Proteomic Charting of Imipenem Adaptive Responses in a Highly Carbapenem Resistant Clinical Enterobacter roggenkampii Isolate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolate
2.2. Growth Media
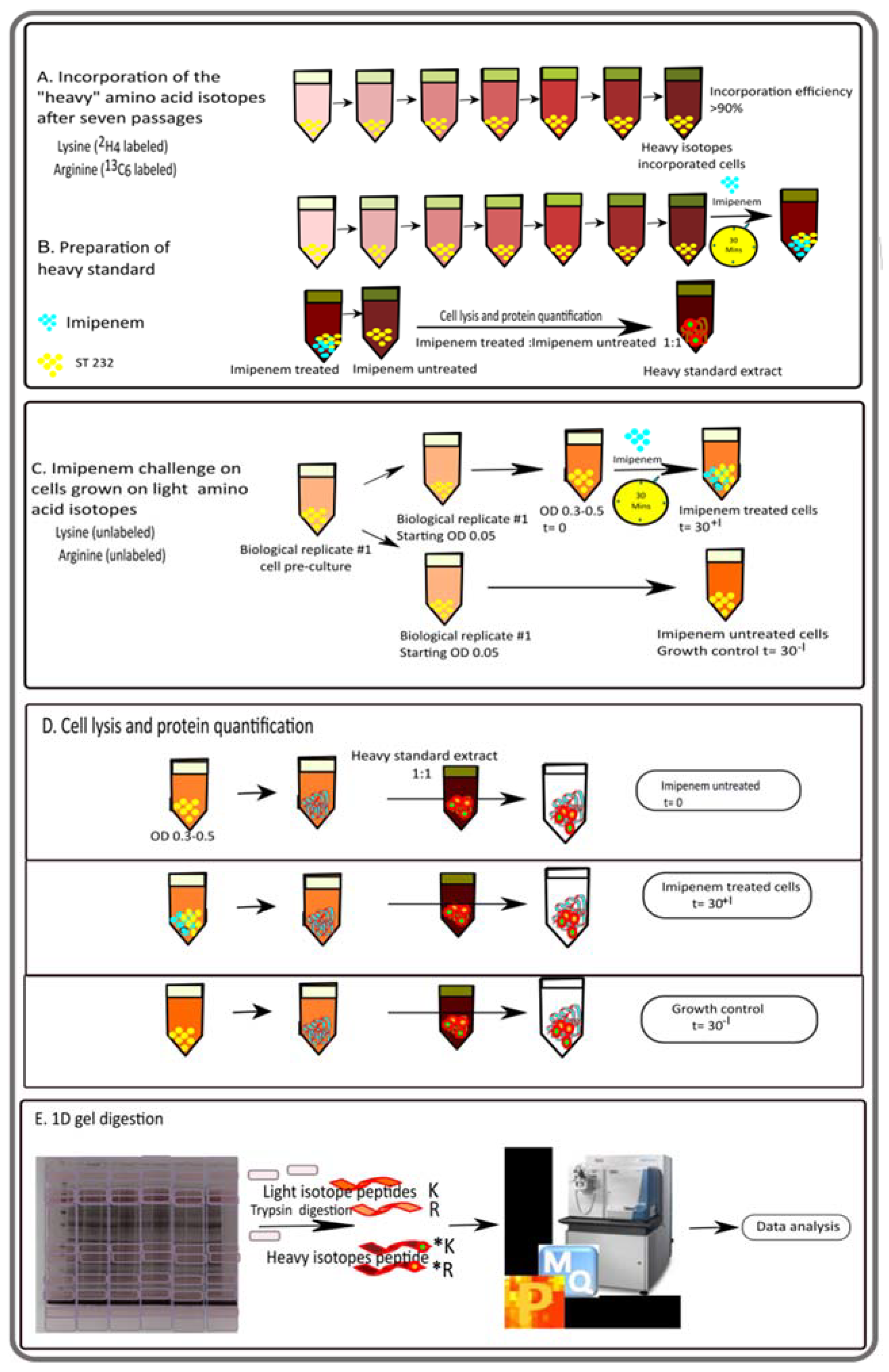
2.3. SILAC and Protein Sample Preparation for Mass Spectrometry
2.4. Liquid Chromatography and Mass Spectrometry of SILAC Samples
2.5. SILAC Data Analysis
2.6. In Silico Functional Proteome Annotation
2.7. Pathway Analyses
2.8. Identification of Fur Boxes in the E. roggenkampii Genome Sequence
2.9. Inductively Coupled Plasma Mass Spectrometry (ICP-MS) Analyses
2.10. Biological and Chemical Safety
2.11. Data Availability
3. Results and Discussion
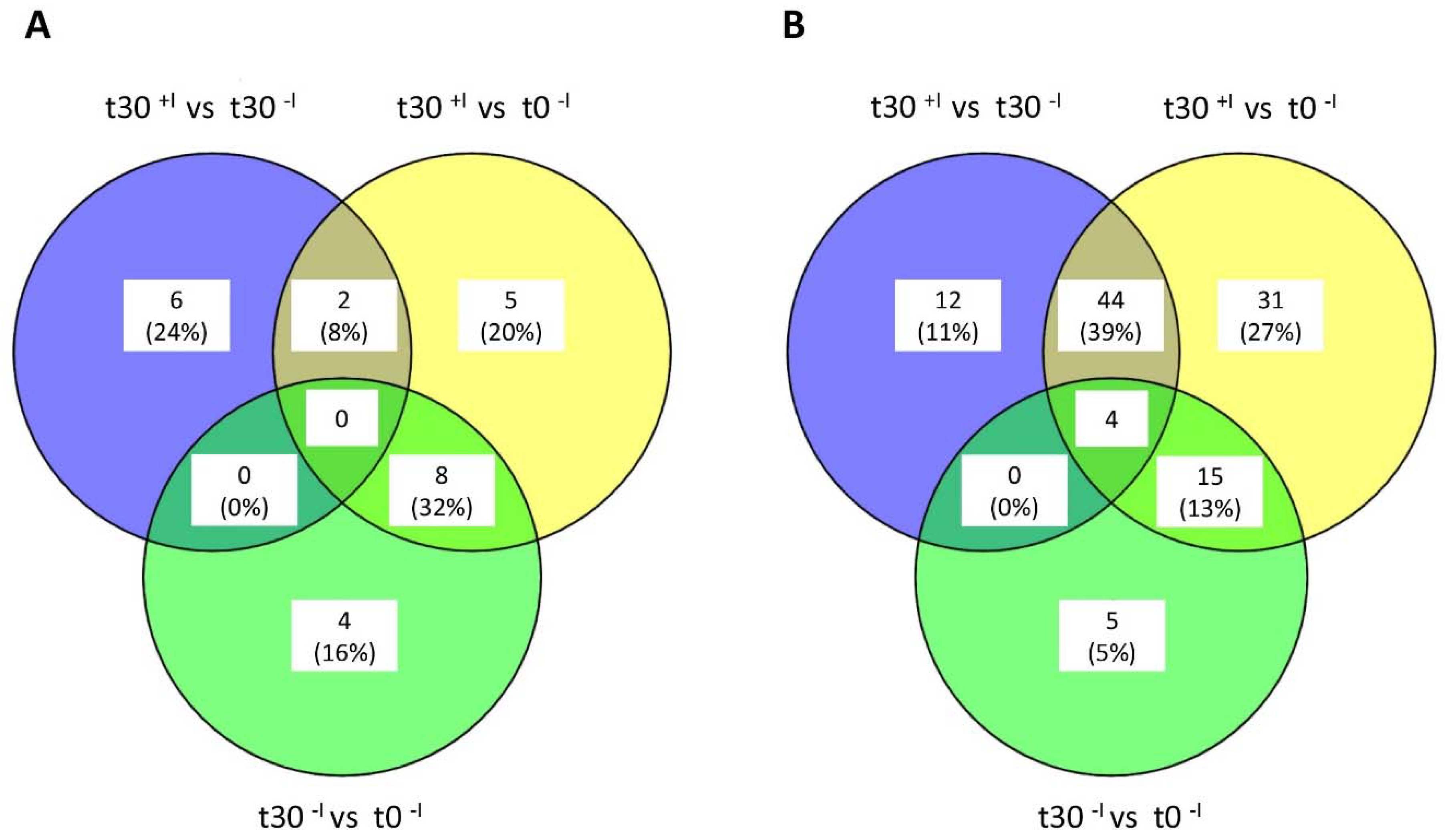
3.1. Analysis of the Imipenem Stress Response of E. roggenkampii ST232 by Stable Isotope Labeling
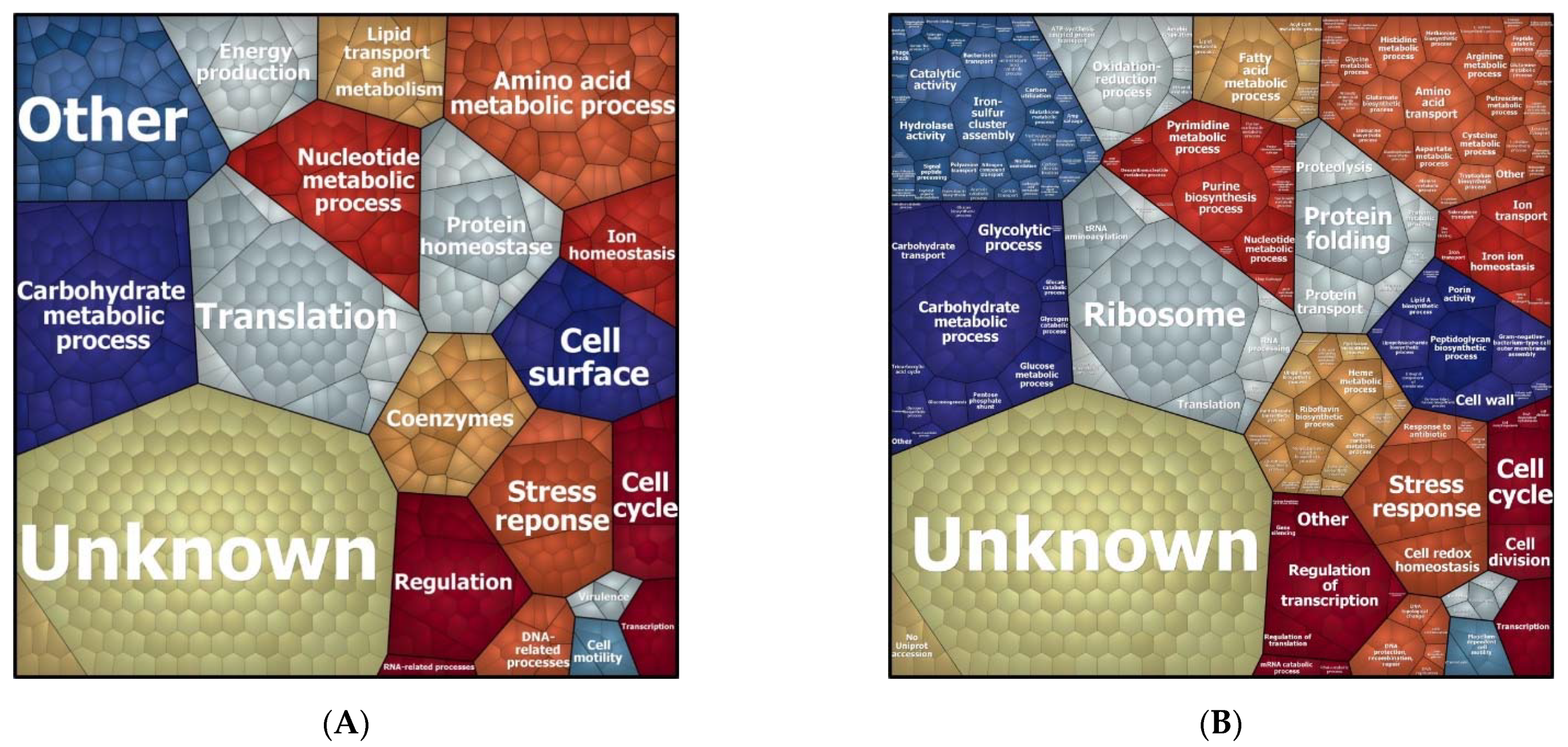
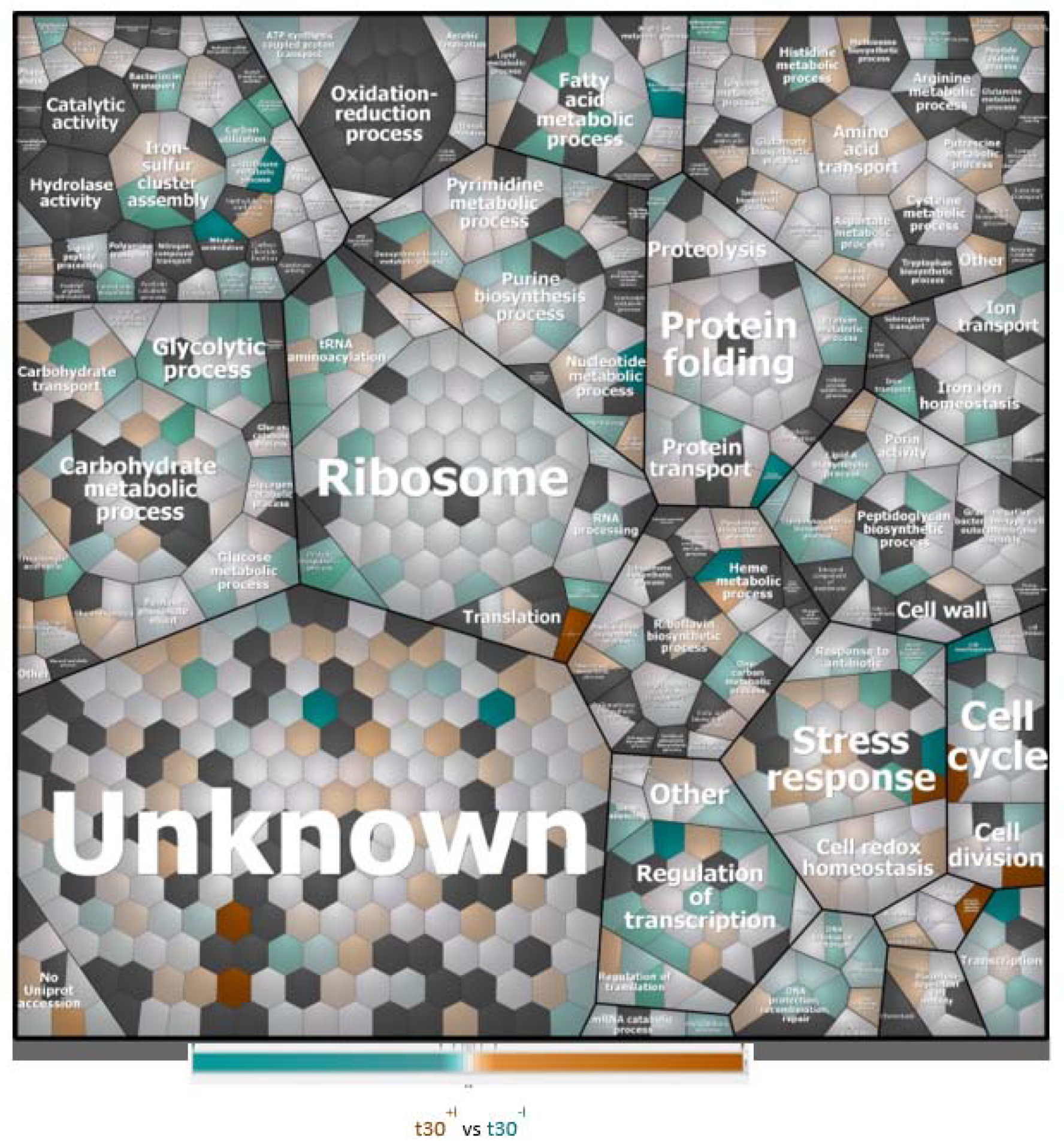
3.2. Biological Processes Adapted in Response to Imipenem Challenge
3.3. The AmpC MIR17 Is Hyper-Expressed, but Not Inducible by Imipenem
3.4. Function of Proteins Differentially Expressed in the Presence of Imipenem
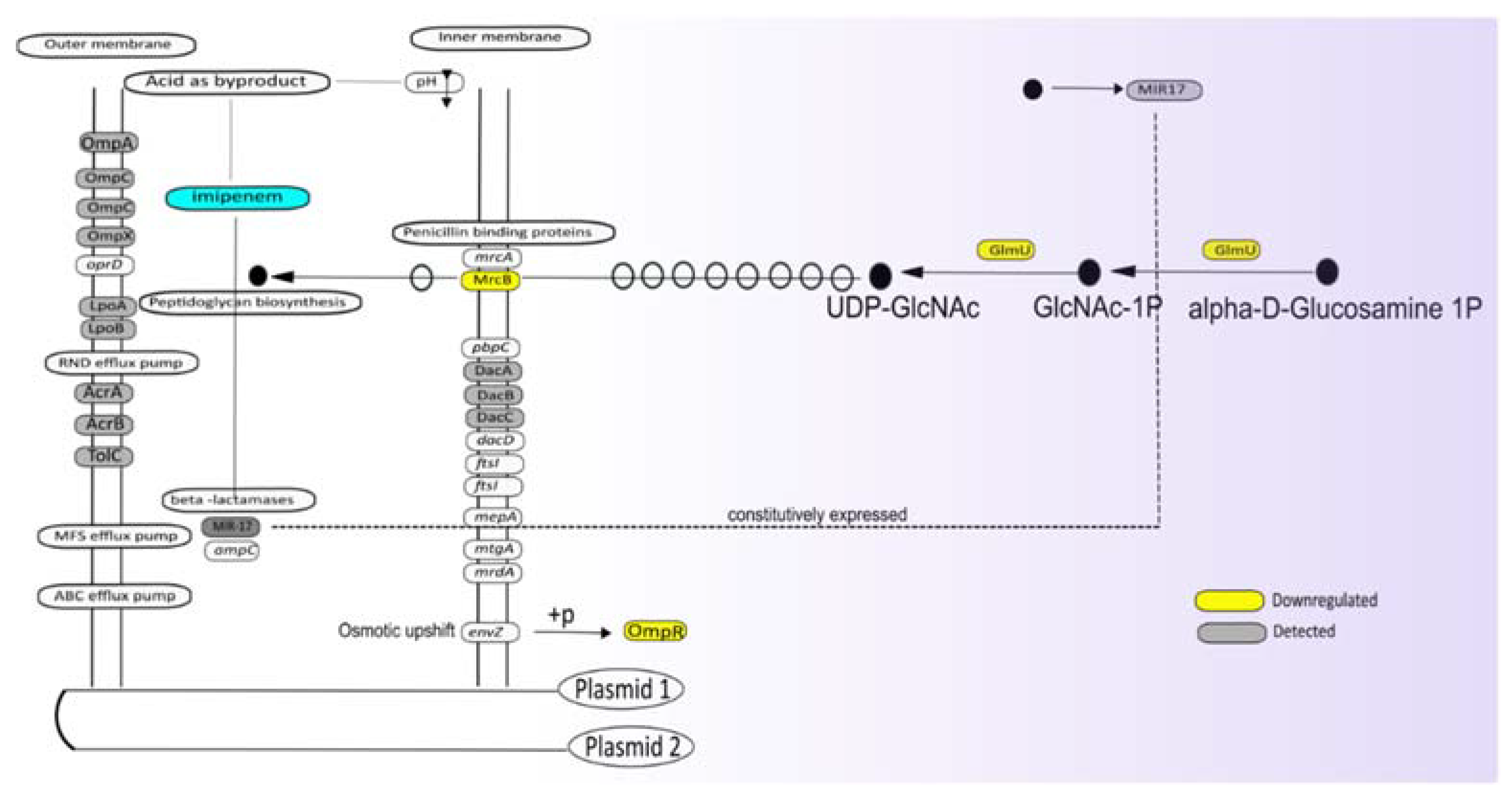
3.5. Potential Impact of Imipenem on Cell Wall Biogenesis
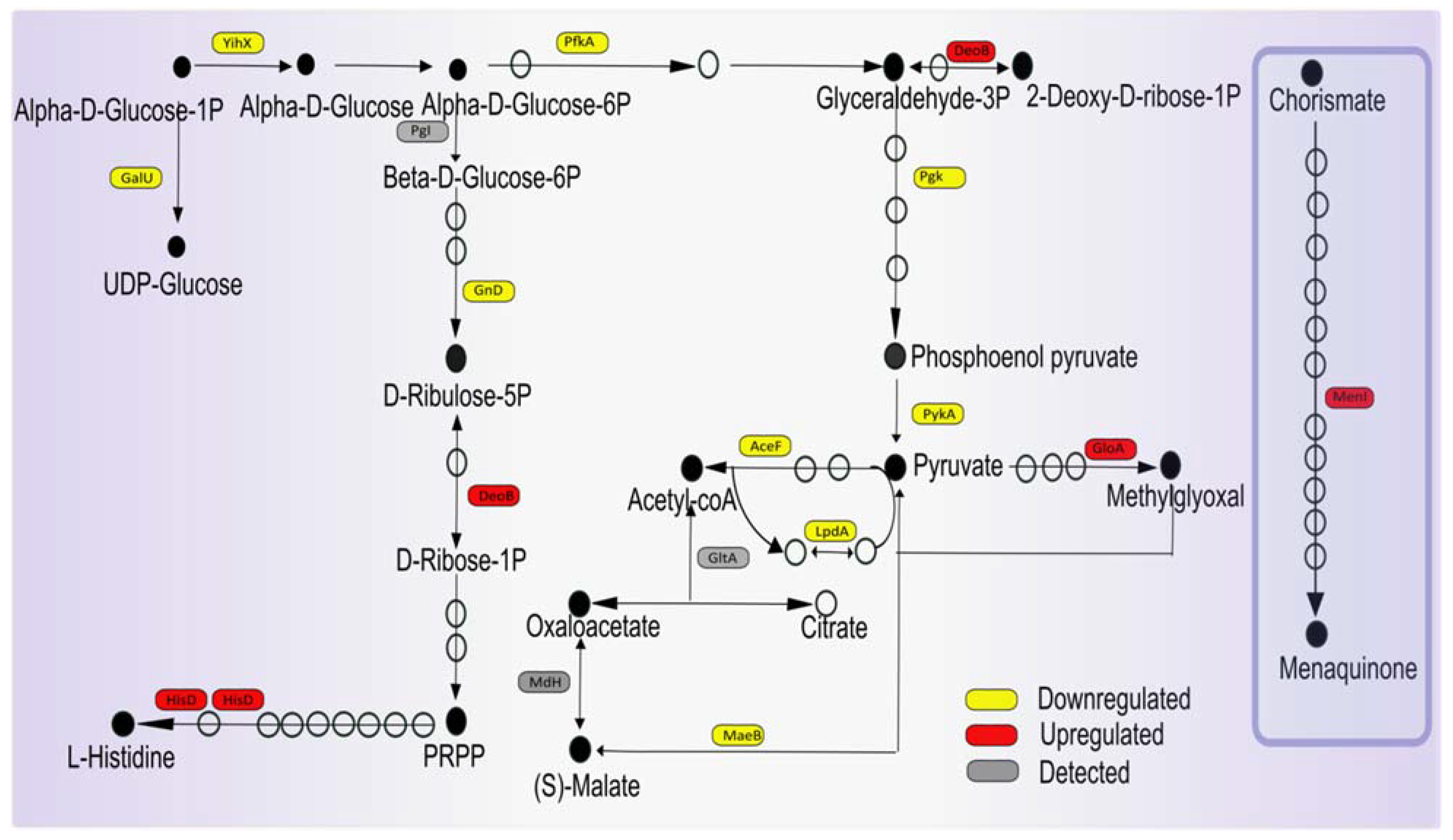
3.6. Potential Impact of Imipenem on Central Carbon Metabolism and Respiration
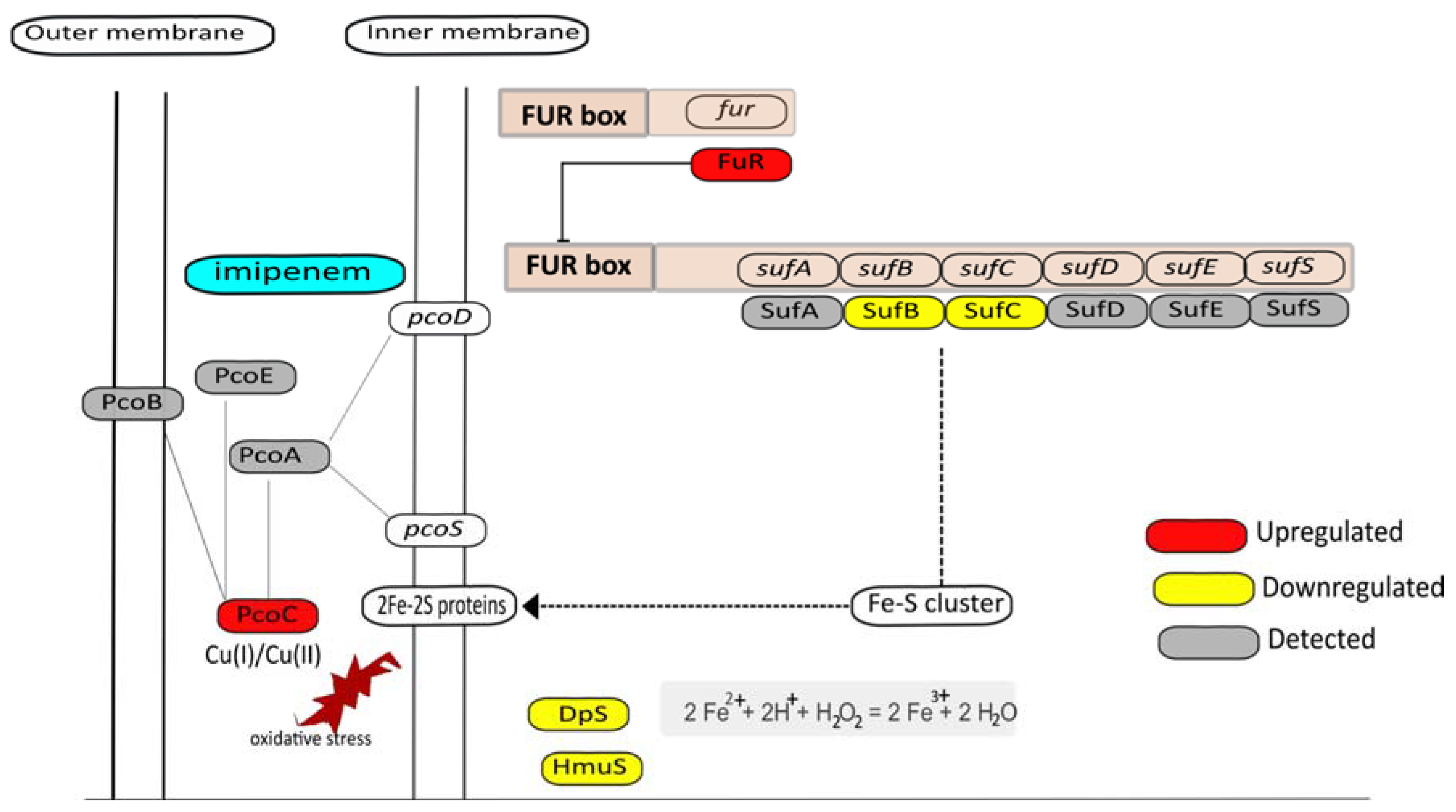
3.7. Metal Homeostasis
3.8. Downregulation of Proteins Involved in Transcription and Translation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSL-2 | biosafety level 2 |
| COG | clusters of orthologous groups |
| ICP-MS | inductively coupled plasma mass spectrometry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LDS | lithium dodecyl sulphate |
| MS | mass spectrometry |
| OD600 | optical density at 600 nm |
| PAGE | polyacrylamide gel electrophoresis |
| PBPs | penicillin binding proteins |
| PSSM | position specific scoring matrix |
| ROS | reactive oxygen species |
| RPMI | Roswell Park Memorial Institute 1640 medium |
| SILAC | stable isotope labeling of amino acids in cell culture |
| ST | sequence type |
| TCA | tricarboxylic acid |
References
- Neu, H.C. Carbapenems: Special properties contributing to their activity. Am. J. Med. 1985, 78, 33–40. [Google Scholar] [CrossRef]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [Green Version]
- Burke, T.P. The Unexpected Effects of the Combination of Antibiotics and Immunity. Cell 2018, 172, 891–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasz, A. The Mechanism of the Irreversible Antimicrobial Effects of Penicillins: How the Beta-Lactam Antibiotics Kill and Lyse Bacteria. Annu. Rev. Microbiol. 1979, 33, 113–137. [Google Scholar] [CrossRef]
- Kawai, Y.; Mercier, R.; Mickiewicz, K.; Serafini, A.; Sório de Carvalho, L.P.; Errington, J. Crucial role for central carbon metabolism in the bacterial L-form switch and killing by β-lactam antibiotics. Nat. Microbiol. 2019, 4, 1716–1726. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Belenky, P.; Ye, J.D.; Porter, C.B.M.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, R.; Adachi, Y.; Imamura, H.; Yamada, K.; Hashizume, T.; Morishima, H. Carbapenem derivatives as potential inhibitors of various β-lactamases, including class B metallo-β-lactamases. Antimicrob. Agents Chemother. 1999, 43, 2497–2503. [Google Scholar] [CrossRef] [Green Version]
- Nordmann, P. Carbapenemase-producing Enterobacteriaceae: Overview of a major public health challenge. Med. Mal. Infect. 2014, 44, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.F.; Yan, J.J.; Lei, H.Y.; Teng, C.H.; Wang, M.C.; Tseng, C.C.; Wu, J.J. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect. Immun. 2012, 80, 1815–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, H.; Scorciapino, M.A.; Moynie, L.; Page, M.G.P.; Naismith, J.H.; Ceccarelli, M.; Winterhalter, M. Molecular Basis of filtering carbapenems by porins from βlactam-resistant clinical strains of Escherichia coli. J. Biol. Chem. 2016, 291, 2837–2847. [Google Scholar] [CrossRef] [Green Version]
- Majewski, P.; Wieczorek, P.; Ojdana, D.; Sienko, A.; Kowalczuk, O.; Sacha, P.; Niklinski, J.; Tryniszewska, E. Altered outer membrane transcriptome balance with AmpC overexpression in carbapenem-resistant enterobacter cloacae. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Nepal, S.; Bonn, F.; Grasso, S.; Stobernack, T.; De Jong, A.; Zhou, K.; Wedema, R.; Rosema, S.; Becher, D.; Otto, A.; et al. An ancient family of mobile genomic islands introducing cephalosporinase and carbapenemase genes in enterobacteriaceae. Virulence 2018, 9, 1377–1389. [Google Scholar] [CrossRef] [Green Version]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Bonn, F.; Bartel, J.; Büttner, K.; Hecker, M.; Otto, A.; Becher, D. Picking vanished proteins from the void: How to collect and ship/share extremely dilute proteins in a reproducible and highly efficient manner. Anal. Chem. 2014, 86, 7421–7427. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Gris, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Schwanhüusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.-Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. EGGNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [Green Version]
- Goto, S.; Kanehisa, M. Kyoto Encylopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar]
- Prajapat, M.K.; Saini, S. Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium. Comput. Biol. Med. 2012, 42, 1133–1140. [Google Scholar] [CrossRef]
- Beckstette, M.; Homann, R.; Giegerich, R.; Kurtz, S. Significant speedup of database searches with HMMs by search space reduction with PSSM family models. Bioinformatics 2009, 25, 3251–3258. [Google Scholar] [CrossRef] [Green Version]
- Bartel, J.; Charkiewicz, E.; Bartz, T.; Bartel, J.; Schmidt, D.; Grbavac, I.; Kyriakopoulos, A. Metalloproteome of the prostate: Carcinoma cell line DU-145 in comparison to healthy rat tissue. Cancer Genom. Proteom. 2010, 7, 81–86. [Google Scholar]
- Wojdyr, M. Fityk: A general-purpose peak fitting program. J. Appl. Crystallogr. 2010, 43, 1126–1128. [Google Scholar] [CrossRef]
- Mäder, U.; Nicolas, P.; Depke, M.; Pané-Farré, J.; Debarbouille, M.; van der Kooi-Pol, M.M.; Guérin, C.; Dérozier, S.; Hiron, A.; Jarmer, H.; et al. . Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions. PLoS Genet. 2016, 12, 1–32. [Google Scholar] [CrossRef]
- Verpooten, G.; Verbist, L.; Buntinx, A.P.; Entwistle, L.A.; Jones, K.H.; De Broe, M.E. The pharmacokinetics of imipenem (thienamycin-formamidine) and the renal dehydropeptidase inhibitor cilastatin sodium in normal subjects and patients with renal failure. Br. J. Clin. Pharmacol. 1984, 18, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Guérin, F.; Isnard, C.; Cattoir, V.; Giard, J.C. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother. 2015, 59, 7753–7761. [Google Scholar] [CrossRef] [Green Version]
- Tipper, D.J.; Strominger, J.L. Mechanism of action of penicillins: A proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc. Natl. Acad. Sci. USA 1965, 54, 1133–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.J.; Inouye, M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002, 277, 24155–24161. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.; Mercier, R.; Wu, L.J.; Domínguez-Cuevas, P.; Oshima, T.; Errington, J. Cell Growth of Wall-Free L-Form Bacteria Is Limited by Oxidative Damage. Curr. Biol. 2015, 25, 1613–1618. [Google Scholar] [CrossRef] [Green Version]
- Van Acker, H.; Coenye, T. The Role of Reactive Oxygen Species in Antibiotic-Mediated Killing of Bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, E.; Proudfoot, M.; Gonzalez, C.F.; Brown, G.; Omelchenko, M.V.; Borozan, I.; Carmel, L.; Wolf, Y.I.; Mori, H.; Savchenko, A.V.; et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 2006, 281, 36149–36161. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, M.; Wildberger, P.; Nidetzky, B. Yihx-encoded haloacid dehalogenase-like phosphatase HAD4 from Escherichia coli is a specific α-d-glucose 1-phosphate hydrolase useful for substrate-selective sugar phosphate transformations. J. Mol. Catal. B Enzym. 2014, 110, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.M.; Akhtar, M.S.; Ansari, A.Z.; Landick, R.; Kiley, P.J. The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally. PLoS Genet. 2013, 9, e1003839. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Wang, B.; Ding, H.; Zhang, J.; Li, S. The role of NADP-malic enzyme in plants under stress. Plant. Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef]
- Tedeschi, P.M.; Bansal, N.; Kerrigan, J.E.; Abali, E.E.; Scotto, K.W.; Bertino, J.R. NAD+ kinase as a therapeutic target in cancer. Clin. Cancer Res. 2016, 22, 5189–5195. [Google Scholar] [CrossRef] [Green Version]
- Vervoort, L.M.T.; Ronden, J.E.; Thijssen, H.H.W. The potent antioxidant activity of the vitamin K cycle in microsomal lipid peroxidation. Biochem. Pharmacol. 1997, 54, 871–876. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, T.; Happe, T.; Lu, Y.; Sawyer, A. Iron-sulphur cluster biogenesis: Via the SUF pathway. Metallomics 2018, 10, 1038–1052. [Google Scholar] [CrossRef]
- Yang, L.; Mih, N.; Anand, A.; Park, J.H.; Tan, J.; Yurkovich, J.T.; Monk, J.M.; Lloyd, C.J.; Sandberg, T.E.; Seo, S.W.; et al. Cellular responses to reactive oxygen species are predicted from molecular mechanisms. Proc. Natl. Acad. Sci. USA 2019, 116, 14368–14373. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.M.; Jones, H.A.; Perry, R.D. Molecular Characterization of the Hemin Uptake Locus (hmu) from Yersinia pestis and Analysis ofhmu Mutants for Hemin and Hemoprotein Utilization. Infect. Immun. 1999, 67, 3879–3892. [Google Scholar] [CrossRef] [Green Version]
- Chillappagari, S.; Seubert, A.; Trip, H.; Kuipers, O.P.; Marahiel, M.A.; Miethke, M. Copper Stress Affects Iron Homeostasis by Destabilizing Iron-Sulfur Cluster Formation in Bacillus subtilis. J. Bacteriol. 2010, 192, 2512–2524. [Google Scholar] [CrossRef] [Green Version]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.; Yang, J.; Li, T.; Zhao, J.; Sun, S.; Li, X.; Lin, C.; Li, J.; Zhou, H.; Lyu, J.; et al. Anaerobic Copper Toxicity and Iron-Sulfur Cluster Biogenesis in Escherichia coli. Appl. Environ. Microbiol. 2017, 83, 16. [Google Scholar] [CrossRef] [Green Version]
- Peariso, K.; Huffman, D.L.; Penner-Hahn, J.E.; O’Halloran, T.V. The PcoC Copper Resistance Protein Coordinates Cu(I) via Novel S-Methionine Interactions. J. Am. Chem. Soc. 2003, 125, 342–343. [Google Scholar] [CrossRef]
- Ling, J.; Söll, D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. USA 2010, 107, 4028–4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nepal, S.; Maaß, S.; Grasso, S.; Cavallo, F.M.; Bartel, J.; Becher, D.; Bathoorn, E.; van Dijl, J.M. Proteomic Charting of Imipenem Adaptive Responses in a Highly Carbapenem Resistant Clinical Enterobacter roggenkampii Isolate. Antibiotics 2021, 10, 501. https://doi.org/10.3390/antibiotics10050501
Nepal S, Maaß S, Grasso S, Cavallo FM, Bartel J, Becher D, Bathoorn E, van Dijl JM. Proteomic Charting of Imipenem Adaptive Responses in a Highly Carbapenem Resistant Clinical Enterobacter roggenkampii Isolate. Antibiotics. 2021; 10(5):501. https://doi.org/10.3390/antibiotics10050501
Chicago/Turabian StyleNepal, Suruchi, Sandra Maaß, Stefano Grasso, Francis M. Cavallo, Jürgen Bartel, Dörte Becher, Erik Bathoorn, and Jan Maarten van Dijl. 2021. "Proteomic Charting of Imipenem Adaptive Responses in a Highly Carbapenem Resistant Clinical Enterobacter roggenkampii Isolate" Antibiotics 10, no. 5: 501. https://doi.org/10.3390/antibiotics10050501
APA StyleNepal, S., Maaß, S., Grasso, S., Cavallo, F. M., Bartel, J., Becher, D., Bathoorn, E., & van Dijl, J. M. (2021). Proteomic Charting of Imipenem Adaptive Responses in a Highly Carbapenem Resistant Clinical Enterobacter roggenkampii Isolate. Antibiotics, 10(5), 501. https://doi.org/10.3390/antibiotics10050501





