Multiresistant Bacteria Isolated from Intestinal Faeces of Farm Animals in Austria
Abstract
:1. Introduction
2. Results
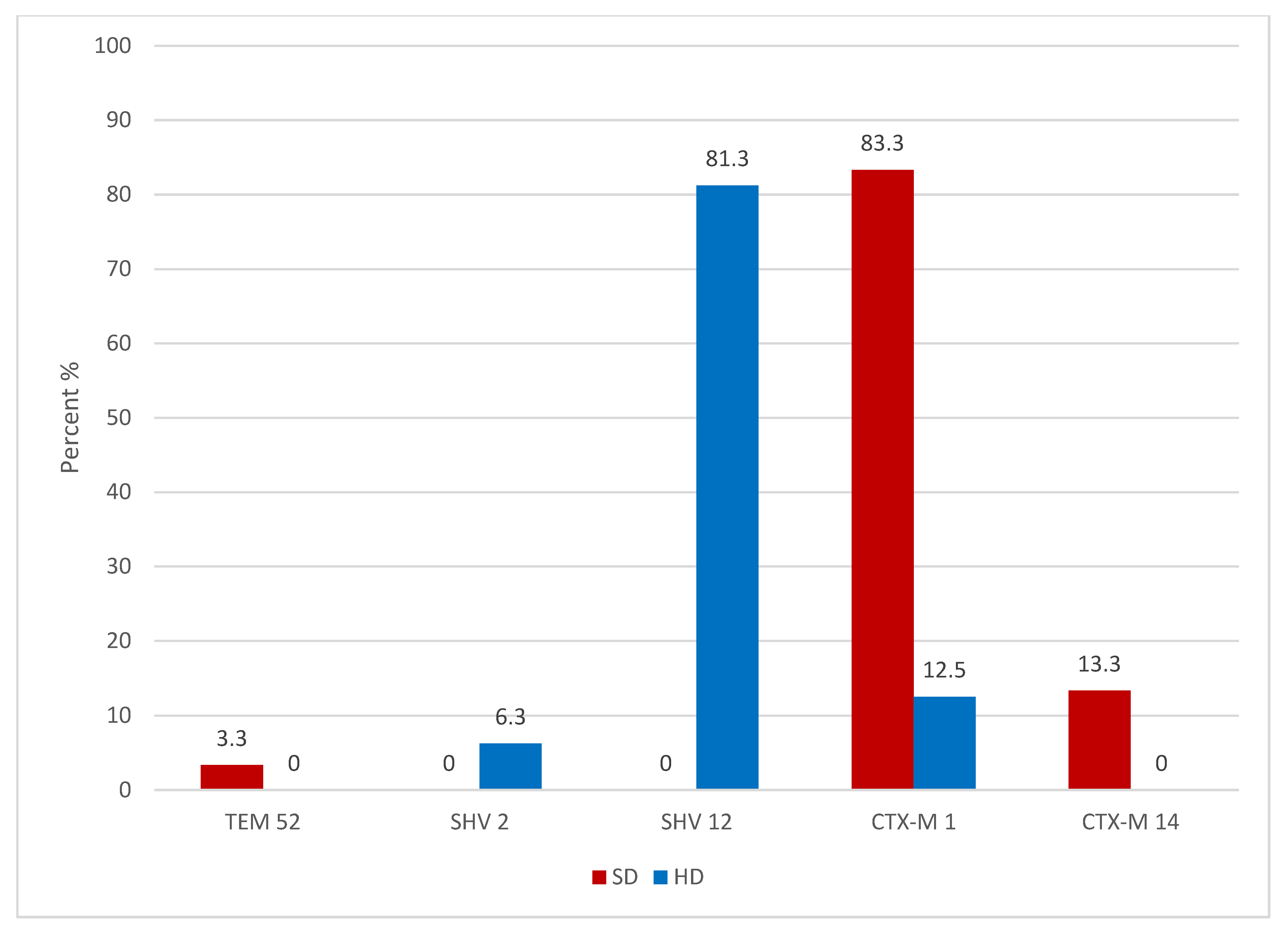
2.1. Swine ESBL Enterobacteriaceae
2.2. Swine VRE
2.3. Broiler ESBL Enterobacteriaceae
2.4. Broiler VRE
3. Discussion
4. Material and Methods
4.1. Samples
4.2. Strain Isolation and Detection
4.3. Antimicrobial Susceptibility Testing
4.4. Detection of Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Livermore, D.M. Fourteen years in resistance. Int. J. Antimicrob. Agents 2012, 39, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Lester, C.H.; Heuer, O.E. Antimicrobial-resistant enterococci in animals and meat: A human health hazard? Foodborne Pathog. Dis. 2010, 7, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Ghidan, A.; Dobay, O.; Kaszanyitzky, E.J.; Samu, P.; Amyes, S.G.B.; Nagy, K.; Rozgonyi, F. Vancomycin Resistant Enterococci (Vre) Still Persist in Slaughtered Poultry in Hungary 8 Years After the Ban on Avoparcin. Acta Microbiol. Immunol. Hung 2008, 55, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, B.; Orendi, U.; Much, P.; Hoger, G.; Ruppitsch, W.; Krziwanek, K.; Metz-Gercek, S.; Mittermayer, H. Methicillin-resistant Staphylococcus aureus: A new zoonotic agent? Wien Klin. Wochenschr. 2009, 121, 86–90. [Google Scholar] [CrossRef]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.L.E.; Canica, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria Among Food-Producing Animals: Health Implications of Extended Spectrum beta-lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Unal, N.; Bal, E.; Karagoz, A.; Altun, B.; Kocak, N. Detection of vancomycin-resistant enterococci in samples from broiler flocks and houses in Turkey. Acta Vet. Hung 2020, 68, 117–122. [Google Scholar] [CrossRef]
- Kock, R.; Herr, C.; Kreienbrock, L.; Schwarz, S.; Tenhagen, B.A.; Walther, B. Multiresistant Gram-Negative Pathogens-A Zoonotic Problem. Dtsch. Arztebl. Int. 2021, 118. [Google Scholar] [CrossRef]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 2006, 34, S20–S28, discussion S64–S73. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Carattoli, A.; Karisik, E.; Underwood, A.; Ellington, M.J.; Livermore, D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009, 53, 4472–4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.M.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007, 59, 165–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, A.; Stephan, R.; Nuesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016, 541, 667–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudin, M.A.; Nykasenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-Associated Methicillin Resistant and Methicillin Susceptible Staphylococcus aureus Sequence Type (CC)1 in European Farmed Animals: High Genetic Relatedness of Isolates from Italian Cattle Herds and Humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarfel, G.; Krziwanek, K.; Johler, S.; Hoenigl, M.; Leitner, E.; Kittinger, C.; Masoud, L.; Feierl, G.; Grisold, A.J. Virulence and antimicrobial resistance genes in human MRSA ST398 isolates in Austria. Epidemiol. Infect. 2013, 141, 888–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Cleef, B.A.; Monnet, D.L.; Voss, A.; Krziwanek, K.; Allerberger, F.; Struelens, M.; Zemlickova, H.; Skov, R.L.; Vuopio-Varkila, J.; Cuny, C.; et al. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg. Infect. Dis. 2011, 17, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Moon, D.C.; Mechesso, A.F.; Kang, H.Y.; Song, H.J.; Na, S.H.; Choi, J.H.; Yoon, S.S.; Lim, S.K. Nationwide Surveillance on Antimicrobial Resistance Profiles of Staphylococcus aureus Isolated from Major Food Animal Carcasses in South Korea During 2010–2018. Foodborne Pathog. Dis. 2021. [Google Scholar] [CrossRef]
- Shenoy, E.S.; Paras, M.L.; Noubary, F.; Walensky, R.P.; Hooper, D.C. Natural history of colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE): A systematic review. BMC Infect. Dis. 2014, 14, 177. [Google Scholar] [CrossRef]
- Gastmeier, P.; Schroder, C.; Behnke, M.; Meyer, E.; Geffers, C. Dramatic increase in vancomycin-resistant enterococci in Germany. J. Antimicrob. Chemother. 2014, 69, 1660–1664. [Google Scholar] [CrossRef] [Green Version]
- Bager, F.; Madsen, M.; Christensen, J.; Aarestrup, F.M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 1997, 31, 95–112. [Google Scholar] [CrossRef]
- Nilsson, O. Vancomycin resistant enterococci in farm animals—Occurrence and importance. Infect. Ecol. Epidemiol. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Mutters, N.T.; Mersch-Sundermann, V.; Mutters, R.; Brandt, C.; Schneider-Brachert, W.; Frank, U. Control of the spread of vancomycin-resistant enterococci in hospitals: Epidemiology and clinical relevance. Dtsch. Arztebl. Int. 2013, 110, 725–731. [Google Scholar] [PubMed] [Green Version]
- Doi, Y.; Paterson, D.L.; Egea, P.; Pascual, A.; Lopez-Cerero, L.; Navarro, M.D.; Adams-Haduch, J.M.; Qureshi, Z.A.; Sidjabat, H.E.; Rodriguez-Bano, J. Extended-spectrum and CMY-type beta-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin. Microbiol. Infect. 2010, 16, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1216–1222. [Google Scholar] [CrossRef]
- Egea, P.; Lopez-Cerero, L.; Torres, E.; Gomez-Sanchez Mdel, C.; Serrano, L.; Navarro Sanchez-Ortiz, M.D.; Rodriguez-Bano, J.; Pascual, A. Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int. J. Food Microbiol. 2012, 159, 69–73. [Google Scholar] [CrossRef]
- Zelendova, M.; Dolejska, M.; Masarikova, M.; Jamborova, I.; Vasek, J.; Smola, J.; Manga, I.; Cizek, A. CTX-M-producing Escherichia coli in pigs from a Czech farm during production cycle. Lett. Appl. Microbiol. 2020, 71, 369–376. [Google Scholar]
- Belmar Campos, C.; Fenner, I.; Wiese, N.; Lensing, C.; Christner, M.; Rohde, H.; Aepfelbacher, M.; Fenner, T.; Hentschke, M. Prevalence and genotypes of extended spectrum beta-lactamases in Enterobacteriaceae isolated from human stool and chicken meat in Hamburg, Germany. Int. J. Med. Microbiol. 2014, 304, 678–684. [Google Scholar] [CrossRef]
- Kola, A.; Kohler, C.; Pfeifer, Y.; Schwab, F.; Kuhn, K.; Schulz, K.; Balau, V.; Breitbach, K.; Bast, A.; Witte, W.; et al. High prevalence of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012, 67, 2631–2634. [Google Scholar] [CrossRef] [Green Version]
- Paivarinta, M.; Latvio, S.; Fredriksson-Ahomaa, M.; Heikinheimo, A. Whole genome sequence analysis of antimicrobial resistance genes, multilocus sequence types and plasmid sequences in ESBL/AmpC Escherichia coli isolated from broiler caecum and meat. Int. J. Food Microbiol. 2020, 315, 108361. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Kant, A.; van Essen-Zandbergen, A.; Dierikx, C.; Hordijk, J.; Wit, B.; Mevius, D.J.; Veldman, K.T. Diversity of Plasmids and Genes Encoding Resistance to Extended Spectrum Cephalosporins in Commensal Escherichia coli From Dutch Livestock in 2007–2017. Front. Microbiol. 2019, 10, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Zarfel, G.; Galler, H.; Feierl, G.; Haas, D.; Kittinger, C.; Leitner, E.; Grisold, A.J.; Mascher, F.; Posch, J.; Pertschy, B.; et al. Comparison of extended-spectrum-beta-lactamase (ESBL) carrying Escherichia coli from sewage sludge and human urinary tract infection. Environ. Pollut. 2013, 173, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Petternel, C.; Galler, H.; Zarfel, G.; Luxner, J.; Haas, D.; Grisold, A.J.; Reinthaler, F.F.; Feierl, G. Isolation and characterization of multidrug-resistant bacteria from minced meat in Austria. Food Microbiol. 2014, 44, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Zarfel, G.; Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Kittinger, C.; Grisold, A.J.; Pless, P.; Feierl, G. Multiresistant bacteria isolated from chicken meat in Austria. Int. J. Environ. Res. Public Health 2014, 11, 12582–12593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, B.; Bruckner, K. Characterization of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli from raw meat and comparison to human isolates. Wien Tierarztl. Monatsschr. 2012, 99, 44–50. [Google Scholar]
- Sting, R.; Richter, A.; Popp, C.; Hafez, H.M. Occurrence of vancomycin-resistant enterococci in turkey flocks. Poult. Sci. 2013, 92, 346–351. [Google Scholar] [CrossRef]
- Anonymous. CLSI, Clinical and Laboratory Standards Institute, 2008: Performance Standards for Antimicrobial Susceptibility Testing: 18th Informational Supplement; CLSI document M100-S18; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Eckert, C.; Gautier, V.; Saladin-Allard, M.; Hidri, N.; Verdet, C.; Ould-Hocine, Z.; Barnaud, G.; Delisle, F.; Rossier, A.; Lambert, T.; et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 2004, 48, 1249–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grisold, A.J.; Kessler, H.H. Use of hybridization probes in a real-time PCR assay on the LightCycler for the detection of methicillin-resistant Staphylococcus aureus. Methods Mol. Biol. 2006, 345, 79–89. [Google Scholar]
- Koh, T.H.; Deepak, R.N.; Se-Thoe, S.Y.; Lin, R.V.; Koay, E.S. Experience with the Roche LightCycler VRE detection kit during a large outbreak of vanB2/B3 vancomycin-resistant Enterococcus faecium. J. Antimicrob. Chemother. 2007, 60, 182–183. [Google Scholar] [CrossRef] [Green Version]
| Isolate a | Species | Sample b | Encoding Resistance | Resistance Pattern c |
|---|---|---|---|---|
| SD 3/1–100a | E. coli | sw_01 | CTX–M1 | AM, CN, CXM, SXT, FEP, TET |
| SD 3/2–100a | E. coli | sw_02 | CTX–M1 | AM, CN, CXM, FOX, CTX, SXT, FEP, TET, NA, C |
| SD 3/4–100c | E. coli | sw_03 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP |
| SD 3/4–100d | E. coli | sw_04 | CTX–M1 | AM, CN, CXM, CTX, SXT, CAZ, FEP, TET |
| SD 3/5 –100b | E. coli | sw_05 | TEM–52 | AM, CXM, CTX, SXT, CAZ, TET |
| SD 3/5 –100c | E. coli | sw_06 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET |
| SD 3/5 –100e | E. coli | sw_07 | CTX–M1 | AM, CN, CXM, CTX, SXT, CAZ, FEP |
| SD 4/2 –100a | E. coli | sw_08 | CTX–M1 | AM, CN, CXM, CTX, GM, SXT, FEP, C |
| SD 4/4 –100a | E. coli | sw_09 | CTX–M1 | AM, CN, CXM, CTX, SXT, FEP, TET |
| SD 5/1 –100a | E. coli | sw_10 | CTX–M14 | AM, CN, CXM, CTX, TET |
| SD 5/1 –100b | E. coli | sw_11 | CTX–M1 | AM, CN, CXM, CTX, SXT, CAZ, FEP |
| SD 5/2 –100a | E. coli | sw_12 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP |
| SD 5/2 –100d | E. coli | sw_13 | CTX–M14 | AM, CN, CXM, CTX, CAZ, FEP, TET |
| SD 5/3 –100a | E. coli | sw_14 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP, TET |
| SD 5/5 –100a | E. coli | sw_15 | CTX–M1 | AM, CN, CXM, CTX, SXT, CAZ, FEP |
| SD 6/2 –100a | E. coli | sw_16 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET, NA |
| SD 6/2 –100d | E. coli | sw_17 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP, TET, NA |
| SD 6/4 –100a | E. coli | sw_18 | CTX–M14 | AM, CN, CXM, CTX, TET, NA |
| SD 6/4 –100c | E. coli | sw_19 | CTX–M14 | AM, CN, CXM, CTX, CAZ, FEP, TET, NA |
| SD 10/1–100b | E. coli | sw_20 | CTX–M1 | AM, CN, CXM, CTX, SXT, CAZ, FEP, TET |
| SD 10/4–100a | E. coli | sw_21 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET |
| SD 10/5–100a | E. coli | sw_22 | CTX–M1 | AM, CN, CXM, CTX, GM, SXT, FEP, TET, C |
| SD 11/4–100a | E. coli | sw_23 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP, TET, NA |
| SD 11/5–100a | E. coli | sw_24 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP, TET, NA |
| SD 15/1–100b | E. coli | sw_25 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP, TET |
| SD 15/2–100a | E. coli | sw_26 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET |
| SD 15/3–100a | E. coli | sw_27 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET, NA |
| SD 15/5–100a | E. coli | sw_28 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET |
| SD 15/6–100a | E. coli | sw_29 | CTX–M1 | AM, CN, CXM, CTX, FEP, TET |
| SD 15/10–100a | E. coli | sw_30 | CTX–M1 | AM, CN, CXM, CTX, CAZ, FEP |
| HD 1/1 100a Th | E. coli | bs_31 | SHV–12 | AM, CXM, CTX, SXT, CAZ, TET, NA, C |
| HD 1/1 100b Th | E. coli | bs_32 | SHV–12 | AM, CXM, CTX, SXT, CAZ, TET, NA, C |
| HD 1/1 100c Th | E. coli | bs_33 | SHV–12 | AM, CXM, CTX, SXT, CAZ, TET, NA, C |
| HD 1/2 100a Th | E. coli | bs_34 | CTX–M1 | AM, CN, CXM, CTX, CAZ, TET, NA, C |
| HD 1/2 100b Th | E. coli | bs_35 | SHV–12 | AM, CN, CXM, CTX, CAZ, TET, NA, C |
| HD 1/2 100c Th | E. coli | bs_36 | CTX–M1 | AM, CN, CXM, CTX, SXT, MXF, CAZ, TET, NA, C |
| HD 1/2 100d Th | E. coli | bs_37 | SHV–12 | AM, CTX, MXF, CAZ, TET, NA, C |
| HD 2/9–0a | E. coli | bs_38 | SHV–12 | AM, CXM, CTX, CAZ, TET, NA, C |
| HD 3/2 100a | E. coli | bs_39 | SHV–12 | AM, CTX, SXT, MXF, CAZ, TET, NA, C |
| HD 3/3–100a | E. coli | bs_40 | SHV–12 | AM, CTX, SXT, CAZ, TET, NA, C |
| HD 3/4–0a | E. coli | bs_41 | SHV–12 | AM, CTX, SXT, CAZ, TET, NA, C |
| HD 3/5–0a | E. coli | bs_42 | SHV–12 | AM, CTX, SXT, CAZ, TET, NA, C |
| HD 8/2–100a | E. coli | bs_43 | SHV–2 | AM, CTX |
| HD 9/2–0b | E. coli | bs_44 | SHV–12 | AM, CTX, CAZ, TET, C |
| HD 9/2–100b | E. coli | bs_45 | SHV–12 | AM, CTX, SXT, CAZ, TET, NA, C |
| HD 3/10–0c | E. coli | bs_46 | SHV–12 | AM, CTX, SXT, CAZ, TET, NA, C |
| HD 6/1–1a | E. faecium | bs_47 | VanA | AM, VA, TEC |
| HD 5/3–2a | E. faecium | bs_48 | VanA | AM, VA, TEC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Habib, J.; Haas, D.; Kittinger, C.; Pless, P.; Feierl, G.; Zarfel, G. Multiresistant Bacteria Isolated from Intestinal Faeces of Farm Animals in Austria. Antibiotics 2021, 10, 466. https://doi.org/10.3390/antibiotics10040466
Galler H, Luxner J, Petternel C, Reinthaler FF, Habib J, Haas D, Kittinger C, Pless P, Feierl G, Zarfel G. Multiresistant Bacteria Isolated from Intestinal Faeces of Farm Animals in Austria. Antibiotics. 2021; 10(4):466. https://doi.org/10.3390/antibiotics10040466
Chicago/Turabian StyleGaller, Herbert, Josefa Luxner, Christian Petternel, Franz F. Reinthaler, Juliana Habib, Doris Haas, Clemens Kittinger, Peter Pless, Gebhard Feierl, and Gernot Zarfel. 2021. "Multiresistant Bacteria Isolated from Intestinal Faeces of Farm Animals in Austria" Antibiotics 10, no. 4: 466. https://doi.org/10.3390/antibiotics10040466
APA StyleGaller, H., Luxner, J., Petternel, C., Reinthaler, F. F., Habib, J., Haas, D., Kittinger, C., Pless, P., Feierl, G., & Zarfel, G. (2021). Multiresistant Bacteria Isolated from Intestinal Faeces of Farm Animals in Austria. Antibiotics, 10(4), 466. https://doi.org/10.3390/antibiotics10040466






