect. Dis. 2014, 1, ofu028. [Google Scholar] [CrossRef] [Green Version]
- Shon, A.S.; Russo, T.A. Hypervirulent Klebsiella pneumoniae: The next superbug? Future Microbiol. 2012, 7, 669–671. [Google Scholar] [CrossRef]
- Piperaki, E.-T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atzori, A.; Malloci, G.; Cardamone, F.; Bosin, A.; Vargiu, A.V.; Ruggerone, P. Molecular Interactions of Carbapenem Antibiotics with the Multidrug Efflux Transporter AcrB of Escherichia coli. Int. J. Mol. Sci. 2020, 21, 860. [Google Scholar] [CrossRef] [Green Version]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Bengoechea, J.A.; Pessoa, J.S. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019, 43, 123–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Prakash, P.; Singh, R.; Nandy, N.; Firdaus, Z.; Bansal, M.; Singh, R.K.; Srivastava, A.; Roy, J.K.; Mishra, B.; et al. Curcumin Quantum Dots Mediated Degradation of Bacterial Biofilms. Front. Microbiol. 2017, 8, 1517. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Yadav, S.; Sharma, K.; Firdaus, Z.; Aditi, P.; Neogi, K.; Bansal, M.; Gupta, M.K.; Shanker, A.; Singh, R.K.; et al. Quantum curcumin mediated inhibition of gingipains and mixed-biofilm of Porphyromonas gingivalis causing chronic periodontitis. RSC Adv. 2018, 8, 40426–40445. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-J.; Pan, M.-H.; Cheng, A.-L.; Lin, L.-I.; Ho, Y.-S.; Hsieh, C.-Y.; Lin, J.-K. Stability of curcumin in buffer solutions and characterization of its degradation products. J. Pharm. Biomed. Anal. 1997, 15, 1867–1876. [Google Scholar] [CrossRef]
- Krausz, A.E.; Adler, B.L.; Cabral, V.; Navati, M.; Doerner, J.; Charafeddine, R.A.; Chandra, D.; Liang, H.; Gunther, L.; Clendaniel, A.; et al. Curcumin-encapsulated nanoparticles as innovative antimicrobial and wound healing agent. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Mishra, H.; Firdaus, Z.; Yadav, S.; Aditi, P.; Nandy, N.; Sharma, K.; Bose, P.; Pandey, A.K.; Chauhan, B.S.; et al. MoS2-Modified Curcumin Nanostructures: The Novel Theranostic Hybrid Having Potent Antibacterial and Antibiofilm Activities against Multidrug-Resistant Hypervirulent Klebsiella pneumoniae. Chem. Res. Toxicol. 2019, 32, 1599–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, S.; Singh, A.K.; Agrahari, A.K.; Sharma, K.; Singh, A.S.; Gupta, M.K.; Tiwari, V.K.; Prakash, P. Making of water soluble curcumin to potentiate conventional antimicrobials by inducing apoptosis-like phenomena among drug-resistant bacteria. Sci. Rep. 2020, 10, 14204. [Google Scholar] [CrossRef]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.L.; Da Silva, B.C.M.; Rezende, G.S.; Nakamura-Silva, R.; Pitondo-Silva, A.; Campanini, E.B.; Brito, M.C.A.; Da Silva, E.M.L.; Freire, C.C.D.M.; Da Cunha, A.F.; et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2019, 9, 3198. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Bag, S.; Ghosh, T.S.; Dey, P.; Dayal, M.; Saha, B.; Verma, J.; Pant, A.; Saxena, S.; Desigamani, A.; et al. Molecular Insights into Antimicrobial Resistance Traits of Multidrug Resistant Enteric Pathogens isolated from India. Sci. Rep. 2017, 7, 14468. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Pagès, J.-M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef] [Green Version]
- Routh, M.D.; Zalucki, Y.; Su, C.-C.; Long, F.; Zhang, Q.; Shafer, W.M.; Yu, E.W. Efflux Pumps of the Resistance-Nodulation-Division Family: A Perspective of their Structure, Function, and Regulation in Gram-Negative Bacteria. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 109–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, K. Efflux-Mediated Resistance to Fluoroquinolones in Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2000, 44, 2233–2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppé, É.; Woerther, P.-L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensiv. Care 2015, 5, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppe, U.; Von Laer, A.; Kroll, L.E.; Noll, I.; Feig, M.; Schneider, M.; Claus, H.; Eckmanns, T.; Abu Sin, M. Carbapenem non-susceptibility of Klebsiella pneumoniae isolates in hospitals from 2011 to 2016, data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob. Resist. Infect. Control 2018, 7, 71. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Nordmann, P.; Poirel, L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef] [Green Version]
- Wilson, L.A.; Sharp, P.M. Enterobacterial Repetitive Intergenic Consensus (ERIC) Sequences in Escherichia coli: Evolution and Implications for ERIC-PCR. Mol. Biol. Evol. 2006, 23, 1156–1168. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.-C.; Yang, S.-L.; Peng, H.-L.; Chang, H.-Y. Identification of Genes Present Specifically in a Virulent Strain of Klebsiella pneumoniae. Infect. Immun. 2000, 68, 7149–7151. [Google Scholar] [CrossRef] [Green Version]
- Sidjabat, H.; Nimmo, G.R.; Walsh, T.R.; Binotto, E.; Htin, A.; Hayashi, Y.; Li, J.; Nation, R.L.; George, N.; Paterson, D.L. Carbapenem Resistance in Klebsiella pneumoniae Due to the New Delhi Metallo-β-lactamase. Clin. Infect. Dis. 2011, 52, 481–484. [Google Scholar] [CrossRef]
- Veeraraghavan, B.; Shankar, C.; Karunasree, S.; Kumari, S.; Ravikar, R.; Ralph, R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog. Glob. Health 2017, 111, 240–246. [Google Scholar] [CrossRef]
- Thomson, K.S. Extended-Spectrum-β-Lactamase, AmpC, and Carbapenemase Issues. J. Clin. Microbiol. 2010, 48, 1019–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrill, H.J.; Pogue, J.M.; Kaye, K.S.; Laplante, K.L. Treatment Options for Carbapenem-Resistant Enterobacteriaceae Infections. Open Forum Infect. Dis. 2015, 2, ofv050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veeraraghavan, B.; Pragasam, A.K.; Bakthavatchalam, Y.D.; Anandan, S.; Swaminathan, S.; Sundaram, B. Colistin-Sparing Approaches with Newer Antimicrobials to Treat Carbapenem-Resistant Organisms: Current Evidence and Future Prospects. Indian J. Med. Microbiol. 2019, 37, 72–90. [Google Scholar] [CrossRef]
- Murphy, C.N.; Mortensen, M.S.; Krogfelt, K.A.; Clegg, S. Role of Klebsiella pneumoniae Type 1 and Type 3 Fimbriae in Colonizing Silicone Tubes Implanted into the Bladders of Mice as a Model of Catheter-Associated Urinary Tract Infections. Infect. Immun. 2013, 81, 3009–3017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.-R.; Lin, T.-L.; Chen, Y.-C.; Chou, H.-C.; Wang, J.-T. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011, 157, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [Green Version]
- Schillaci, D.; Spanò, V.; Parrino, B.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G.; Cascioferro, S. Pharmaceutical Approaches to Target Antibiotic Resistance Mechanisms. J. Med. Chem. 2017, 60, 8268–8297. [Google Scholar] [CrossRef]
- Cannon, J.P.; Lee, T.A.; Clark, N.M.; Setlak, P.; Grim, S.A. The risk of seizures among the carbapenems: A meta-analysis. J. Antimicrob. Chemother. 2014, 69, 2043–2055. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; Van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Wasfi, R.; Elkhatib, W.F.; Ashour, H.M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci. Rep. 2016, 6, 38929. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Piddock, L.J.V. How to Measure Export via Bacterial Multidrug Resistance Efflux Pumps. mBio 2016, 7, e00840-16. [Google Scholar] [CrossRef] [Green Version]
- Bohnert, J.A.; Schuster, S.; Szymaniak-Vits, M.; Kern, W.V. Determination of Real-Time Efflux Phenotypes in Escherichia coli AcrB Binding Pocket Phenylalanine Mutants Using a 1,2′-Dinaphthylamine Efflux Assay. PLoS ONE 2011, 6, e21196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Yadav, S.; Chauhan, B.S.; Nandy, N.; Singh, R.; Neogi, K.; Roy, J.K.; Srikrishna, S.; Singh, R.K.; Prakash, P. Classification of Clinical Isolates of Klebsiella pneumoniae Based on Their in vitro Biofilm Forming Capabilities and Elucidation of the Biofilm Matrix Chemistry With Special Reference to the Protein Content. Front. Microbiol. 2019, 10, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sfeir, M.M.; Hayden, J.A.; Fauntleroy, K.A.; Mazur, C.; Johnson, J.K.; Simner, P.J.; Das, S.; Satlin, M.J.; Jenkins, S.G.; Westblade, L.F. EDTA-Modified Carbapenem Inactivation Method: A Phenotypic Method for Detecting Metallo-β-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, e01757-18. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.-M.; Wang, S.; Chiu, H.-C.; Kao, C.-Y.; Wen, L.-L. Combination of modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) for phenotypic detection of carbapenemase-producing Enterobacteriaceae. BMC Microbiol. 2020, 20, 315. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, P.C.S.; Monteiro, A.S.; Marques, S.G.; Monteiro, S.G.; Monteiro-Neto, V.; Coqueiro, M.M.M.; Marques, A.C.G.; de Jesus Gomes Turri, R.; Santos, S.G.; Bomfim, M.R.Q. Phenotypic and molecular detection of the blaKPC gene in clinical isolates from inpatients at hospitals in São Luis, MA, Brazil. BMC Infect. Dis. 2016, 16, 737. [Google Scholar] [CrossRef] [Green Version]
- Meletiadis, J.; Pournaras, S.; Roilides, E.; Walsh, T.J. Defining Fractional Inhibitory Concentration Index Cutoffs for Additive Interactions Based on Self-Drug Additive Combinations, Monte Carlo Simulation Analysis, and In Vitro-In Vivo Correlation Data for Antifungal Drug Combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010, 54, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Mosca, A.; Miragliotta, L.; Del Prete, R.; Tzakis, G.; Dalfino, L.; Bruno, F.; Pagani, L.; Migliavacca, R.; Piazza, A.; Miragliotta, G. Rapid and sensitive detection of blaKPC gene in clinical isolates of Klebsiella pneumoniae by a molecular real-time assay. SpringerPlus 2013, 2, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghadas, A.J.; Kalantari, F.; Sarfi, M.; Shahhoseini, S.; Mirkalantari, S. Evaluation of Virulence Factors and Antibiotic Resistance Patterns in Clinical Urine Isolates of Klebsiella pneumoniae in Semnan, Iran. Jundishapur J. Microbiol. 2018, 11, e63637. [Google Scholar] [CrossRef] [Green Version]
- Bialek-Davenet, S.; Marcon, E.; Leflon-Guibout, V.; Lavigne, J.-P.; Bert, F.; Moreau, R.; Nicolas-Chanoine, M.-H. In Vitro Selection of ramR and soxR Mutants Overexpressing Efflux Systems by Fluoroquinolones as Well as Cefoxitin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2011, 55, 2795–2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.-W.; Onishi, M.; Kishino, T.; Matsuo, T.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Properties and Expression of a Multidrug Efflux Pump AcrAB-KocC from Klebsiella pneumoniae. Biol. Pharm. Bull. 2008, 31, 577–582. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, H.; Zhang, Y.; Wang, Q.; Zhao, C.; Li, H.; He, W.; Zhang, F.; Wang, Z.; Li, S.; et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: Role of the global regulator RamA and its local repressor RamR. Int. J. Antimicrob. Agents 2015, 45, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Kumar, V.; Gedda, M.R.; Singh, A.K.; Singh, V.K.; Singh, S.P.; Singh, R.K. Identification and Characterization of miRNAs in Response to Leishmania donovani Infection: Delineation of Their Roles in Macrophage Dysfunction. Front. Microbiol. 2017, 8, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAuley, S.; Huynh, A.; Czarny, T.; Brown, E.D.; Nodwell, J.R. Membrane activity profiling of small molecule B. subtilis growth inhibitors utilizing novel duel-dye fluorescence assay. MedChemComm 2018, 9, 554–561. [Google Scholar] [CrossRef]





| Condition | Number of Isolates (n = 157) | Interpretation |
|---|---|---|
| mCIM positive, eCIM positive | 65 (41.40%) | Member of Ambler Class B |
| mCIM positive, eCIM negative | 31 (19.51%) | Member of Ambler Class A or D |
| mCIM negative, eCIM negative | 61 (38.85%) | Non-carbapenemase producer |
| Drug Concentration | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drugs | <1 µg/mL | 1 µg/mL | 2 µg/mL | 4 µg/mL | 8 µg/mL | 16 µg/mL | 32 µg/mL | 64 µg/mL | 128 µg/mL | 256 µg/mL | 512 µg/mL | 1024 µg/mL | >1024 µg/mL | |
| Mero | 7 | 2 | - | 6 | 18 | 16 | 18 | 17 | 11 | 1 | - | - | - | |
| Imi | 5 | 6 | - | 13 | 20 | 19 | 14 | 8 | 9 | 2 | - | - | - | |
| Erta | 7 | 6 | 5 | 4 | 22 | 15 | 16 | 13 | 8 | - | - | - | - | |
| CCCP | - | - | - | - | - | 12 | 43 | 37 | 4 | - | - | - | - | |
| Vera | - | - | - | - | - | - | - | - | - | 42 | 31 | 23 | - | |
| Vali | - | - | 44 | 29 | 17 | 6 | - | - | - | - | - | - | - | |
| Curaq | - | - | - | - | - | 14 | 82 | - | - | - | - | - | - | |
| Fold Reduction in MIC | Meropenem + CCCP (No of Isolates) | Meropenem + Verapamil (No of Isolates) | Meropenem + Valinomycin (No of Isolates) | Meropenem + Soluble Curcumin (No of Isolates) |
|---|---|---|---|---|
| No Change | 13 | 20 | 3 | - |
| Δ 2 | 3 | 36 | 21 | 3 |
| Δ 4 | 8 | 27 | 42 | 24 |
| Δ 6 | 19 | 11 | 23 | 51 |
| Δ 8 | 26 | 2 | 6 | 13 |
| Δ 10 | 17 | - | 1 | 5 |
| Δ 12 | 8 | - | - | - |
| Δ 14 | 2 | - | - | - |
| Meropenem + CCCP (No of Isolates) | Meropenem + Verapamil (No of Isolates) | Meropenem + Valinomycin (No of Isolates) | Meropenem + Soluble Curcumin (No of Isolates) | |
|---|---|---|---|---|
| No of meropenem sensitive isolates without the addition | 9 (9.4%) | 9 (9.4%) | 9 (9.4%) | 9 (9.4%) |
| No of meropenem sensitive isolates after addition | 59 (60.82%) | 36 (37.11%) | 74 (76.28%) | 85 (87.62%) |
| Parameters | Untreated | [I] = 4.46 µM | [I] = 9.34 µM | [I] = 18.69 µM | [I] = 37.38 µM |
|---|---|---|---|---|---|
| Km (µM) | 4.702 | 4.702 | 4.702 | 4.702 | 4.702 |
| Vmax (µM/Sec) | 0.7742 | 0.727 | 0.628 | 0.659 | 0.647 |
| Ki (µM) | - | 8.741 | 9.063 | 23.02 | 45.75 |
| R2 | - | 0.984 | 0.981 | 0.984 | 0.980 |
| % Inhibition | - | 33.7% | 50% | 44.86% | 44.96% |
| Gene | Sequence (5’→3’) | Amplicon Size (bp) | Melting Temperature (Tm in °C) | Reference |
|---|---|---|---|---|
| bla_KPC | F: TGTCACTGTATCGCCGTC R: CTCAGTGCTCTACAGAAAACC | 1010 | 60 | [49] |
| RmpA | F: ACTGGGCTACCTCTGCTTCA R: CTTGCATGAGCCATCTTTCA | 435 | 53 | [50] |
| FimH-1 | F: GCCAACGTCTACGTTAACCTG R: ATATTTCACGGTGCCTGAAAA | 180 | 43 | [41] |
| AcrAB | F: ATCAGCGGCCGGATTGGTAAA R: CGGGTTCGGGAAAATAGCGCG | 312 | 53 | [41] |
| TolC | F: ATCAGCAACCCCGATCTGCGT R: CCGGTGACTTGACGCAGTCCT | 527 | 51 | [41] |
| rpoB | F: AAGGCGAATCCAGCTTGTTCAGC R: GACGTTGCATGTTCGCACCCATCA | For Real-time | [51] | |
| Acr A | F: GTCCTCAGGTCAGTGGCATTA R: ATTGCTCTGCTGCGCCGTT | For Real-time | [52] | |
| Acr B | F: AAACTTCGCCACTACGTCATA R: AGCTTAACGCCTCGATCAT | For Real-time | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, S.; Singh, A.K.; Agrahari, A.K.; Pandey, A.K.; Gupta, M.K.; Chakravortty, D.; Tiwari, V.K.; Prakash, P. Galactose-Clicked Curcumin-Mediated Reversal of Meropenem Resistance among Klebsiella pneumoniae by Targeting Its Carbapenemases and the AcrAB-TolC Efflux System. Antibiotics 2021, 10, 388. https://doi.org/10.3390/antibiotics10040388
Yadav S, Singh AK, Agrahari AK, Pandey AK, Gupta MK, Chakravortty D, Tiwari VK, Prakash P. Galactose-Clicked Curcumin-Mediated Reversal of Meropenem Resistance among Klebsiella pneumoniae by Targeting Its Carbapenemases and the AcrAB-TolC Efflux System. Antibiotics. 2021; 10(4):388. https://doi.org/10.3390/antibiotics10040388
Chicago/Turabian StyleYadav, Shivangi, Ashish Kumar Singh, Anand K. Agrahari, Akhilesh Kumar Pandey, Munesh Kumar Gupta, Dipshikha Chakravortty, Vinod Kumar Tiwari, and Pradyot Prakash. 2021. "Galactose-Clicked Curcumin-Mediated Reversal of Meropenem Resistance among Klebsiella pneumoniae by Targeting Its Carbapenemases and the AcrAB-TolC Efflux System" Antibiotics 10, no. 4: 388. https://doi.org/10.3390/antibiotics10040388
APA StyleYadav, S., Singh, A. K., Agrahari, A. K., Pandey, A. K., Gupta, M. K., Chakravortty, D., Tiwari, V. K., & Prakash, P. (2021). Galactose-Clicked Curcumin-Mediated Reversal of Meropenem Resistance among Klebsiella pneumoniae by Targeting Its Carbapenemases and the AcrAB-TolC Efflux System. Antibiotics, 10(4), 388. https://doi.org/10.3390/antibiotics10040388