Honey as an Ecological Reservoir of Antibacterial Compounds Produced by Antagonistic Microbial Interactions in Plant Nectars, Honey and Honey Bee
Abstract
:1. Introduction
2. Antagonistic Interspecies Interactions as a Source of Antimicrobial Compounds
3. Discovery of Antibacterial Activity of Honey-Associated Microbiota
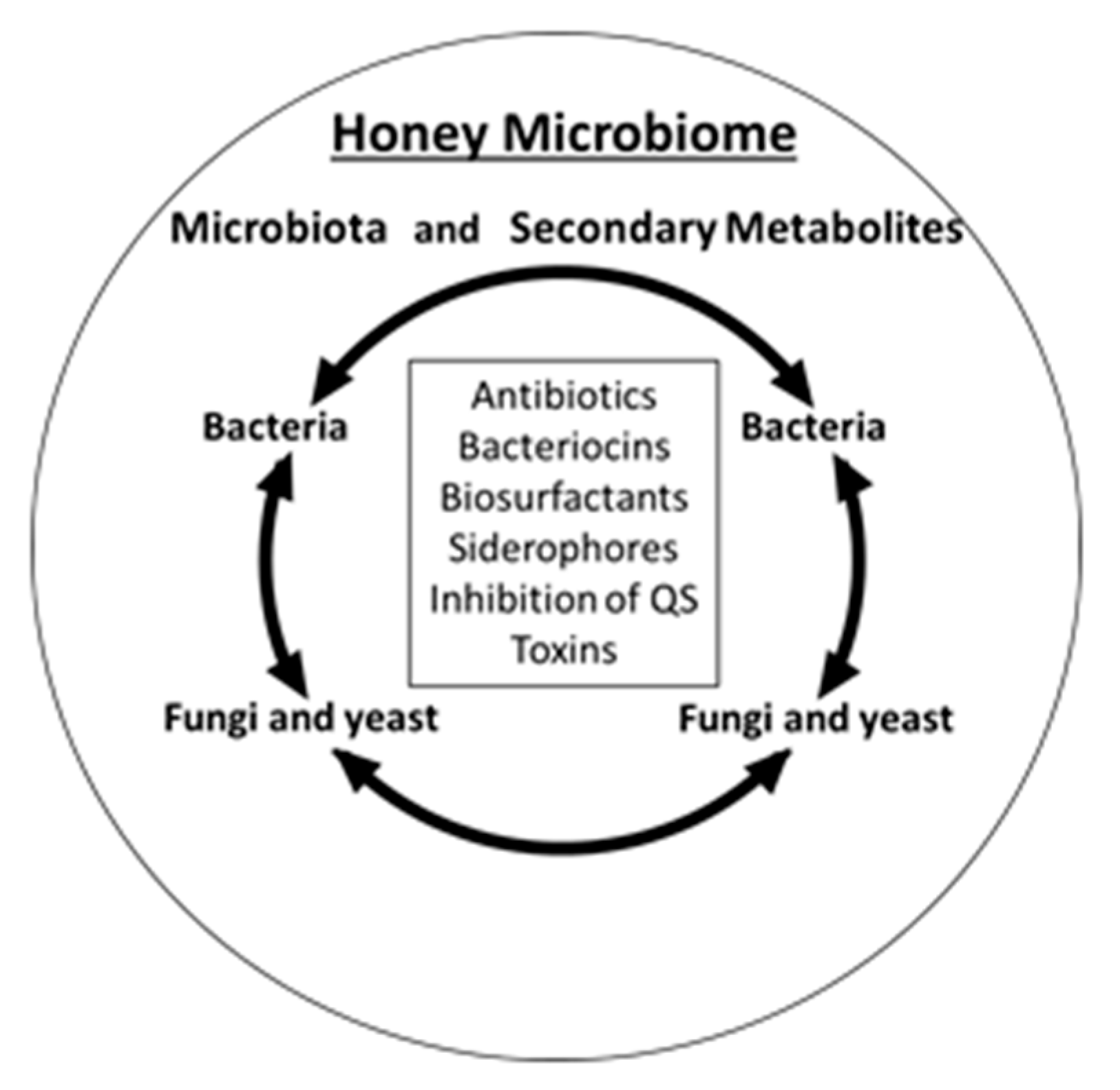
4. Honey Microbiome
5. The Core Bacteria of Honey
5.1. The Composition Lactic Acid Bacteria in Honey
5.2. The Composition of the Family Bacillaceae in Honey
5.3. Fungal Composition of Nectar and Honey
6. The Overview of Antimicrobial Compounds Produced by Honey Microbiota
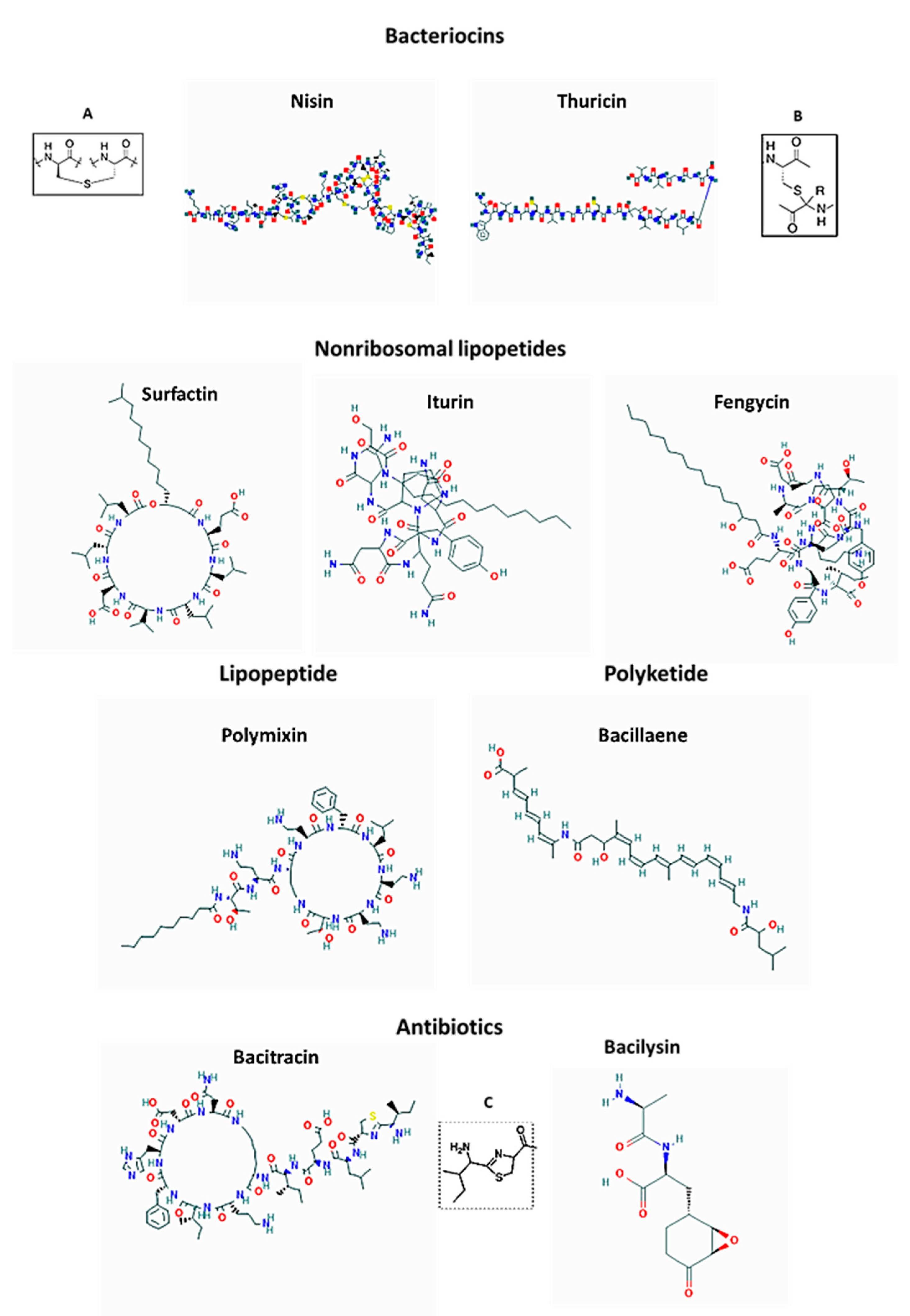
6.1. LAB Bacteriocins
6.1.1. Kunkecin A
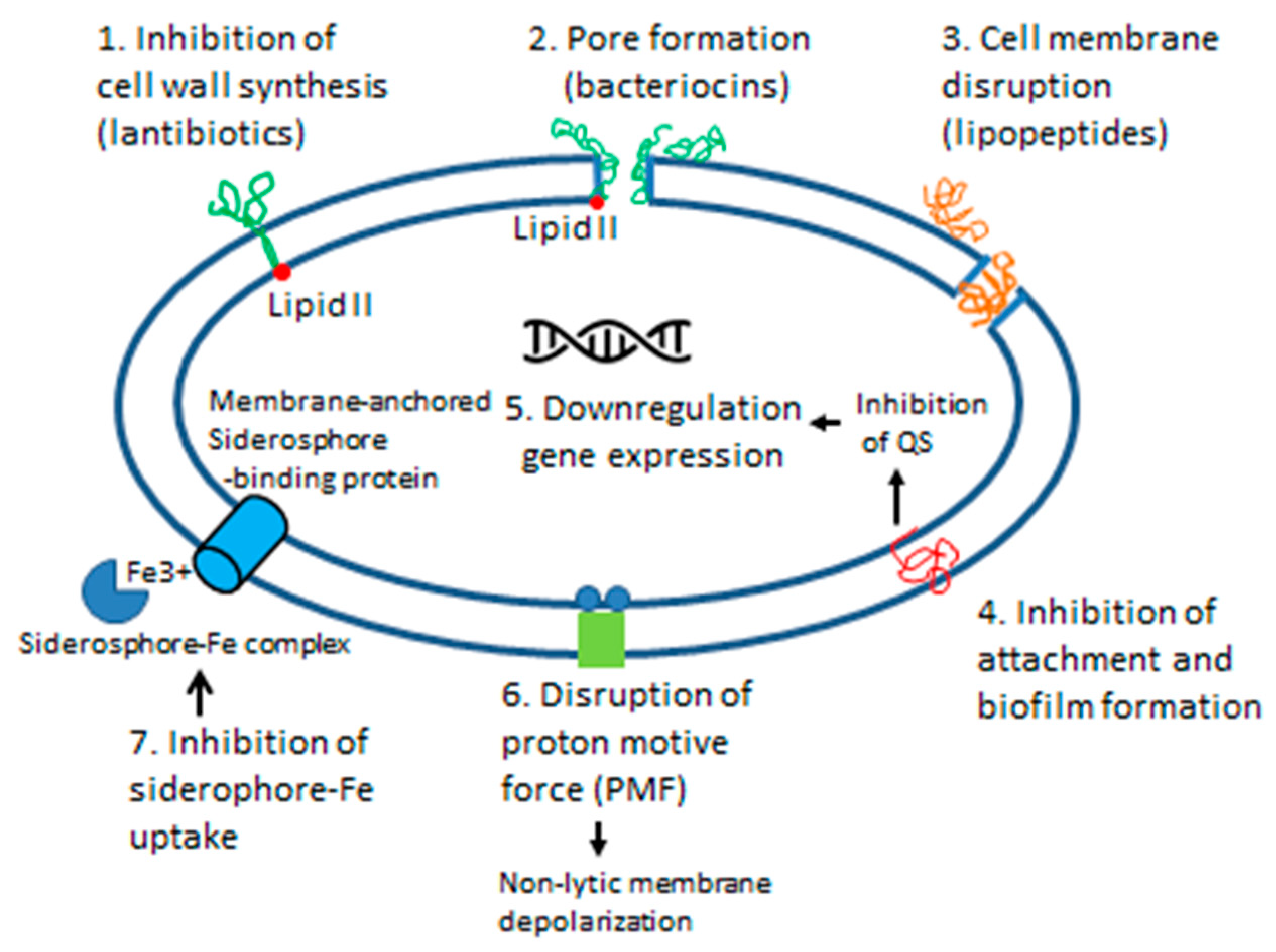
6.1.2. Mode of Action
6.1.3. Spectrum of Antimicrobial Activity
6.2. LAB Surfactants, Modes of Anti-Biofilm Action and Spectrum of Activity
6.2.1. Mode of Action
6.2.2. Spectrum of Activity
7. Antimicrobial Compounds Produced by Bacillus spp.
7.1. Bacteriocins of Bacillus Species
Bacteriocins and Autolysins
| Species | Ribosomal Peptides | Nonribosomal Peptides | Target | Ref. | |||
|---|---|---|---|---|---|---|---|
| Antibiotics | Lipopetides | Siderophores | Polyketides | ||||
| B. subtilis | subtilin | [64,110,111,112] | |||||
| subtilosin A | Gram+ L.monocytogenes Gardnerella vaginalis S. agalactiae | [64,110,111,112] | |||||
| sublancin | B. cereus S. pyogenes S. aureus | [64,110,111] | |||||
| surfactin | Bacteria, viruses fungi | [64] | |||||
| fengycin | fungi | ||||||
| bacillomycin | bacteria | [64] | |||||
| bacillibactin | [111] | ||||||
| bacitracin | Gram+ PP synthesis C55-PP carrier | [64,111] | |||||
| bacilysin | Gram+, PP synthesis fungi | ||||||
| bacillaene | |||||||
| B. licheniformis | lichenin | bacitracin | PP synthesis, Gram+ | [110,111] | |||
| lichenicidin | L. monocytogenes MRSA VRE | [110] | |||||
| lychenisin | [111] | ||||||
| B. amyloliquefa-ciens | amylolysin | iturin | bacillaene | ||||
| bacilysin | S. aureus | [111] | |||||
| Subtiliosin | fengycin | ||||||
| surfactin | |||||||
| B. cereus | cereins | B. cereus, B.coagulans, B. subtilis, B. pumilus | [110,111] | ||||
| bacillibactin | |||||||
| thuricin | [110,111] | ||||||
| B. thuringiensis | thuricin 17 | B.thuringiensis, B. cereus E. coli MM294 | [110,111] | ||||
| thurincin H | B. cereus, B. subtilis, B. megaterium, L. monocytogenes, L. innocua, L. ivanovii, S. aureus, Carnobacterim psicola, Geobacillus stearothermophillus | [115] | |||||
| thuricin CD | C. difficile | [117] | |||||
| B. mycoides | |||||||
| B. pumilis | pumilicin | surfactin | |||||
| bacilysin | [110] | ||||||
| Pumilacidin | [112] | ||||||
| bacitracin | |||||||
| B. safensis | |||||||
| B. altitudinis | |||||||
| B. mojavensis | |||||||
| B. megaterium | megacin | surfactin | |||||
| fengycin | |||||||
| bacillomycins | [112] | ||||||
| B. aerius | |||||||
| B. altitudinis | |||||||
| P. alvei | |||||||
| P. larvae | paenibacterin | ||||||
| P. polymyxa | paeniba-cillin | Bacillus spp., C. sporogenes, Lactobacillus spp., L. lactis, Leuconostoc mesenteroides, Listeria spp., Pediococcus cerevisiae, S. aureus S. agalactiae | [110] | ||||
| bacillibactin | |||||||
| bacillaene | [111] | ||||||
| polymyxin | Gram-positive Gram-negative | [111] | |||||
| paenima-crolidin | S. aureus | [111] | |||||
| B. brevis | gramicidin | [112] | |||||
7.2. Non-Ribosomal Peptide Antibiotics of Bacillus spp.
7.2.1. Antibiotics
7.2.2. Lipopetide Surfactants
7.2.3. Siderophores
7.3. Paenibacillus
7.4. Antimicrobial Compounds of Fungal Origin and Their Potential Contribution to Honey Antimicrobial Activity
7.4.1. Mycotoxins
7.4.2. β-lactams
7.4.3. Surfactants
7.4.4. Siderophores
8. The Antagonistic Interactions between Microbes at the Ecological Level
9. Pathogenesis-Related Proteins of Plants
10. Honey Bee Antimicrobial Peptides of Honey
11. Conclusions
- Cell wall damaging compounds:
- Bacteriocins originating from Bacillus and Lactobacillus species
- Antimicrobial peptides originated from bee: defensins, hymenoptaecins, jelleins
- Antimicrobial peptides originating from plants: thionins and thaumatin-like peptides
- Antibiotics; bacilysin and bacillaene of bacterial origin
- Biosurfactants; lipopetides of bacterial origin (surfactin, iturin, fengycin, polymyxins)
- Biosurfactants of plant origin; lipid transfer proteins
- Biosurfactants of fungal origin
- Anti-fungal enzymes of plant origin: chitinases, glucanases and lysozymes hydrolyzing peptidoglycan (PG) of bacterial cell envelope
- Inhibitors of peptidoglycan synthesis:
- I.
- Antibiotics; β-lactams of fungal origin and bacitracin of bacterial origin
- J.
- Lantibiotic bacteriocins of bacterial origin
- Siderophores of bacterial and fungal origin
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Nolan, V.C.; Harrison, J.; Cox, J.A.G. Dissecting the Antimicrobial Composition of Honey. Antibiotics 2019, 8, 251. [Google Scholar] [CrossRef] [Green Version]
- Combarros-Fuertes, P.; Fresno, J.M.; Estevinho, M.M.; Sousa-Pimenta, M.; Tornadijo, M.E.; Estevinho, L.M. Honey: Another Alternative in the Fight against Antibiotic-Resistant Bacteria? Antibiotics 2020, 9, 774. [Google Scholar] [CrossRef]
- Molan, P.C. The Antibacterial Activity of Honey. 2.Variation in the potency of the antibacterial activity. Bee World 1992, 73, 59–76. [Google Scholar] [CrossRef]
- Brudzynski, K. A current perspective on hydrogen peroxide production in honey. A review. Food Chem. 2020, 332, 127229. [Google Scholar] [CrossRef] [PubMed]
- Mavric, E.; Wittmann, S.; Barth, G.; Henle, T. Identification and quantification of methylglyoxal as the dominant antibacterial constituent of Manuka (Leptospermum scoparium) honeys from New Zealand. Mol. Nutr. Food Res. 2008, 52, 483–489. [Google Scholar] [CrossRef]
- Imlay, J.A.; Linn, S. DNA damage and oxygen radical toxicity. Science 1998, 240, 1302–1309. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From target to networks. Nat. Rev. 2010, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Brudzynski, K.; Abubaker, K.; Wang, T. Powerful bacterial killing by buckwheat honeys is concentration-dependent, involves complete DNA degradation and requires hydrogen peroxide. Front. Microbiol. 2012, 3, 242. [Google Scholar] [CrossRef] [Green Version]
- Adams, C.J.; Boult, C.H.; Deadman, B.J.; Farr, J.M.; Grainger, M.N.; Manley-Harris, M.; Snow, M.J. Isolation by HPLC and characterisation of the bioactive fraction of New Zealand Manuka (Leptospermum scoparium) honey. Carbohydr. Res. 2008, 343, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Lamb, A.J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef]
- Covan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar]
- Nicolson, S.W.; Thornburg, R.W. Nectar chemistry. In Nectaries and Nectar; Nicolson, S.W., Nepi, M., Pacini, E., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 215–249. [Google Scholar] [CrossRef]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman, J.D. Natural Cinnamic Acids, Synthetic Derivatives and Hybrids with Antimicrobial Activity. Molecules 2014, 19, 19292–19349. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Deighton, M.; Livanos, G.; Morrison, P.D.; Pang, E.C.K.; Mantri, N. Antimicrobial activity of Agastache honey and characterization of its bioactive compounds in comparison with important commercial honeys. Front. Microbiol. 2019, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Blair, S.E.; Cokcetin, N.N.; Harry, E.J.; Carter, D.A. The unusual antibacterial activity of medical-grade Leptospermum honey: Antibacterial spectrum, resistance and transcriptome analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R. Using honey to inhibit wound pathogens. Nurs. Times 2008, 104, 46–49. [Google Scholar] [PubMed]
- Henriques, A.F.; Jenkins, R.E.; Burton, N.F.; Cooper, R.A. The intracellular effects of manuka honey on Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Henriques, A.F.; Jenkins, R.E.; Burton, N.F.; Cooper, R.A. The effect of manuka honey on the structure of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 167–171. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.; Burton, N.; Cooper, R. Manuka honey inhibits cell division in methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2011, 66, 2536–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brudzynski, K.; Sjaarda, C. Antibacterial Compounds of Canadian Honeys Target Bacterial Cell Wall Inducing Phenotype Changes, Growth Inhibition and Cell Lysis That Resemble Action of β-Lactam Antibiotics. PLoS ONE 2014, 9, e106967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasfi, R.; Elkhatib, W.F.; Khairalla, A.S. Effects of selected Egyptian honeys on the cellular ultrastructure and the gene expression profile of Escherichia coli. PLoS ONE 2016, 11, e0150984. [Google Scholar]
- Pajor, M.; Xiong, Z.R.; Worobo, R.W.; Szweda, P. Paenibacillus alvei MP1 as a Producer of the Proteinaceous Compound with Activity against Important Human Pathogens, Including Staphylococcus aureus and Listeria monocytogenes. Pathogens 2020, 9, 319. [Google Scholar] [CrossRef] [PubMed]
- Combarros-Fuertes, P.; Estevinho, L.M.; Teixeira-Santos, R.; Rodrigues, A.G.; Pina-Vaz, C.; Fresno, J.M.; Tornadijo, M.E. Evaluation of Physiological Effects Induced by Manuka Honey Upon Staphylococcus aureus and Escherichia coli. Microorganisms 2019, 7, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hibbing, M.; Fuqua, C.; Parsek, M.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, A.E.; Robinson, C.J.; Peterson, S.B.; Raffa, K.F.; Handelsman, J. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 2008, 62, 375–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; Straight, P.D. Multifaceted interfaces of bacterial competition. J. Bacteriol. 2016, 198, 2145–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snowdon, J.A.; Cliver, D.O. Microorganisms in honey. Int. J. Food Microbiol. 1996, 31, 1–26. [Google Scholar] [CrossRef]
- Olaitan, P.B.; Adeleke, O.E.; Iyabo, O.O. Honey: A reservoir for microorganisms and an inhibitory agent for microbes. Afr. Health Sci. 2007, 7, 159–165. [Google Scholar]
- Alvarez-Pérez, S.; Herrera, C.M.; de Vega, C. Zooming-in on floral nectar: A first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 2012, 80, 591–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundo, M.A.; Padilla-Zakour, O.I.; Worobo, R.W. Growth inhibition of foodborne pathogens and food spoilage organisms by select raw honeys. Int. J. Food Microbiol. 2004, 97, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Churey, J.J.; Worobo, R.W. Purification and structural characterization of bacillomycin F produced by a bacterial honey isolate active against Byssochlamys fulva H25. J. App. Microbiol. 2008, 105, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Churey, J.J.; Worobo, R.W. Antimicrobial activity of bacterial isolates from different floral sources of honey. Int. J. Food Microbiol. 2008, 126, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Champomier Vergès, M.-C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Aizenberg-Gershtein, Y.; Izhaki, I.; Halpern, M. Do Honeybees Shape the Bacterial Community Composition in Floral Nectar? PLoS ONE 2013, 8, e67556. [Google Scholar] [CrossRef] [Green Version]
- Fridman, S.; Izhaki, I.; Gerchman, Y.; Halpern, M.; Samuni-Blank, M.; Izhaki, I.; Laviad, S.; Bar-Massada, A.; Gerchman, Y.; Halpern, M. The Role of Abiotic Environmental Conditions and Herbivory in Shaping Bacterial Community Composition in Floral Nectar. PLoS ONE 2014, 9, e99107. [Google Scholar]
- Ruiz-Argueso, T.; Rodriguez-Navarro, A. Microbiology of ripening honey. Appl. Microbiol. 1975, 30, 893–896. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, L.; Jin, Y.; Zhang, J.; Su, L.; Zhang, X.; Zhou, J.; Li, Y. The microbial community dynamics during the vitex honey ripening process in the honeycomb. Front. Microbiol. 2017, 8, 1649. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef] [Green Version]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef] [Green Version]
- Bovo, S.; Ribani, A.; Utzeri, V.J.; Schiavo, G.; Bertolini, F.; Fontanesi, L. Shotgun metagenomics of honey DNA: Evaluation of a methodological approach to describe a multi-kingdom honey bee derived environmental DNA signature. PLoS ONE 2018, 13, e0205575. [Google Scholar] [CrossRef] [PubMed]
- Manirajan, A.B.; Ratering, S.; Rusch, V.; Schwiertz, A.; Geissler-Plaum, R.; Cardinale, M.; Schnell, S. Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 2016, 18, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Futagawa-Endo, Y.; Dicks, L.M. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst. Appl. Microbiol. 2009, 32, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Salminen, S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 2013, 36, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Neveling, D.P.; Endo, A.; Dicks, L.M. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and beehives. Curr. Microbiol. 2012, 65, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Vásquez, A. Detection and Identification of a Novel Lactic Acid Bacterial Flora within the Honey Stomach of the Honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, T.C.; Alsterfjord, M.; Nilson, B.; Butler, È.; Vásquez, A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 9, 3109–3119. [Google Scholar] [CrossRef] [PubMed]
- Gustaw, K.; Michalak, M.; Polak-Berecka, M.; Wasko, A. Isolation and characterization of a new fructophilic Lactobacillus plantarum FPL strain from honeydew. Ann. Microbiol. 2018, 68, 459–470. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.C.; Vásquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2009, 41, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Ramos, O.Y.; Basualdo, M.; Libonatti, C.; Vega, M.F. Current status and application of lactic acid bacteria in animal production systems with a focus on bacteria from honey bee colonies. J. Appl. Microbiol. 2020, 128, 1248–1260. [Google Scholar] [CrossRef]
- Aween, M.M.; Hassan, Z.; Muhialdin, B.J.; Noor, H.M.; Eljamel, Y.A. Evaluation on antibacterial activity of Lactobacillus acidophilus strains isolated from honey. Am. J. Appl. Sci. 2012, 9, 807–817. [Google Scholar] [CrossRef] [Green Version]
- Berríos, P.; Fuentes, J.A.; Salas, D.; Carreño, A.; Aldea, P.; Fernández, F.; Trombert, A.N. Inhibitory effect of biofilm-forming Lactobacillus kunkeei strains against virulent Pseudomonas aeruginosa in vitro and in honeycomb moth (Galleria mellonella) infection model. Benef. Microbes 2018, 9, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Chaven, S. Honey, Confectionery and Bakery Products. In Food Safety Management; Motarjemi, Y., Lelieveld, H., Eds.; Academic Press: Cambridge, MA, USA, 2014; Chapter 11; pp. 283–299. [Google Scholar] [CrossRef]
- Muhialdin, B.J.; Hassan, Z.; Saari, N. In vitro antifungal activity of lactic acid bacteria low molecular peptides against spoilage fungi of bakery products. Ann. Microbiol. 2018, 68, 557–567. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Yan, B.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Lactic Acid Bacteria as Antifungal and Anti-Mycotoxigenic Agents: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1403–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinacori, M.; Francesca, N.; Alfonzo, A.; Cruciata, M.; Sannino, C.; Settanni, L.; Moschetti, G. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 2014, 38, 284–294. [Google Scholar] [CrossRef] [Green Version]
- Pajor, M.; Worobo, R.W.; Milewski, S.; Szweda, P. The Antimicrobial Potential of Bacteria Isolated from Honey Samples Produced in the Apiaries Located in Pomeranian Voivodeship in Northern Poland. Int. J. Environ. Res. Public Health 2018, 15, 2002. [Google Scholar] [CrossRef] [Green Version]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef] [Green Version]
- Brudzynski, K.; Flick, R. Accumulation of soluble menaquinones MK-7 in honey coincides with death of Bacillus spp. present in honey. Food Chem. X 2019, 1, 100008. [Google Scholar] [CrossRef]
- Jack, R.W.; Tagg, J.R.; Ray, B. Bacteriocins of Gram-positive bacteria. Microbiol. Rev. 1995, 59, 171. [Google Scholar] [CrossRef] [PubMed]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grecka, K.; Kus, P.M.; Worobo, R.W.; Szweda, P. Study of the anti-staphylococcal potential of honeys produced in Northern Poland. Molecules 2018, 23, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, C.M.; De Vega, C.; Canto, A.; Pozo, M.I. Yeasts in floral nectar: A quantitative survey. Ann. Bot. 2009, 103, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Andrade, E.; Stchigel, A.M.; Terrab, A.J.; Guarro, A.J.; Cano-Lira, J.F. Diversity of xerotolerant and xerophilic fungi in honey. IMA Fungus 2019, 10, 20. [Google Scholar] [CrossRef]
- Kačániová, M.; Kňazovická, V.; Felšöciová, S.; Rovná, K. Microscopic fungi recovered from honey and their toxinogenity. J. Environ. Sci. Health Part A. 2012, 47, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Golubev, W.I. Antagonistic interactions among yeasts. In Biodiversity and Ecophysiology of Yeasts; Rosa, C.A., Peter., G., Eds.; Springer: Berlin, Germany, 2006; pp. 197–219. [Google Scholar]
- Deshmukh, S.K.; Verekar, S.A.; Bhave, S.V. Endophytic fungi: A reservoir of antibacterials. Front. Microbiol. 2015, 5, 715. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.J.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Agriopoulou, S.; Stamatelopoulou, E.; Varzakas, T. Advances in Occurrence, Importance, and Mycotoxin Control Strategies: Prevention and Detoxification in Foods. Foods 2020, 9, 137. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef]
- Zacharof, M.P.; Lovitt, R.W. Bacteriocins produced by lactic acid bacteria. A review article. APCBEE Proc. 2012, 2, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaenhammer, T.R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- McAuliffe, O.R.; Ross, P.; Hill, C. Lantibiotics: Structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zendo, T.; Ohashi, C.; Maeno, S.; Piao, X.; Salminen, S.; Sonomoto, K.; Endo, A. Kunkecin A, a New Nisin Variant Bacteriocin Produced by the Fructophilic Lactic Acid Bacterium, Apilactobacillus kunkeei FF30-6 Isolated From Honey Bees. Front. Microbiol. 2020, 11, 571903. [Google Scholar] [CrossRef]
- Nissen-Meyer, J.; Holo, H.; Håvarstein, L.S.; Sletten, K.; Nes, I.F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J. Bacteriol. 1992, 174, 5686–5692. [Google Scholar] [CrossRef] [Green Version]
- Breukink, E.; Wiedemann, I.; van Kraaij, C.; Kuipers, O.P.; Sahl, H.G.; de Kruijff, B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef] [Green Version]
- Van Heusden, H.E.; de Kruijff, B.; Breukink, E. Lipid II induces a transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 2002, 41, 12171–12178. [Google Scholar] [CrossRef]
- Tagg, J.R.; Dajani, A.S.; Wannamaker, L.W. Bacteriocins of gram positive bacteria. Bacteriol. Rev. 1976, 40, 722–756. [Google Scholar] [CrossRef]
- Moll, G.N.; Konings, W.N.; Driessen, A.J. Bacteriocins: Mechanism of membrane insertion and pore formation. Antonie Van Leeuwenhoek 1999, 76, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, I.E.; Breukink, C.; van Kraaij, O.P.; Kuipers, G.; Bierbaum, B.; Bierbaum, G.; De Kruijff, B.; Sahl, H. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [Green Version]
- Parada, J.L.; Caron, C.R.; Medeiros, A.B.P.; Soccol, C.R. Bacteriocins from lactic acid bacteria: Purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 2007, 50, 521–542. [Google Scholar] [CrossRef] [Green Version]
- Schneck, E.; Schubert, T.; Konovalov, O.V.; Quinn, B.E.; Gutsmann, T.; Brandenburg, K.; Oliveira, R.G.; Pink, D.A.; Tanaka, M. Quantitative Determination of Ion Distributions in Bacterial Lipopolysaccharide Membranes by Grazing-Incidence X-ray Fluorescence. Proc. Natl. Acad. Sci. USA 2010, 107, 9147–9151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boziaris, I.; Adams, M. Effect of chelators and nisin produced in situ on inhibition and inactivation of Gram-negatives. Int. J. Food Microbiol. 1999, 53, 105–113. [Google Scholar] [CrossRef]
- Butler, É.; Oien, R.F.; Lindholm, C.; Olofsson, T.C.; Nilson, B.; Vásquez, A. A pilot study investigating lactic acid bacterial symbionts from the honeybee in inhibiting human chronic wound pathogens. Int. Wound J. 2016, 13, 729–737. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic acid bacterial symbionts in honeybees—An unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 2014, 13, 668–679. [Google Scholar] [CrossRef]
- Oscáriz, J.C.; Pisabarro, A.G. Classification and mode of action of membrane-active bacteriocins produced by gram-positive bacteria. Int. Microbiol. 2001, 4, 13–19. [Google Scholar] [CrossRef]
- Chen, H.; Hoover, D.G. Bacteriocins and their food applications. Compr. Rev. Food Sci. Food Saf. 2003, 2, 83–97. [Google Scholar]
- Gong, H.S.; Meng, X.C.; Wang, H. Mode of action of plantaricin MG, a bacteriocin active against Salmonella typhimurium. J. Basic Microbiol. 2010, 50, S37–S45. [Google Scholar] [CrossRef]
- Alegría, Á.; Delgado, S.; Roces, C.; López, B.; Mayo, B. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 2010, 143, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, L.R.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef]
- Carvalho, F.M.; Teixeira-Santos, R.; Mergulhão, F.J.M.; Gomes, L.C. The Use of Probiotics to Fight Biofilms in Medical Devices: A Systematic Review and Meta-Analysis. Microorganisms 2020, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Velraeds, M.M.; Van Der Mei, H.C.; Reid, G.; Busscher, H.J. Physicochemical and biochemical characterization of biosurfactants released by Lactobacillus strains. Colloids Surf. B Biointerfaces 1996, 8, 51–61. [Google Scholar] [CrossRef]
- Satpute, S.K.; Kulkarni, G.R.; Banpurkar, A.G.; Banat, I.M.; Mone, N.S.; Patil, R.H.; Cameotra, S.S. Biosurfactant/s from Lactobacilli species: Properties, challenges and potential biomedical applications. J. Basic Microbiol. 2016, 56, 1140–1158. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Fernandes, E.C.; Teixeira, J.A.; Rodrigues, L.R. Antimicrobial and anti-adhesive activities of cell-bound biosurfactant from Lactobacillus agilis CCUG31450. RSC Adv. 2015, 5, 90960–90968. [Google Scholar] [CrossRef] [Green Version]
- Barzegari, A.; Kheyrolahzadeh, K.; Khatibi, S.M.H.; Sharifi, S.; Memar, M.Y.; Vahed, Z.S. The Battle of Probiotics and Their Derivatives against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-H.; Tian, X. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Saharan, B.S. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Rep. 2016, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Tatsaporn, T.; Kornkanok, K. Using Potential Lactic Acid Bacteria Biofilms and their Compounds to Control Biofilms of Foodborne Pathogens. Biotechnol. Rep. 2020, 26, e00477. [Google Scholar] [CrossRef]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Ibarreche, M.; Castellano, P.; Leclercq, A.; Vignolo, G. Control of Listeria monocytogenes biofilms on industrial surfaces by the bacteriocin-producing Lactobacillus sakei CRL1862. FEMS Microbiol. Lett. 2016, 363, fnw118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Abriouel, H.; Franz, C.M.A.P.; Omar, N.B.; Gálvez, A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 2011, 35, 201–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Sumi, C.D.; Yang, B.W.; Yeo, I.C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef]
- Kawulka, K.; Sprules, T.; McKay, R.T.; Mercier, P.; Diaper, C.M.; Zuber, P.; Vederas, J.C. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 2003, 125, 4726–4727. [Google Scholar] [CrossRef] [PubMed]
- Balty, C.; Guillot, A.; Fradale, L.; Brewee, C.; Boulay, M.; Kubiak, X.; Benjdia, A.; Berteau, O. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J. Biol. Chem. 2019, 294, 14512–14525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favret, M.E.; Yousten, A.A. Thuricin: The bacteriocin produced by Bacillus thuringiensis. J. Invertebr. Pathol. 1989, 53, 206–216. [Google Scholar] [CrossRef]
- Lee, H.; Churey, J.J.; Worobo, R.W. Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol. Lett. 2009, 299, 205–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rea, M.C.; Sit, C.S.; Clayton, E.; O’Connor, P.M.; Whittal, R.M.; Zheng, J.; Vederas, J.C.; Ross, R.P.; Hill, C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. USA 2010, 107, 9352–9357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vollmer, W.; Joris, B.; Charlier, P.; Foster, S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008, 32, 259–286. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.J.; Blackman, S.A.; Foster, S.J. Autolysins of Bacillus subtilis: Multiple enzymes with multiple functions. Microbiology 2000, 146 Pt 2, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.; Uehara, T.; Bernhardt, T.G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 2014, 159, 1300–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storm, D.R.; Strominger, J.L. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 1973, 248, 3940–3945. [Google Scholar] [CrossRef]
- Kenig, M.; Abraham, E. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 1976, 94, 37–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef]
- Straight, P.D.; Fischbach, M.A.; Walsh, C.T.; Rudner, D.Z.; Kolter, R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 305–310. [Google Scholar] [CrossRef] [Green Version]
- Butcher, R.A.; Schroeder, F.C.; Fischbach, M.A.; Straight, P.D.; Kolter, R.; Walsh, C.T.; Clardy, J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 1506–1509. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.S.; Huang, S.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P.; Mayerl, F. Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: Production, taxonomy, isolation, physico-chemical characterization and biological activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [Green Version]
- Carrillo, C.; Teruel, J.A.; Aranda, F.J.; Ortiz, A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta 2003, 1611, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Deleu, M.; Lorent, J.; Lins, L.; Brasseur, R.; Braun, N.; El Kirat, K.; Nylander, T.; Dufrêne, Y.F.; Mingeot-Leclercq, M.P. Effects of surfactin on membrane models displaying lipid phase separation. Biochim. Biophys. Acta 2013, 1828, 801–815. [Google Scholar] [CrossRef] [Green Version]
- Vlamakis, H.; Chai, Y.; Beauregard, P.; Losick, R.; Kolter, R. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11, 157–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janek, T.; Drzymała, K.; Dobrowolski, A. In vitro efficacy of the lipopeptide biosurfactant surfactin-C15 and its complexes with divalent counterions to inhibit Candida albicans biofilm and hyphal formation. Biofouling 2020, 36, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Ceresa, C.; Rinaldi, M.; Chiono, V.; Carmagnola, I.; Allegrone, G.; Fracchia, L. Lipopeptides from Bacillus subtilis AC7 inhibit adhesion and biofilm formation of Candida albicans on silicone. Antonie Van Leeuwenhoek 2016, 109, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, J.D.; Jumarie, C.; Cooper, D.G.; Laprade, R. Ionic channels induced by surfactin in planar lipid bilayer membranes. Biochim. Biophys. Acta 1991, 1064, 13–23. [Google Scholar] [CrossRef]
- Zakharova, A.A.; Efimova, S.S.; Malev, V.V.; Ostroumova, O. Fengycin induces ion channels in lipid bilayers mimicking target fungal cell membranes. Sci. Rep. 2019, 9, 16034. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zhou, Z.J.; Han, Y.; Wang, Z.Z.; Fan, J.; Xiao, H.Z. Isolation and identification of antifungal peptides from Bacillus BH072, a novel bacterium isolated from honey. Microbiol. Res. 2013, 168, 598–606. [Google Scholar] [CrossRef]
- Miethke, M.; Klotz, O.; Linne, U.; May, J.J.; Beckering, C.L.; Marahiel, M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006, 61, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Doshi, H.V.; Thakur, M.C. Bacillus spp.: A Prolific Siderophore Producer. In Bacilli and Agrobiotechnology; Islam, M., Rahman, M., Pandey, P., Jha, C., Aeron, A., Eds.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Cendrowski, S.; MacArthur, W.; Hanna, P. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 2004, 51, 407–417. [Google Scholar]
- Hertlein, G.; Mülle, S.; Garcia-Gonzalez, E.; Poppinga, L.; Süssmuth, R.D.; Genersch, E. Production of the Catechol Type Siderophore Bacillibactin by the Honey Bee Pathogen Paenibacillus larvae. PLoS ONE 2014, 9, e108272. [Google Scholar] [CrossRef] [Green Version]
- Keller, A.; Brandel, A.; Becker, M.C.; Balles, R.; Abdelmohsen, U.R.; Ankenbrand, M.J.; Sickel, W. Wild bees and their nests host Paenibacillus bacteria with functional potential of avail. Microbiome 2018, 6, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katznelson, H. Bacillus apiarius, n. sp., an aerobic spore-forming organism isolated from honeybee larvae. J. Bacteriol. 1955, 70, 635–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genersch, E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010, 103, S10–S19. [Google Scholar] [CrossRef]
- Lee, H.; Churey, J.J.; Worobo, R.W. Isolation and characterization of a protective bacterial culture isolated from honey active against American Foulbrood disease. FEMS Microbiol. Lett. 2009, 296, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakonyi, T.; Derakhshifar, I.; Grabensteiner, E.; Nowotny, N. Development and evaluation of PCR assays for the detection of Paenibacillus larvae in honey samples: Comparison with isolation and biochemical characterization. Appl. Environ. Microbiol. 2003, 69, 1504–1510. [Google Scholar] [CrossRef] [Green Version]
- Huang, E.; Yousef, A.E. Biosynthesis of paenibacillin, a lantibiotic with N-terminal acetylation, by Paenibacillus polymyxa. Microbiol. Res. 2015, 181, 15–210. [Google Scholar] [CrossRef]
- Huang, E.; Yousef, A.E. The lipopeptide antibiotic paenibacterin binds to the bacterial outer membrane and exerts bactericidal activity through cytoplasmic membrane damage. Appl. Environ. Microbiol. 2014, 80, 2700–2704. [Google Scholar] [CrossRef] [Green Version]
- Mokhtar, N.F.K.; Hashim, A.M.; Hanish, I.; Zulkarnain, A.; Raja Nhari, R.M.H.; Abdul Sani, A.A.; Abbasiliasi, S.; Ariff, A.; Mustafa, S.; Rahim, R.A. The Discovery of New Antilisterial Proteins From Paenibacillus polymyxa Kp10 via Genome Mining and Mass Spectrometry. Front. Microbiol. 2020, 11, 960. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Hu, Y.; Shou, L.; Song, M. Isolation and partial characterization of cyclic lipopeptide antibiotics produced by Paenibacillus ehimensis B7. BMC Microbiol. 2013, 13, 87. [Google Scholar] [CrossRef] [Green Version]
- Pozo, M.I.; Lachanc, M.-A.; Herrera, C.M. Nectar yeasts of two southern Spanish plants: The roles of immigration and physiological traits in community assembly. FEMS Microbiol. Ecol. 2012, 80, 281–293. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.L.; Chi, Z.; Wang, G.Y.; Wang, Z.P.; Li, Y.; Chi, Z.M. Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 2015, 35, 222–234. [Google Scholar] [CrossRef]
- Magliani, W.; Conti, S.; Gerloni, M.; Bertolotti, D.; Polonelli, L. Yeast Killer Systems. Clin. Microbiol. Rev. 1997, 30, 369–400. [Google Scholar] [CrossRef]
- Niu, G.; Johnson, R.M.; Berenbaum, M.R. Toxicity of mycotoxins to honeybees and its amelioration by propolis. Apidologie 2011, 42, 79. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, N.; Keller, N.P. Mycotoxins in Conversation with Bacteria and Fungi. Front. Microbiol. 2019, 10, 403. [Google Scholar] [CrossRef]
- Pfliegler, W.P.; Pócsi, I.; Győri, Z.; Pusztahelyi, T. The Aspergilli and Their Mycotoxins: Metabolic Interactions with Plants and the Soil Biota. Front. Microbiol. 2020, 10, 2921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilliam, M.; Prest, D.B.; Lorenz, B.J. Microbiology of pollen and bee bread: Taxonomy and enzymology of molds. Apidologie 1989, 20, 53–68. [Google Scholar] [CrossRef] [Green Version]
- Hilldrup, J.A.; Eadie, T.; Llewellyn, G.C. Fungal Growth and Aflatoxin Production on Apiarian Substrates. J. AOAC 1977, 60, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Strohl, W.R. Antibiotics from filamentous fungi. In Biotechnology of Antibiotics; Strohl, W.R., Ed.; Marcel Dekker: New York, NY, USA, 1997; pp. 1–47. [Google Scholar]
- Laich, F.; Fierro, F.; Martín, J.F. Production of penicillin by fungi growing on food products: Identification of a complete penicillin gene cluster in Penicillium griseofulvum and a truncated cluster in Penicillium verrucosum. Appl. Environ. Microbiol. 2002, 68, 1211–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraszkiewicz, K.; Moryl, M.; Płaza, G.; Bhagat, D.; Satpute, S.K.; Bernat, P. Surfactants of microbial origin as antibiofilm agents. Int. J. Environ. Health Res. 2019, 31, 401–420. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Turnbull, L.; Burke, C.M.; Liu, M.; Carter, D.A.; Schlothauer, R.C.; Whitchurch, C.B.; Harry, E.J. Manuka-type honeys can eradicate biofilms produced by Staphylococcus aureus strains with different biofilm-forming abilities. PeerJ 2014, 2, e326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Cokcetin, N.N.; Burke, C.M.; Turnbull, L.; Liu, M.; Carter, D.; Whitchurch, C.B.; Harry, E.J. Honey can inhibit and eliminate biofilms produced by Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 18160. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis Related Proteins in Plant Defense Response. In Plant Defence: Biological Control. Progress in Biological Control; Mérillon, J., Ramawat, K., Eds.; Springer: Dordrecht, The Netherlands, 2011; Volume 12. [Google Scholar] [CrossRef]
- Garcia-Olmedo, F.; Molina, A.; Alamillo, J.M.; Rodriguez-Palenzuela, P. Plant defense peptides. Biopolymers 1998, 47, 479–491. [Google Scholar] [CrossRef]
- Carter, C.; Thornburg, R.W. Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 2004, 9, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Erban, T.; Shcherbachenko, E.; Talacko, P.; Harant, K. The Unique Protein Composition of Honey Revealed by Comprehensive Proteomic Analysis: Allergens, Venom-like Proteins, Antibacterial Properties, Royal Jelly Proteins, Serine Proteases, and Their Inhibitors. J. Nat. Prod. 2019, 82, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Rossano, R.; Larocca, M.; Polito, T.; Perna, A.M.; Padula, M.C.; Martelli, G.; Riccio, P. What Are the Proteolytic Enzymes of Honey and What They Do Tell Us? A Fingerprint Analysis by 2-D Zymography of Unifloral Honeys. PLoS ONE 2012, 7, e49164. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Casteels, P.; Ampe, C.; Jacobs, F.; Vaek, M.; Tempst, P. Apidaecins: Antibacterial peptides from honey bees. EMBO J. 1989, 8, 2387–2391. [Google Scholar] [CrossRef]
- Casteels, P.; Ampe, C.; Riviere, L.; Damme, J.V.; Elicone, C.; Fleming, M.; Jacobs, F.; Tempst, P. Isolation and characterization of abaecin, a major antibacterial peptide in the honey bees (Apis mellifera). Eur. J. Biochem. 1990, 187, 381–386. [Google Scholar] [CrossRef]
- Casteels-Josson, K.; Zhang, W.; Capaci, T.; Casteels, P.; Tempst, P. Acute transcriptional response of the honey bees peptide-antibiotics gene repertoire, required posttranslational conversion of the precursor structures. J. Biol. Chem. 1994, 269, 28569–28575. [Google Scholar] [CrossRef]
- Klaudiny, J.; Albert, S.; Bachanová, K.; Kopernický, J.; Šimúth, J. Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera. Insect Biochem. Mol. Biol. 2005, 35, 11–22. [Google Scholar] [CrossRef]
- Casteels, P.; Ampe, C.; Jacobs, F.; Tempst, P. Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infection inducible in the honey bees (Apis mellifera). J. Biol. Chem. 1993, 268, 7044–7054. [Google Scholar] [CrossRef]
- Di Girolamo, F.; D’Amato, A.; Righetti, P.G.J. Assessment of the floral origin of honey via proteomic tools. Proteomics 2012, 75, 3688–3693. [Google Scholar] [CrossRef]
- Kwakman, P.H.S.; te Velde, A.A.; de Boer, L.; Speijer, D.; Vandenbroucke-Grauls, C.M.J.E.; Zaat, S.A.J. How honey kills bacteria. FASEB J. 2010, 24, 2576–2582. [Google Scholar] [CrossRef] [Green Version]
- Qu, N.; Jiang, J.; Sun, L.; Lai, C.; Sun, L.; Wu, X. Proteomic Characterization of Royal Jelly Proteins in Chinese (Apis cerana cerana), European (Apis mellifera) Honey bees. Biochemistry 2008, 1, 1–12. [Google Scholar]
- Fujiwara, S.; Imai, J.; Fujiwara, M.; Yaeshima, T.; Kawashima, T.; Kobayashi, K. A potent antibacterial protein in royal jelly. Purification and determination of the primary structure of royalisin. J. Biol. Chem. 1990, 265, 11333–11337. [Google Scholar] [CrossRef]
- Bilikova, K.; Gusui, W.; Simuth, J. Isolation of a peptide fraction from honey bees royal jelly as a potential antifoulbrood factor. Apidologie 2001, 32, 275–283. [Google Scholar] [CrossRef] [Green Version]
- Bachanova, K.; Klaudiny, J.; Kopernicky, J.; Simuth, J. Identification of honeybee peptide active against Paenibacillus larvae larvae through bacterial growth-inhibition assay on polyacrylamide gel. Apidologie 2002, 33, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Shi, M.; Chen, X.X. Antimicrobial peptide evolution in the Asiatic honey bee Apis cerana. PLoS ONE 2009, 4, e4239. [Google Scholar] [CrossRef] [Green Version]
- Brudzynski, K.; Sjaarda, C. Honey Glycoproteins Containing Antimicrobial Peptides, Jelleins of the Major Royal Jelly Protein 1, Are Responsible for the Cell Wall Lytic and Bactericidal Activities of Honey. PLoS ONE 2015, 10, e0120238. [Google Scholar] [CrossRef] [Green Version]
- Fontana, R.; Mendes, M.A.; Monson de Souza, B.; Konno, K.; Cesar, L.M.M.; Malaspina, O.; Palma, M.S. Jelleines: A family of antimicrobial peptides from the Royal Jelly of honeybees (Apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef]
- Brudzynski, K.; Sjaarda, C.; Lannigan, R. MRJP1-containing glycoproteins isolated from honey, a novel antibacterial drug candidate with broad spectrum activity against multi-drug resistant clinical isolates. Front. Microbiol. 2015, 6, 711. [Google Scholar] [CrossRef] [Green Version]
- Brudzynski, K.; Sjaarda, C. Colloidal structure of honey and its influence on antibacterial activity. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2063–2080. [Google Scholar] [CrossRef] [PubMed]

| Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|
| Proteobacteria | Alphaproteobacteria | Rhodospirillales | Acetobacteraceae | Gluconobacter |
| Gammaproteobacteria | Pseudomonadales | Psedomonadaceae | Pseudomonas | |
| Enerobacteriales | Enetrobacteriacea | Enterobacter | ||
| Escherichia | ||||
| Klebsiella | ||||
| Actinobacteria | Actinomycetales | Micrococcaceae | Micrococcus | |
| Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | ||
| Microbacteriaceae | Microbacterium | |||
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| Paenibacillaceae | Paenibacillus | |||
| Staphylococcacea | Staphylococcus | |||
| Lactobacillales | Lactobacillaceae | Lactobacillus | ||
| Leuconostocacea | Fructobacillus | |||
| Leuconstoc | ||||
| Oenococcus | ||||
| Weisella | ||||
| Streptococcaceae | Lactococcus | |||
| Streptococcus | ||||
| Enterococcacea | Melissococcus | |||
| Clostridiales | Clostridiaceae | Clostridium |
| Order | Family | Genus | Species |
|---|---|---|---|
| Lactobacillales | Lactobacillaceae | Lactobacillus | L. acidophilus |
| L. apis | |||
| L. apinorum | |||
| L.jensenii | |||
| L. brevis | |||
| L. florum | |||
| L. helsingborgensis | |||
| L. johnsonii | |||
| L. kefiranofaciens | |||
| L. kimbladii | |||
| L. kullabergensis | |||
| L. mellifer | |||
| L. mellis | |||
| L. melliventris | |||
| L. kunkeei | |||
| L. plantarum | |||
| L. rossiae | |||
| L. versmoldensis | |||
| Leuconostocaceae | Fructobacillus | F. fructosus | |
| Leuconostoc | |||
| Oenococcus | |||
| Weissella | |||
| Pediococcaceae | Pediococcus | ||
| Streptococcaceae | Streptococcus | ||
| Enterococcaceae | Enterococcus | E. faecalis | |
| E.faecium | |||
| Aerococcus | |||
| Carnobacterium |
| Family | Genus | Species |
|---|---|---|
| Bacillaceae | Bacillus | B. subtilis |
| B.methylotrophicus | ||
| B. atrophaeus | ||
| B. licheniformis | ||
| B. amyloliquefaciens | ||
| B. cereus | ||
| B. thuringiensis | ||
| B. mycoides | ||
| B. pseudomycoides | ||
| B. weihenstephanen | ||
| B. pumilis | ||
| B. safensis | ||
| B. altitudinis | ||
| B. mojavensis | ||
| B. anthracis | ||
| B. aerius | ||
| B. xiamenensis | ||
| B. wiedmannii | ||
| B. proteolyticus | ||
| B. tropicus | ||
| B. circulans | ||
| B. flexus | ||
| B. zhangzhouensis | ||
| Lysinibacillus | L. fusiformis | |
| L. macroides | ||
| L. pakistanensis | ||
| L. boronitolerans | ||
| Oceanobacillus | ||
| Paenibacillaceae | Paenibacillus | P. alvei |
| P. larvae | ||
| P. polymyxa | ||
| P. apiarius | ||
| Brevibacillus | B. brevis | |
| B. limnophilus | ||
| Listeriaceae | L. monocytogenes | |
| Staphylococcaceae | S. epidermidis | |
| S. caprae | ||
| S. pasteuri |
| Division | Class | Oder | Family | Genus | Species |
|---|---|---|---|---|---|
| Ascomycota | Eurotiomycetes | Eurotiales | Trichocomaceae | Aspergillus | A. pseudoglaucus |
| A. asperescens | |||||
| A. montevidensis | |||||
| A. flavus | |||||
| A. versicolor | |||||
| A. niger | |||||
| A. fumigatus | |||||
| Penicillium | P. camemberti | ||||
| P. citrinum | |||||
| P. corylophilum | |||||
| P. cravenianum | |||||
| P. apimei | |||||
| Talaromyces | |||||
| Monascaceae | Monascus | M. pilosus | |||
| M. mellicola | |||||
| M. purpureus | |||||
| M. ruber | |||||
| Ascosphaerales | Ascosphaeracea | Bettsia | B. alvei | ||
| Ascosphaera apis | |||||
| Onygenales | Myxotrichaceae | Skoua | Skoua fertilis | ||
| Oidiodendron | |||||
| Eremascaceae | Eremascus | Ermascus albus | |||
| Ascosphaeriacea | Ascosphaera | Ascosphaera atra | |||
| Ascosphaera apis | |||||
| Spiromastigaceae | |||||
| Schizosaccharomy-cetales | Schizosaccharomyce-taceae | Schizosaccharomyces | S. octosporus | ||
| Saccharomycetes | Saccharomycetales | Saccharomycetaceae | Zygosaccharomyces | Z. favi | |
| Z. mellis | |||||
| Z. richteri | |||||
| Z. rouxii | |||||
| Z. siamensis | |||||
| Candida | C. lundiana | ||||
| C. magnoliae | |||||
| C. sorbosivorans | |||||
| C. suthepensis | |||||
| Saccharomyces | S. cerevisiae | ||||
| Cyberlindnera | C. jadinii (Torula) | ||||
| Starmerella | |||||
| Metschnikowiaceae | Metschnikowia | ||||
| Dothideomycetes | Capnodiales | Davidiellaceae | Cladoisporium | ||
| Pleosporales | Pleosporaceae | Alternaria | A. multiformis | ||
| Stemphylium | |||||
| Sordariomycetes | Hypocreales | Nectriaceae | Fusarium | ||
| Mucoromy-cota | Mucorales | Mucor | M. ruber | ||
| M. plumbeus |
| Species | Bacteriocins | Target | Ref. |
|---|---|---|---|
| L. acidophilus | acidocin | Lactobacillus sp. Listeria monocytogenes Enterococcus faecalis | [85,90] |
| lactacins | Lactobacillus fermentum Enterococcus faecalis Lactobacillus delbrueckii Lactobacillus helveticus Lactobacillus debrweckii Lactobacillus helveticus Lactobacillus.bulgaricus. Lactococcus lactis. | [90] | |
| L. helveticus | helveticin J | Lactobacillus Lactobacillus bulgaricus Lactococcus lactis | [85,91] |
| lactocin 27 | [90] | ||
| L. johnsonii | lactacin F | [77] | |
| L. kunkeei | kunkicin | ||
| L. plantarum | plantaricin | Listeria monocytogenes Staphylococcus aureus, Salmonella typhimurium and Escherichia coli | [92] |
| Bacillus cereus, B. pumilus, B. megaterium, Pediococcus, Carnobacteria, Clostiridia and Propionobacteria | |||
| L. lactis | nisin | Staphylococcus aureus, Listeria innocua, Lactobacillus sakei, Lactobacillus plantarum, Bacillus spp. Micrococcus spp. Clostridium spp. | [85,93] |
| lacticin 3147 | Clostridium sp. Listeria monocytogenes Staphylococcus aureus MRSA VRE Enterococcus faecalis Propionibacterium acne Streptococcus mutans | [85] | |
| Pedicoccus pentosaceus | pediocin | Listeria monocytogenes Lactobacillus Lactococcus Leuconostoc Pediococcus Staphylococcus Enterococcus Listeria Clostridium | [63,85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brudzynski, K. Honey as an Ecological Reservoir of Antibacterial Compounds Produced by Antagonistic Microbial Interactions in Plant Nectars, Honey and Honey Bee. Antibiotics 2021, 10, 551. https://doi.org/10.3390/antibiotics10050551
Brudzynski K. Honey as an Ecological Reservoir of Antibacterial Compounds Produced by Antagonistic Microbial Interactions in Plant Nectars, Honey and Honey Bee. Antibiotics. 2021; 10(5):551. https://doi.org/10.3390/antibiotics10050551
Chicago/Turabian StyleBrudzynski, Katrina. 2021. "Honey as an Ecological Reservoir of Antibacterial Compounds Produced by Antagonistic Microbial Interactions in Plant Nectars, Honey and Honey Bee" Antibiotics 10, no. 5: 551. https://doi.org/10.3390/antibiotics10050551
APA StyleBrudzynski, K. (2021). Honey as an Ecological Reservoir of Antibacterial Compounds Produced by Antagonistic Microbial Interactions in Plant Nectars, Honey and Honey Bee. Antibiotics, 10(5), 551. https://doi.org/10.3390/antibiotics10050551






