The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health
Abstract
1. Introduction
2. Results
2.1. Expansion of the DfrB Family
2.2. Identification of Bacterial Sequences
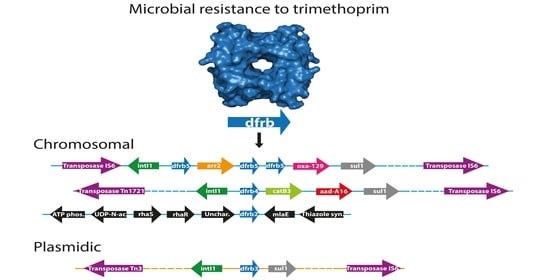
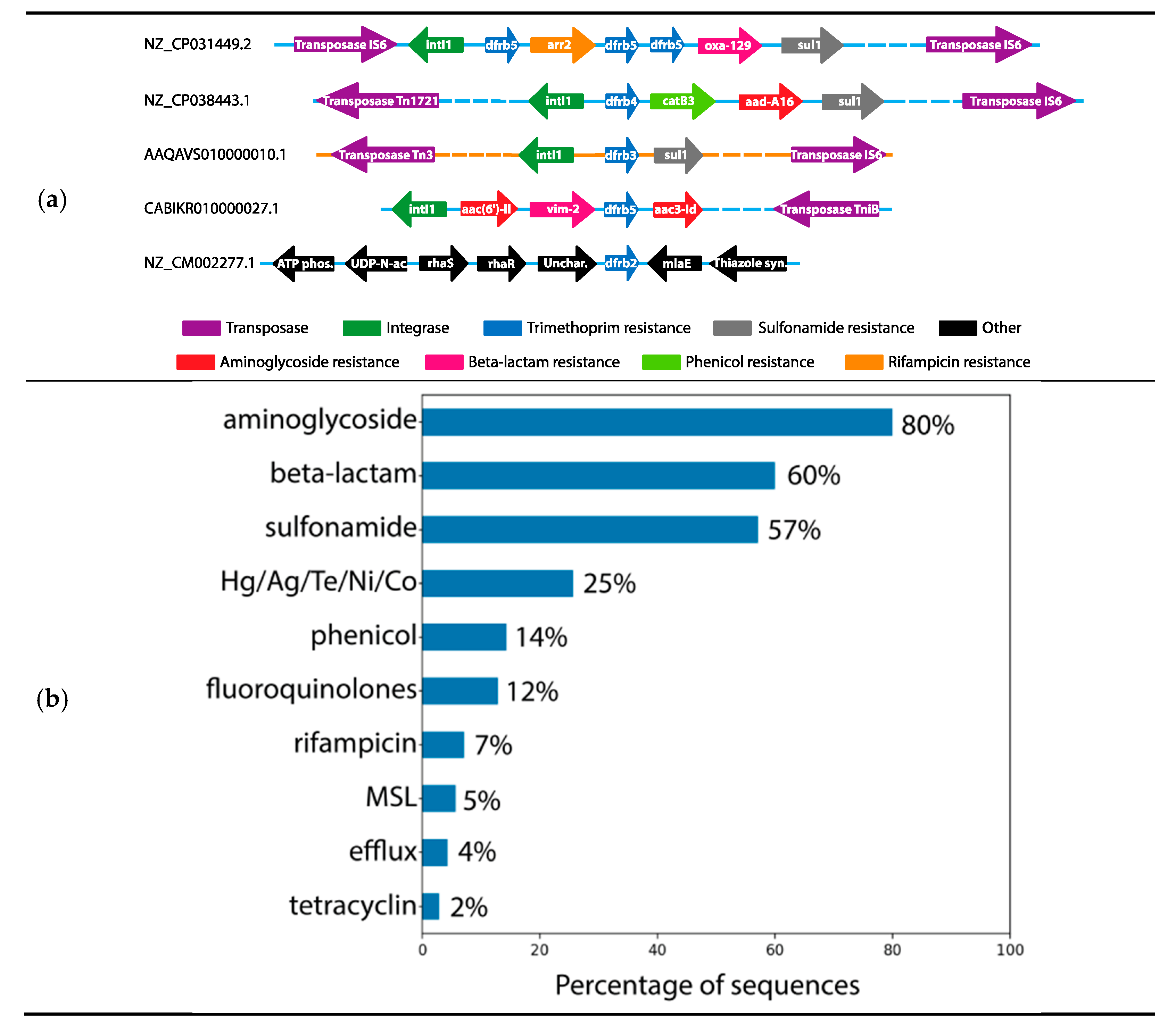
2.3. Analysis of the Genomic Context
3. Discussion
4. Materials and Methods
4.1. Identification of Putative Type B Dihydrofolate Reductases
4.2. Subcloning of dfrb10 and dfrb11
4.3. Minimal Inhibitory Concentration
4.4. Download of Genomes
4.5. Protein Database Constructions
4.6. Annotation
4.7. Classification of Sequences as Chromosomal or Plasmidic
4.8. Identification of Pathogenic Hosts
4.9. Phylogenetic Tree
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Stone, S.R.; Morrison, J.F. Mechanism of Inhibition of Dihydrofolate Reductases from Bacterial and Vertebrate Sources by Various Classes of Folate Analogues. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzymol. 1986, 869, 275–285. [Google Scholar] [CrossRef]
- Eliopoulos, G.M.; Huovinen, P. Resistance to Trimethoprim-Sulfamethoxazole. Clin. Infect. Dis. 2001, 32, 1608–1614. [Google Scholar] [CrossRef]
- Caron, F.; Wehrle, V.; Etienne, M. The Comeback of Trimethoprim in France. Med. Mal. Infect. 2017, 47, 253–260. [Google Scholar] [CrossRef]
- World Health Organization. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Hsia, Y.; Lee, B.R.; Versporten, A.; Yang, Y.; Bielicki, J.; Jackson, C.; Newland, J.; Goossens, H.; Magrini, N.; Sharland, M.; et al. Use of the WHO Access, Watch, and Reserve Classification to Define Patterns of Hospital Antibiotic Use (AWaRe): An Analysis of Paediatric Survey Data from 56 Countries. Lancet Glob. Health 2019, 7, e861–e871. [Google Scholar] [CrossRef]
- Cuong, N.V.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J.J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low- and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef]
- World Health Organization Department of Food Safety and Zoonoses; World Health Organization. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-155013-0. [Google Scholar]
- Sánchez-Osuna, M.; Cortés, P.; Llagostera, M.; Barbé, J.; Erill, I. Exploration into the Origins and Mobilization of Di-Hydrofolate Reductase Genes and the Emergence of Clinical Resistance to Trimethoprim. Microb. Genom. 2020, 6. [Google Scholar] [CrossRef]
- Howell, E.E. Searching Sequence Space: Two Different Approaches to Dihydrofolate Reductase Catalysis. ChemBioChem 2005, 6, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Faltyn, M.; Alcock, B.; McArthur, A. Evolution and Nomenclature of the Trimethoprim Resistant Dihydrofolate (Dfr) Reductases. Preprints 2019. [Google Scholar] [CrossRef]
- Kim, Y.B. Improved Trimethoprim-Resistance Cassette for Prokaryotic Selections. J. Biosci. Bioeng. 2009, 108, 441–445. [Google Scholar] [CrossRef]
- Pattishall, K.H.; Acar, J.; Burchall, J.J.; Goldstein, F.W.; Harvey, R.J. Two Distinct Types of Trimethoprim-Resistant Dihydrofolate Reductase Specified by R-Plasmids of Different Compatibility Groups. J. Biol. Chem. 1977, 252, 2319–2323. [Google Scholar] [CrossRef]
- Krahn, J.M.; Jackson, M.R.; DeRose, E.F.; Howell, E.E.; London, R.E. Crystal Structure of a Type II Dihydrofolate Reductase Catalytic Ternary Complex †. Biochemistry 2007, 46, 14878–14888. [Google Scholar] [CrossRef] [PubMed]
- Bhojane, P.P.; Duff, M.R.; Bafna, K.; Agarwal, P.; Stanley, C.; Howell, E.E. Small Angle Neutron Scattering Studies of R67 Dihydrofolate Reductase, a Tetrameric Protein with Intrinsically Disordered N-Termini. Biochemistry 2017, 56, 5886–5899. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.C.; Morley, K.L.; Volpato, J.P.; Schmitzer, A.R.; Pelletier, J.N. Asymmetric Mutations in the Tetrameric R67 Dihydrofolate Reductase Reveal High Tolerance to Active-Site Substitutions: Asymmetric Mutations in R67 Dihydrofolate Reductase. Protein Sci. 2015, 24, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Schmitzer, A.R.; Lépine, F.; Pelletier, J.N. Combinatorial Exploration of the Catalytic Site of a Drug-Resistant Dihydrofolate Reductase: Creating Alternative Functional Configurations. Protein Eng. Des. Sel. 2004, 17, 809–819. [Google Scholar] [CrossRef]
- Martinez, M.A.; Pezo, V.; Marlière, P.; Wain-Hobson, S. Exploring the Functional Robustness of an Enzyme by In Vitro Evolution. EMBO J. 1996, 15, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Toulouse, J.L.; Shi, G.; Lemay-St-Denis, C.; Ebert, M.C.; Deon, D.; Gagnon, M.; Ruediger, E.; Saint-Jacques, K.; Forge, D.; Vanden Eynde, J.J.; et al. Dual-Target Inhibitors of the Folate Pathway Inhibit Intrinsically Trimethoprim-Resistant DfrB Dihydrofolate Reductases. ACS Med. Chem. Lett. 2020. [Google Scholar] [CrossRef] [PubMed]
- Toulouse, J.L.; Yachnin, B.J.; Ruediger, E.H.; Deon, D.; Gagnon, M.; Saint-Jacques, K.; Ebert, M.C.; Forge, D.; Bastien, D.; Colin, D.Y.; et al. Structure-Based Design of Dimeric Bisbenzimidazole Inhibitors to an Emergent Trimethoprim-Resistant Type II Dihydrofolate Reductase Guides the Design of Monomeric Analogues. ACS Omega 2019, 4, 10056–10069. [Google Scholar] [CrossRef]
- Toulouse, J.L.; Abraham, S.M.J.; Kadnikova, N.; Bastien, D.; Gauchot, V.; Schmitzer, A.R.; Pelletier, J.N. Investigation of Classical Organic and Ionic Liquid Cosolvents for Early-Stage Screening in Fragment-Based Inhibitor Design with Unrelated Bacterial and Human Dihydrofolate Reductases. Assay Drug Dev. Technol. 2017, 15, 141–153. [Google Scholar] [CrossRef]
- Bastien, D.; Ebert, M.C.; Forge, D.; Toulouse, J.; Kadnikova, N.; Perron, F.; Mayence, A.; Huang, T.L.; Vanden Eynde, J.J.; Pelletier, J.N. Fragment-Based Design of Symmetrical Bis-Benzimidazoles as Selective Inhibitors of the Trimethoprim-Resistant, Type II R67 Dihydrofolate Reductase. J. Med. Chem. 2012, 55, 3182–3192. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, gkz935. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Grape, M.; Farra, A.; Kronvall, G.; Sundström, L. Integrons and Gene Cassettes in Clinical Isolates of Co-Trimoxazole-Resistant Gram-Negative Bacteria. Clin. Microbiol. Infect. 2005, 11, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Toulouse, J.L.; Edens, T.J.; Alejaldre, L.; Manges, A.R.; Pelletier, J.N. Integron-Associated DfrB4, a Previously Uncharacterized Member of the Trimethoprim-Resistant Dihydrofolate Reductase B Family, Is a Clinically Identified Emergent Source of Antibiotic Resistance. Antimicrob. Agents Chemother. 2017, 61, e02665-16. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Kehrenberg, C.; Schwarz, S. Molecular Basis of Resistance to Trimethoprim, Chloramphenicol and Sulphonamides in Bordetella Bronchiseptica. J. Antimicrob. Chemother. 2005, 56, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Grape, M.; Motakefi, A.; Pavuluri, S.; Kahlmeter, G. Standard and Real-Time Multiplex PCR Methods for Detection of Trimethoprim Resistance Dfr Genes in Large Collections of Bacteria. Clin. Microbiol. Infect. 2007, 13, 1112–1118. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What Is a Resistance Gene? Ranking Risk in Resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Jaillard, M.; van Belkum, A.; Cady, K.C.; Creely, D.; Shortridge, D.; Blanc, B.; Barbu, E.M.; Dunne, W.M.; Zambardi, G.; Enright, M.; et al. Correlation between Phenotypic Antibiotic Susceptibility and the Resistome in Pseudomonas Aeruginosa. Int. J. Antimicrob. Agents 2017, 50, 210–218. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the Pathogenicity Status of Pseudomonas Aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Sun, F.; Zhou, D.; Wang, Q.; Feng, J.; Feng, W.; Luo, W.; Liu, Y.; Qiu, X.; Yin, Z.; Xia, P. Genetic Characterization of a Novel BlaDIM-2-Carrying Megaplasmid P12969-DIM from Clinical Pseudomonas Putida. J. Antimicrob. Chemother. 2016, 71, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Hernsdorf, A.W.; Amano, Y.; Miyakawa, K.; Ise, K.; Suzuki, Y.; Anantharaman, K.; Probst, A.; Burstein, D.; Thomas, B.C.; Banfield, J.F. Potential for Microbial H2 and Metal Transformations Associated with Novel Bacteria and Archaea in Deep Terrestrial Subsurface Sediments. ISME J. 2017, 11, 1915–1929. [Google Scholar] [CrossRef] [PubMed]
- Workentine, M.L.; Surette, M.G.; Bernier, S.P. Draft Genome Sequence of Burkholderia Dolosa PC543 Isolated from Cystic Fibrosis Airways. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Stone, D.; Smith, S.L. The Amino Acid Sequence of the Trimethoprim-Resistant Dihydrofolate Reductase Specified in Escherichia Coli by R-Plasmid R67. J. Biol. Chem. 1979, 254, 10857–10861. [Google Scholar] [CrossRef]
- Amyes, S.G.B.; Smith, J.T. R-Factor Trimethoprim Resistance Mechanism: An Insusceptible Target Site. Biochem. Biophys. Res. Commun. 1974, 58, 412–418. [Google Scholar] [CrossRef]
- San Millan, A. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends Microbiol. 2018, 26, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting Plasmid Sequences in Metagenomic Data Using Genome Signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef]
- Masters, P.A.; O’Bryan, T.A.; Zurlo, J.; Miller, D.Q.; Joshi, N. Trimethoprim-Sulfamethoxazole Revisited. Arch. Intern. Med. 2003, 163, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Public Health Agency of Canada. Canadian Antimicrobial Resistance Surveillance System Report; Public Health Agency of Canada: Ottawa, ON, Canada, 2020. [Google Scholar]
- Swift, G.; McCarthy, B.J.; Heffron, F. DNA Sequence of a Plasmid-Encoded Dihydrofolate Reductase. Mol. Gen. Genet. MGG 1981, 181, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Brisson, N.; Hohn, T. Nucleotide Sequence of the Dihydrofolate-Reductase Gene Borne by the Plasmid R67 and Conferring Methotrexate Resistance. Gene 1984, 28, 271–274. [Google Scholar] [CrossRef]
- Narayana, N.; Matthews, D.A.; Howell, E.E.; Xuong, N. A Plasmid-Encoded Dihydrofolate Reductase from Trimethoprim-Resistant Bacteria Has a Novel D2-Symmetric Active Site. Nat. Struct. Mol. Biol. 1995, 2, 1018–1025. [Google Scholar] [CrossRef]
- Park, H.; Zhuang, P.; Nichols, R.; Howell, E.E. Mechanistic Studies of R67 Dihydrofolate Reductase. J. Biol. Chem. 1997, 272, 2252–2258. [Google Scholar] [CrossRef]
- West, F.W.; Seo, H.-S.; Bradrick, T.D.; Howell, E.E. Effects of Single-Tryptophan Mutations on R67 Dihydrofolate Reductase †. Biochemistry 2000, 39, 3678–3689. [Google Scholar] [CrossRef]
- Kamath, G.; Howell, E.E.; Agarwal, P.K. The Tail Wagging the Dog: Insights into Catalysis in R67 Dihydrofolate Reductase. Biochemistry 2010, 49, 9078–9088. [Google Scholar] [CrossRef]
- Gillings, M.R. Class 1 Integrons as Invasive Species. Curr. Opin. Microbiol. 2017, 38, 10–15. [Google Scholar] [CrossRef]
- Santos, C.; Caetano, T.; Ferreira, S.; Mendo, S. Tn5090-like Class 1 Integron Carrying BlaVIM-2 in a Pseudomonas Putida Strain from Portugal. Clin. Microbiol. Infect. 2010, 16, 1558–1561. [Google Scholar] [CrossRef]
- Chiu, C.-H.; Lee, H.-Y.; Tseng, L.-Y.; Chen, C.-L.; Chia, J.-H.; Su, L.-H.; Liu, S.-Y. Mechanisms of Resistance to Ciprofloxacin, Ampicillin/Sulbactam and Imipenem in Acinetobacter Baumannii Clinical Isolates in Taiwan. Int. J. Antimicrob. Agents 2010, 35, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Mikami, Y.; Imai, T.; Poirel, L.; Nordmann, P. Characterization of In53, a Class 1 Plasmid- and Composite Transposon-Located Integron of Escherichia Coli Which Carries an Unusual Array of Gene Cassettes. J. Bacteriol. 2001, 183, 235–249. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.-C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. The Inoue Method for Preparation and Transformation of Competent E. Coli: “Ultra-Competent” Cells. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot3944. [Google Scholar] [CrossRef]
- Gobeil, S.M.C.; Gagné, D.; Doucet, N.; Pelletier, J.N. 15N, 13C and 1H Backbone Resonance Assignments of an Artificially Engineered TEM-1/PSE-4 Class A β-Lactamase Chimera and Its Deconvoluted Mutant. Biomol. NMR Assign. 2016, 10, 93–99. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A Curated Non-Redundant Sequence Database of Genomes, Transcripts and Proteins. Nucleic Acids Res. 2005, 33, D501–D504. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 45, D37–D42. [Google Scholar] [CrossRef]
- Zhang, A.N.; Li, L.-G.; Ma, L.; Gillings, M.R.; Tiedje, J.M.; Zhang, T. Conserved Phylogenetic Distribution and Limited Antibiotic Resistance of Class 1 Integrons Revealed by Assessing the Bacterial Genome and Plasmid Collection. Microbiome 2018, 6, 130. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.-Y. ICEberg 2.0: An Updated Database of Bacterial Integrative and Conjugative Elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Boeckmann, B. The SWISS-PROT Protein Sequence Data Bank. Nucleic Acids Res. 1991, 19, 2247–2249. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.Fr: New Generation Phylogenetic Services for Non-Specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]


| New Name | UniprotKB Accession Number | Genbank Accession Number | Closest Characterized DfrB (Protein Identity/ DNA Identity) a | MIC (µg/mL) |
|---|---|---|---|---|
| DfrB10 | A0A2Z1CLP9 | ALZ46148.1 | DfrB3 (92%/93%) | >600 |
| DfrB11 | A0A2N2TNN4 | PKO69073.1 | DfrB3 (90%/87%) | >600 |
| Class/Order/Family/Genus | Strain Count a |
|---|---|
| Betaproteobacteria | 1 |
| Burkholderiales | 1 |
| Burkholderiaceae | 1 |
| Burkholderia | 1 |
| Gammaproteobacteria | 60 (110) |
| Aeromonadales | 2 |
| Aeromonadaceae | 2 |
| Aeromonas | 2 |
| Enterobacterales | 16 (17) |
| Enterobacteriaceae | 14 (15) |
| Citrobacter | 1 |
| Enterobacter | 1 |
| Escherichia | 4 |
| Klebsiella | 4 (5) |
| Salmonella | 4 |
| Morganellaceae | 1 |
| Providencia | 1 |
| Yersiniaceae | 1 |
| Serratia | 1 |
| Pseudomonadales | 42 (91) |
| Moraxellaceae | 1 |
| Acinetobacter | 1 |
| Pseudomonadaceae | 41 (90) |
| Pseudomonas | 41 (90) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemay-St-Denis, C.; Diwan, S.-S.; Pelletier, J.N. The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health. Antibiotics 2021, 10, 433. https://doi.org/10.3390/antibiotics10040433
Lemay-St-Denis C, Diwan S-S, Pelletier JN. The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health. Antibiotics. 2021; 10(4):433. https://doi.org/10.3390/antibiotics10040433
Chicago/Turabian StyleLemay-St-Denis, Claudèle, Sarah-Slim Diwan, and Joelle N. Pelletier. 2021. "The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health" Antibiotics 10, no. 4: 433. https://doi.org/10.3390/antibiotics10040433
APA StyleLemay-St-Denis, C., Diwan, S.-S., & Pelletier, J. N. (2021). The Bacterial Genomic Context of Highly Trimethoprim-Resistant DfrB Dihydrofolate Reductases Highlights an Emerging Threat to Public Health. Antibiotics, 10(4), 433. https://doi.org/10.3390/antibiotics10040433