Technologies for High-Throughput Identification of Antibiotic Mechanism of Action
Abstract
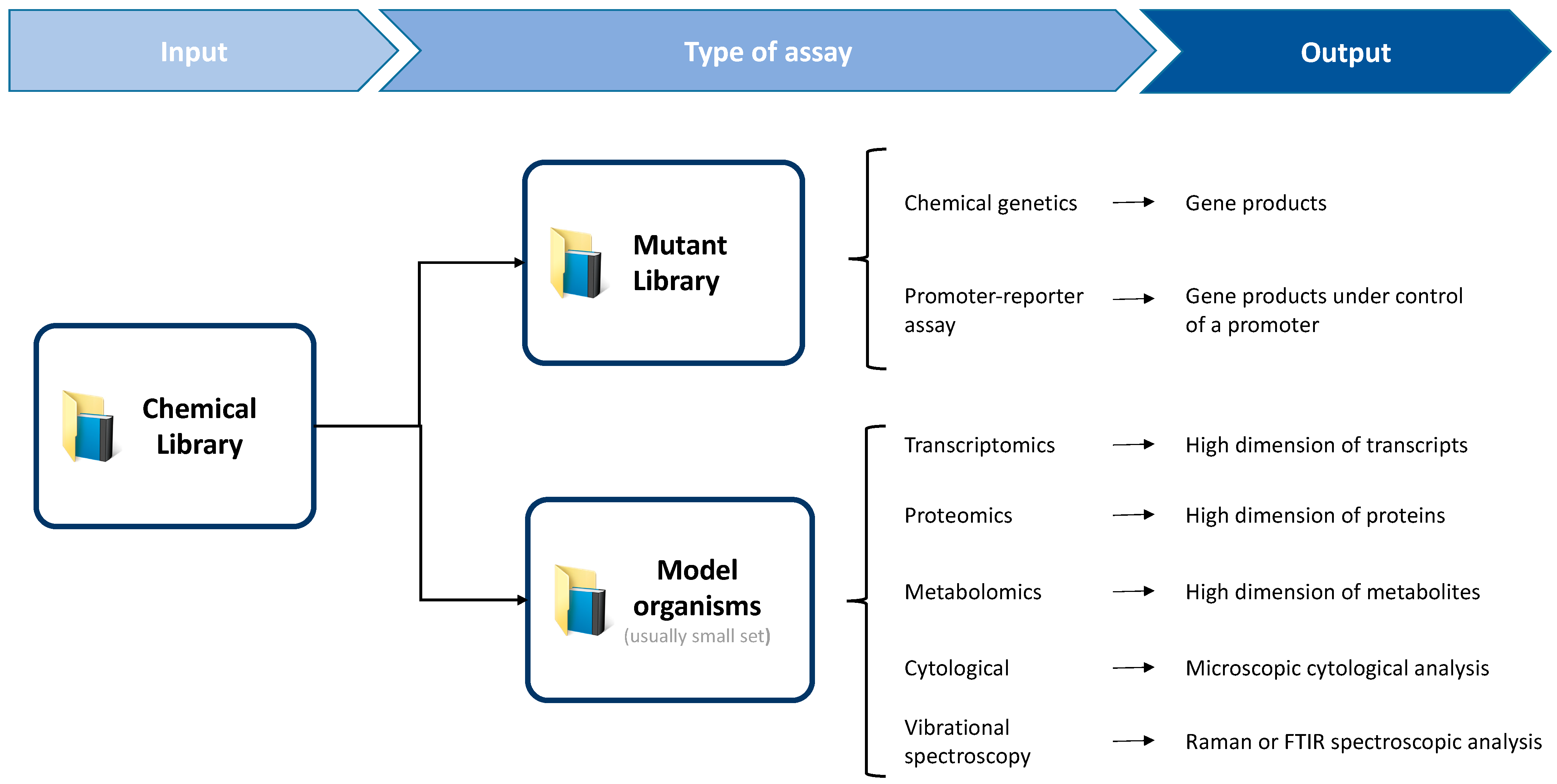
1. Introduction
2. Chemical Genetics
2.1. Overexpression Libraries
2.2. Knockout and Knockdown Collections
3. Promoter-Reporter Libraries
4. Transcriptomics
4.1. Hybridization Assays
4.2. The Uprising of Next-Generation Sequencing
5. Proteomics
5.1. Gel-Based Assays
5.2. Gel-Free Methods
6. Metabolomics
6.1. Nuclear Magnetic Resonance Spectroscopy
6.2. Mass Spectrometry-Based Methods
7. Bacterial Cytological Profiling
8. Vibrational Spectroscopy
8.1. Raman Scattering
8.2. Fourier-Transform Infrared Spectroscopy
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Mol, M.L.; Snoeck, N.; De Maeseneire, S.L.; Soetaert, W.K. Hidden antibiotics: Where to uncover? Biotechnol. Adv. 2018, 36, 2201–2218. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Funabashi, M.; Ogura, Y. Target deconvolution from phenotype-based drug discovery by using chemical proteomics approaches. Biochim. Biophys. Acta-Proteins Proteomics 2019, 1867, 22–27. [Google Scholar] [CrossRef]
- Ohki, Y.; Sakurai, H.; Hoshino, M.; Terashima, H.; Shimizu, H.; Ishikawa, T.; Ogiyama, T.; Muramatsu, Y.; Nakanishi, T.; Miyazaki, S.; et al. Perturbation-Based Proteomic Correlation Profiling as a Target Deconvolution Methodology. Cell Chem. Biol. 2019, 26, 137–143. [Google Scholar] [CrossRef]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of kibdelomycin, a potent new class of bacterial type II topoisomerase inhibitor by chemical-genetic profiling in Staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef]
- Kurita, K.L.; Glassey, E.; Linington, R.G. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc. Natl. Acad. Sci. USA 2015, 112, 11999–12004. [Google Scholar] [CrossRef]
- Birkenstock, T.; Liebeke, M.; Winstel, V.; Krismer, B.; Gekeler, C.; Niemiec, M.J.; Bisswanger, H.; Lalk, M.; Peschel, A. Exometabolome analysis identifies pyruvate dehydrogenase as a target for the antibiotic triphenylbismuthdichloride in multiresistant bacterial pathogens. J. Biol. Chem. 2012, 287, 2887–2895. [Google Scholar] [CrossRef]
- Cho, H.; Uehara, T.; Bernhardt, T.G. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 2014, 159, 1300–1311. [Google Scholar] [CrossRef]
- French, S.; Ellis, M.J.; Coutts, B.E.; Brown, E.D. Chemical genomics reveals mechanistic hypotheses for uncharacterized bioactive molecules in bacteria. Curr. Opin. Microbiol. 2017, 39, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.L.; Kwan, B.P.; Nelson, K.J.; Bensen, D.C.; Shaw, K.J. Distinguishing on-target versus off-target activity in early antibacterial drug discovery using a macromolecular synthesis assay. J. Biomol. Screen. 2013, 18, 1018–1026. [Google Scholar] [CrossRef]
- Bantscheff, M.; Drewes, G. Chemoproteomic approaches to drug target identification and drug profiling. Bioorganic Med. Chem. 2012, 20, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
- Zoffmann, S.; Vercruysse, M.; Benmansour, F.; Maunz, A.; Wolf, L.; Blum Marti, R.; Heckel, T.; Ding, H.; Truong, H.H.; Prummer, M.; et al. Machine learning-powered antibiotics phenotypic drug discovery. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Nonejuie, P.; Burkart, M.; Pogliano, K.; Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. USA 2013, 110, 16169–16174. [Google Scholar] [CrossRef]
- Sato, S.-i.; Murata, A.; Shirakawa, T.; Uesugi, M. Biochemical Target Isolation for Novices: Affinity-Based Strategies. Chem. Biol. 2010, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Nishiya, Y.; Hamada, T.; Abe, M.; Takashima, M.; Tsutsumi, K.; Okawa, K. A new efficient method of generating photoaffinity beads for drug target identification. Bioorganic Med. Chem. Lett. 2017, 27, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Burdine, L.; Thomas, K. Target Identification in Chemical Genetics: The (Often) Missing Link. Chem. Biol. 2004, 11, 593–597. [Google Scholar] [CrossRef]
- Zampieri, M.; Sekar, K.; Zamboni, N.; Sauer, U. Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 2017, 36, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Cacace, E.; Kritikos, G.; Typas, A. Chemical genetics in drug discovery. Curr. Opin. Syst. Biol. 2017, 4, 35–42. [Google Scholar] [CrossRef]
- Barker, C.A.; Farha, M.A.; Brown, E.D. Chemical Genomic Approaches to Study Model Microbes. Chem. Biol. 2010, 17, 624–632. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Zolli-Juran, M.; Cechetto, J.D.; Daigle, D.M.; Wright, G.D.; Brown, E.D. Multicopy Suppressors for Novel Antibacterial Compounds Reveal Targets and Drug Efflux Susceptibility. Chem. Biol. 2004, 11, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Zlitni, S.; Barker, C.; Das, R.; Gerritsma, D.A.; Lebert, J.; Awuah, E.; Melacini, G.; Capretta, F.A.; Brown, E.D. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 2009, 5, 849. [Google Scholar] [CrossRef]
- Barker, C.A.; Allison, S.E.; Zlitni, S.; Nguyen, N.D.; Das, R.; Melacini, G.; Capretta, A.A.; Brown, E.D. Degradation of MAC13243 and studies of the interaction of resulting thiourea compounds with the lipoprotein targeting chaperone LolA. Bioorganic Med. Chem. Lett. 2013, 23, 2426–2431. [Google Scholar] [CrossRef] [PubMed]
- Muheim, C.; Götzke, H.; Eriksson, A.U.; Lindberg, S.; Lauritsen, I.; Nørholm, M.H.H.; Daley, D.O. Increasing the permeability of Escherichia coli using MAC13243. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Nickerson, N.N.; Jao, C.C.; Xu, Y.; Quinn, J.; Skippington, E.; Alexander, M.K.; Miu, A.; Skelton, N.; Hankins, J.V.; Lopez, M.S.; et al. A Novel Inhibitor of the LolCDE ABC Transporter Essential for Lipoprotein Trafficking in Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2018, 62, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Liu, A.; Tran, L.; Becket, E.; Lee, K.; Chinn, L.; Park, E.; Tran, K.; Miller, J.H. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic bar code. Antimicrob. Agents Chemother. 2010, 54, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Sen, S.; Choo, Y.J.; Beltrao, P.; Zietek, M.; Chaba, R.; Lee, S.; Kazmierczak, K.M.; Lee, K.J.; Wong, A.; et al. Phenotypic landscape of a bacterial cell. Cell 2011, 144, 143–156. [Google Scholar] [CrossRef]
- Côté, J.-P.; French, S.; Gehrke, S.S.; MacNair, C.R.; Mangat, C.S.; Bharat, A.; Brown, E.D. The Genome-Wide Interaction Network of Nutrient Stress Genes in Escherichia coli. MBio 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Shiver, A.L.; Osadnik, H.; Kritikos, G.; Li, B.; Krogan, N.; Typas, A.; Gross, C.A. A Chemical-Genomic Screen of Neglected Antibiotics Reveals Illicit Transport of Kasugamycin and Blasticidin S. PLoS Genet. 2016, 12, 1–19. [Google Scholar] [CrossRef]
- Stokes, J.M.; French, S.; Ovchinnikova, O.G.; Bouwman, C.; Whitfield, C.; Brown, E.D. Cold Stress Makes Escherichia coli Susceptible to Glycopeptide Antibiotics by Altering Outer Membrane Integrity. Cell Chem. Biol. 2016, 23, 267–277. [Google Scholar] [CrossRef]
- Deutschbauer, A.M.; Jaramillo, D.F.; Proctor, M.; Kumm, J.; Hillenmeyer, M.E.; Davis, R.W.; Nislow, C.; Giaever, G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 2005, 169, 1915–1925. [Google Scholar] [CrossRef]
- DeVito, J.A.; Mills, J.A.; Liu, V.G.; Agarwal, A.; Sizemore, C.F.; Yao, Z.; Stoughton, D.M.; Cappiello, M.G.; Barbosa, M.D.F.S.; Foster, L.A.; et al. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 2002, 20, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.J.; Miller, J.H.; Suzuki, D.T.; Lewontin, R.C.; Gelbart, W.M. An Introduction to Genetic Analysis, 7th ed.; W. H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Donald, R.G.K.; Skwish, S.; Forsyth, R.A.; Anderson, J.W.; Zhong, T.; Burns, C.; Lee, S.; Meng, X.; LoCastro, L.; Jarantow, L.W.; et al. A Staphylococcus aureus Fitness Test Platform for Mechanism-Based Profiling of Antibacterial Compounds. Chem. Biol. 2009, 16, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, R.A.; Haselbeck, R.J.; Ohlsen, K.L.; Yamamoto, R.T.; Xu, H.; Trawick, J.D.; Wall, D.; Wang, L.; Brown-driver, V.; Froelich, J.M.; et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002, 43, 1387–1400. [Google Scholar] [CrossRef]
- Peters, J.M.; Colavin, A.; Shi, H.; Czarny, T.L.; Larson, M.H.; Wong, S.; Hawkins, J.S.; Lu, C.H.S.; Koo, B.M.; Marta, E.; et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 2016, 165, 1493–1506. [Google Scholar] [CrossRef]
- Liu, X.; Gallay, C.; Kjos, M.; Domenech, A.; Slager, J.; van Kessel, S.P.; Knoops, K.; Sorg, R.A.; Zhang, J.; Veening, J. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 2017, 13, 931. [Google Scholar] [CrossRef]
- Elad, T.; Seo, H.B.; Belkin, S.; Gu, M.B. High-throughput prescreening of pharmaceuticals using a genome-wide bacterial bioreporter array. Biosens. Bioelectron. 2015, 68, 699–704. [Google Scholar] [CrossRef]
- Zaslaver, A.; Bren, A.; Ronen, M.; Itzkovitz, S.; Kikoin, I.; Shavit, S.; Liebermeister, W.; Surette, M.G.; Alon, U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 2006, 3, 623–628. [Google Scholar] [CrossRef]
- Bollenbach, T.; Quan, S.; Chait, R.; Kishony, R. Nonoptimal Microbial Response to Antibiotics Underlies Suppressive Drug Interactions. Cell 2009, 139, 707–718. [Google Scholar] [CrossRef]
- Nagaraj, N.S.; Singh, O.V. Using genomics to develop novel antibacterial therapeutics. Crit. Rev. Microbiol. 2010, 36, 340–348. [Google Scholar] [CrossRef]
- Miller, M.B.; Tang, Y.W. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 2009, 22, 611–633. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, H.I.M.; Myers, T.G.; Copp, B.R.; McNeil, M.R.; Wilson, M.A.; Barry, C.E. The Transcriptional Responses of Mycobacterium tuberculosis to Inhibitors of Metabolism. J. Biol. Chem. 2004, 279, 40174–40184. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Tang, X.; Guo, N.; Zhang, K.; Guo, A.; Wu, X.; Wang, X.; Guan, Z.; Liu, L.; Shen, F.; et al. Genome-wide expression profiling of the response to linezolid in mycobacterium tuberculosis. Curr. Microbiol. 2012, 64, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Bonn, F.; Pané-Farré, J.; Schlüter, R.; Schaffer, M.; Fuchs, S.; Bernhardt, J.; Riedel, K.; Otto, A.; Völker, U.; van Dijl, J.M.; et al. Global analysis of the impact of linezolid onto virulence factor production in S. aureus USA300. Int. J. Med. Microbiol. 2016, 306, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Eklund, A.C.; Szallasi, Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006, 22, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333. [Google Scholar] [CrossRef]
- Hua, X.; Chen, Q.; Li, X.; Yu, Y. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int. J. Antimicrob. Agents 2014, 44, 337–344. [Google Scholar] [CrossRef]
- Wecke, T.; Mascher, T. Antibiotic research in the age of omics: From expression profiles to interspecies communication. J. Antimicrob. Chemother. 2011, 66, 2689–2704. [Google Scholar] [CrossRef]
- Briffotaux, J.; Liu, S.; Gicquel, B. Genome-wide transcriptional responses of Mycobacterium to antibiotics. Front. Microbiol. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Beaume, M.; Harrison, P.F.; Hernandez, D.; Schrenzel, J.; Seemann, T.; Francois, P.; Stinear, T.P. Analysis of the Small RNA Transcriptional Response in Multidrug-Resistant Staphylococcus aureus after Antimicrobial Exposure. Antimicrob. Agents Chemother. 2013, 57, 3864–3874. [Google Scholar] [CrossRef] [PubMed]
- Molina-Santiago, C.; Daddaoua, A.; Gómez-Lozano, M.; Udaondo, Z.; Molin, S.; Ramos, J.L. Differential transcriptional response to antibiotics by Pseudomonas putidaDOT-T1E. Environ. Microbiol. 2015, 17, 3251–3262. [Google Scholar] [CrossRef]
- Hébert, F.O.; Boyle, B.; Levesque, R.C. Direct In Vivo Microbial Transcriptomics During Infection. Trends Microbiol. 2018, 26, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Frantzi, M.; Latosinska, A.; Mischak, H. Proteomics in Drug Development: The Dawn of a New Era? Proteom.-Clin. Appl. 2019, 13, 1–13. [Google Scholar] [CrossRef]
- dos Santos, B.S.; da Silva, L.C.N.; da Silva, T.D.; Rodrigues, J.F.S.; Grisotto, M.A.G.; Correia, M.T.d.S.; Napoleão, T.H.; da Silva, M.V.; Paiva, P.M.G. Application of omics technologies for evaluation of antibacterial mechanisms of action of plant-derived products. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Wu, R.; Jiang, D.; Bai, B.; Tan, D.; Yan, T.; Sun, X.; Zhang, Q.; Wu, Z. Proteomic Analysis of the Antibacterial Mechanism of Action of Juglone against Staphylococcus aureus. Nat. Prod. Commun. 2016, 11, 825–827. [Google Scholar] [CrossRef]
- Bandow, J.E.; Brötz, H.; Leichert, L.I.O.; Labischinski, H.; Hecker, M. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 2003, 47, 948–955. [Google Scholar] [CrossRef]
- Kim, W.; Hendricks, G.L.; Tori, K.; Fuchs, B.B.; Mylonakis, E. Strategies against methicillin-resistant Staphylococcus aureus persisters. Future Med. Chem. 2018, 10, 779–794. [Google Scholar] [CrossRef]
- Wenzel, M.; Kohl, B.; Münch, D.; Raatschen, N.; Albada, H.B.; Hamoen, L.; Metzler-Nolte, N.; Sahl, H.-G.; Bandow, J.E. Proteomic Response of Bacillus subtilis to Lantibiotics Reflects Differences in Interaction with the Cytoplasmic Membrane. Antimicrob. Agents Chemother. 2012, 56, 5749–5757. [Google Scholar] [CrossRef]
- Maaß, S.; Otto, A.; Albrecht, D.; Riedel, K.; Trautwein-Schult, A.; Becher, D. Proteomic Signatures of Clostridium difficile Stressed with Metronidazole, Vancomycin, or Fidaxomicin. Cells 2018, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Brötz-Oesterhelt, H.; Bandow, J.E.; Labischinski, H. Bacterial proteomics and its role in antibacterial drug discovery. Mass Spectrom. Rev. 2005, 24, 549–565. [Google Scholar] [CrossRef]
- Koehler, C.J.; Strozynski, M.; Kozielski, F.; Treumann, A.; Thiede, B. Isobaric peptide termini labeling for MS/MS-based quantitative proteomics. J. Proteome Res. 2009, 8, 4333–4341. [Google Scholar] [CrossRef]
- Bachor, R.; Waliczek, M.; Stefanowicz, P.; Szewczuk, Z. Trends in the design of new isobaric labeling reagents for quantitative proteomics. Molecules 2019, 24, 701. [Google Scholar] [CrossRef]
- Evans, C.; Noirel, J.; Ow, S.Y.; Salim, M.; Pereira-Medrano, A.G.; Couto, N.; Pandhal, J.; Smith, D.; Pham, T.K.; Karunakaran, E.; et al. An insight into iTRAQ: Where do we stand now? Anal. Bioanal. Chem. 2012, 404, 1011–1027. [Google Scholar] [CrossRef]
- Olsen, J.V.; Mann, M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 2013, 12, 3444–3452. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, D.; Wang, X.; Ma, W.; Deng, S.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Proteomics progresses in microbial physiology and clinical antimicrobial therapy. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, D.; Li, G.; Liu, J.; He, G.; Zhang, P.; Yang, L.; Zhu, H.; Xu, N.; Liang, S. Antibacterial mechanism of daptomycin antibiotic against Staphylococcus aureus based on a quantitative bacterial proteome analysis. J. Proteomics 2017, 150, 242–251. [Google Scholar] [CrossRef]
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, H.; Lin, W.; Lin, X. Quantitative proteomic analysis of Edwardsiella tarda in response to oxytetracycline stress in biofilm. J. Proteomics 2017, 150, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Kamath, K.S.; Pascovici, D.; Penesyan, A.; Goel, A.; Venkatakrishnan, V.; Paulsen, I.T.; Packer, N.H.; Molloy, M.P. Pseudomonas aeruginosa Cell Membrane Protein Expression from Phenotypically Diverse Cystic Fibrosis Isolates Demonstrates Host-Specific Adaptations. J. Proteome Res. 2016, 15, 2152–2163. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Gil-Marqués, M.L.; McConnell, M.J. Identifying targets for antibiotic development using omics technologies. Drug Discov. Today 2016, 21, 465–472. [Google Scholar] [CrossRef]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Kozlowska, J.; Vermeer, L.S.; Rogers, G.B.; Rehnnuma, N.; Amos, S.B.T.A.; Koller, G.; McArthur, M.; Bruce, K.D.; Mason, A.J. Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge in Escherichia coli. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Hoerr, V.; Duggan, G.E.; Zbytnuik, L.; Poon, K.K.H.; Große, C.; Neugebauer, U.; Methling, K.; Löffler, B.; Vogel, H.J. Characterization and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 2016, 16, 1–14. [Google Scholar] [CrossRef]
- Fang, M.; Ivanisevic, J.; Benton, H.P.; Johnson, C.H.; Patti, G.J.; Hoang, L.T.; Uritboonthai, W.; Kurczy, M.E.; Siuzdak, G. Thermal Degradation of Small Molecules: A Global Metabolomic Investigation. Anal. Chem. 2015, 87, 10935–10941. [Google Scholar] [CrossRef]
- Schelli, K.; Zhong, F.; Zhu, J. Comparative metabolomics revealing Staphylococcus aureus metabolic response to different antibiotics. Microb. Biotechnol. 2017, 10, 1764–1774. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Szappanos, B.; Buchieri, M.V.; Trauner, A.; Piazza, I.; Picotti, P.; Gagneux, S.; Borrell, S.; Gicquel, B.; Lelievre, J.; et al. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci. Transl. Med. 2018, 10, 1–12. [Google Scholar] [CrossRef]
- Ang, M.L.T.; Pethe, K. Contribution of high-content imaging technologies to the development of anti-infective drugs. Cytom. Part A 2016, 89, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, K.A.; DeBiasio, R.L.; Dunlay, R.T.; Gough, A.; Volosky, J.M.; Zock, J.; Pavlakis, G.N.; Taylor, D.L. High-Content Screening: A New Approach to Easing Key Bottlenecks in the Drug Discovery Process. J. Biomol. Screen. 1997, 2, 249–259. [Google Scholar] [CrossRef]
- Peach, K.C.; Bray, W.M.; Winslow, D.; Linington, P.F.; Linington, R.G. Mechanism of action-based classification of antibiotics using high-content bacterial image analysis. Mol. Biosyst. 2013, 9, 1837–1848. [Google Scholar] [CrossRef]
- Schulze, C.J.; Bray, W.M.; Woerhmann, M.H.; Stuart, J.; Lokey, R.S.; Linington, R.G. “function-first” lead discovery: Mode of action profiling of natural product libraries using image-based screening. Chem. Biol. 2013, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Yang, Y.L.; Xu, Y.; Lamsa, A.; Haste, N.M.; Yang, J.Y.; Ng, J.; Gonzalez, D.; Ellermeier, C.D.; Straight, P.D.; et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2010, 107, 16286–16290. [Google Scholar] [CrossRef]
- Lamsa, A.; Liu, W.T.; Dorrestein, P.C.; Pogliano, K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 2012, 84, 486–500. [Google Scholar] [CrossRef]
- Quach, D.T.; Sakoulas, G.; Nizet, V.; Pogliano, J.; Pogliano, K. Bacterial Cytological Profiling (BCP) as a Rapid and Accurate Antimicrobial Susceptibility Testing Method for Staphylococcus aureus. EBioMedicine 2016, 4, 95–103. [Google Scholar] [CrossRef]
- McLeod, S.M.; Fleming, P.R.; MacCormack, K.; McLaughlin, R.E.; Whiteaker, J.D.; Narita, S.; Mori, M.; Tokuda, H.; Miller, A.A. Small-Molecule Inhibitors of Gram-Negative Lipoprotein Trafficking Discovered by Phenotypic Screening. J. Bacteriol. 2015, 197, 1075–1082. [Google Scholar] [CrossRef]
- Marques, V.; Cunha, B.; Couto, A.; Sampaio, P.; Fonseca, L.P.; Aleixo, S.; Calado, C.R.C. Characterization of gastric cells infection by diverse Helicobacter pylori strains through Fourier-transform infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 210, 193–202. [Google Scholar] [CrossRef]
- López-Díez, E.C.; Winder, C.L.; Ashton, L.; Currie, F.; Goodacre, R. Monitoring the mode of action of antibiotics using raman spectroscopy: Investigating subinhibitory effects of amikacin on Pseudomonas aeruginosa. Anal. Chem. 2005, 77, 2901–2906. [Google Scholar] [CrossRef] [PubMed]
- Athamneh, A.I.M.; Alajlouni, R.A.; Wallace, R.S.; Seleem, M.N.; Sengera, R.S. Phenotypic profiling of antibiotic response signatures in Escherichia coli using raman spectroscopy. Antimicrob. Agents Chemother. 2014, 58, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Han, Y.Y.; Shih, P.H.; Lian, W.N.; Wang, H.H.; Lin, C.H.; Hsueh, P.R.; Wang, J.K.; Wang, Y.L. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Teng, L.; Wang, X.; Wang, X.; Gou, H.; Ren, L.; Wang, T.; Wang, Y.; Ji, Y.; Huang, W.E.; Xu, J. Label-free, rapid and quantitative phenotyping of stress response in E. coli via ramanome. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Germond, A.; Ichimura, T.; Horinouchi, T.; Fujita, H.; Furusawa, C.; Watanabe, T.M. Raman spectral signature reflects transcriptomic features of antibiotic resistance in Escherichia coli. Commun. Biol. 2018, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xuan Nguyen, N.T.; Sarter, S.; Hai Nguyen, N.; Daniel, P. Detection of molecular changes induced by antibiotics in Escherichia coli using vibrational spectroscopy. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2017, 183, 395–401. [Google Scholar] [CrossRef]
- Huleihel, M.; Pavlov, V.; Erukhimovitch, V. The use of FTIR microscopy for the evaluation of anti-bacterial agents activity. J. Photochem. Photobiol. B. 2009, 96, 17–23. [Google Scholar] [CrossRef]
- Moen, B.; Janbu, A.O.; Langsrud, S.; Langsrud, Ø.; Hobman, J.L.; Constantinidou, C.; Kohler, A.; Rudi, K. Global responses of Escherichia coli to adverse conditions determined by microarrays and FT-IR spectroscopy. Can. J. Microbiol. 2009, 55, 714–728. [Google Scholar] [CrossRef]
- Corte, L.; Rellini, P.; Roscini, L.; Fatichenti, F.; Cardinali, G. Development of a novel, FTIR (Fourier transform infrared spectroscopy) based, yeast bioassay for toxicity testing and stress response study. Anal. Chim. Acta 2010, 659, 258–265. [Google Scholar] [CrossRef]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Metabolic fingerprinting with fourier-transform infrared (FTIR) spectroscopy: Towards a high-throughput screening assay for antibiotic discovery and mechanism-of-action elucidation. Metabolites 2020, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Simultaneous elucidation of antibiotic mechanism of action and potency with high-throughput Fourier-transform infrared (FTIR) spectroscopy and machine learning. Appl. Microbiol. Biotechnol. 2021, 1–18. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Cunha, B.R.; Zoio, P.; Fonseca, L.P.; Calado, C.R.C. Technologies for High-Throughput Identification of Antibiotic Mechanism of Action. Antibiotics 2021, 10, 565. https://doi.org/10.3390/antibiotics10050565
da Cunha BR, Zoio P, Fonseca LP, Calado CRC. Technologies for High-Throughput Identification of Antibiotic Mechanism of Action. Antibiotics. 2021; 10(5):565. https://doi.org/10.3390/antibiotics10050565
Chicago/Turabian Styleda Cunha, Bernardo Ribeiro, Paulo Zoio, Luís P. Fonseca, and Cecília R. C. Calado. 2021. "Technologies for High-Throughput Identification of Antibiotic Mechanism of Action" Antibiotics 10, no. 5: 565. https://doi.org/10.3390/antibiotics10050565
APA Styleda Cunha, B. R., Zoio, P., Fonseca, L. P., & Calado, C. R. C. (2021). Technologies for High-Throughput Identification of Antibiotic Mechanism of Action. Antibiotics, 10(5), 565. https://doi.org/10.3390/antibiotics10050565







