Molecular Characterization of KPC-2-Producing Enterobacter cloacae Complex Isolates from Cali, Colombia
Abstract
:1. Introduction
2. Results
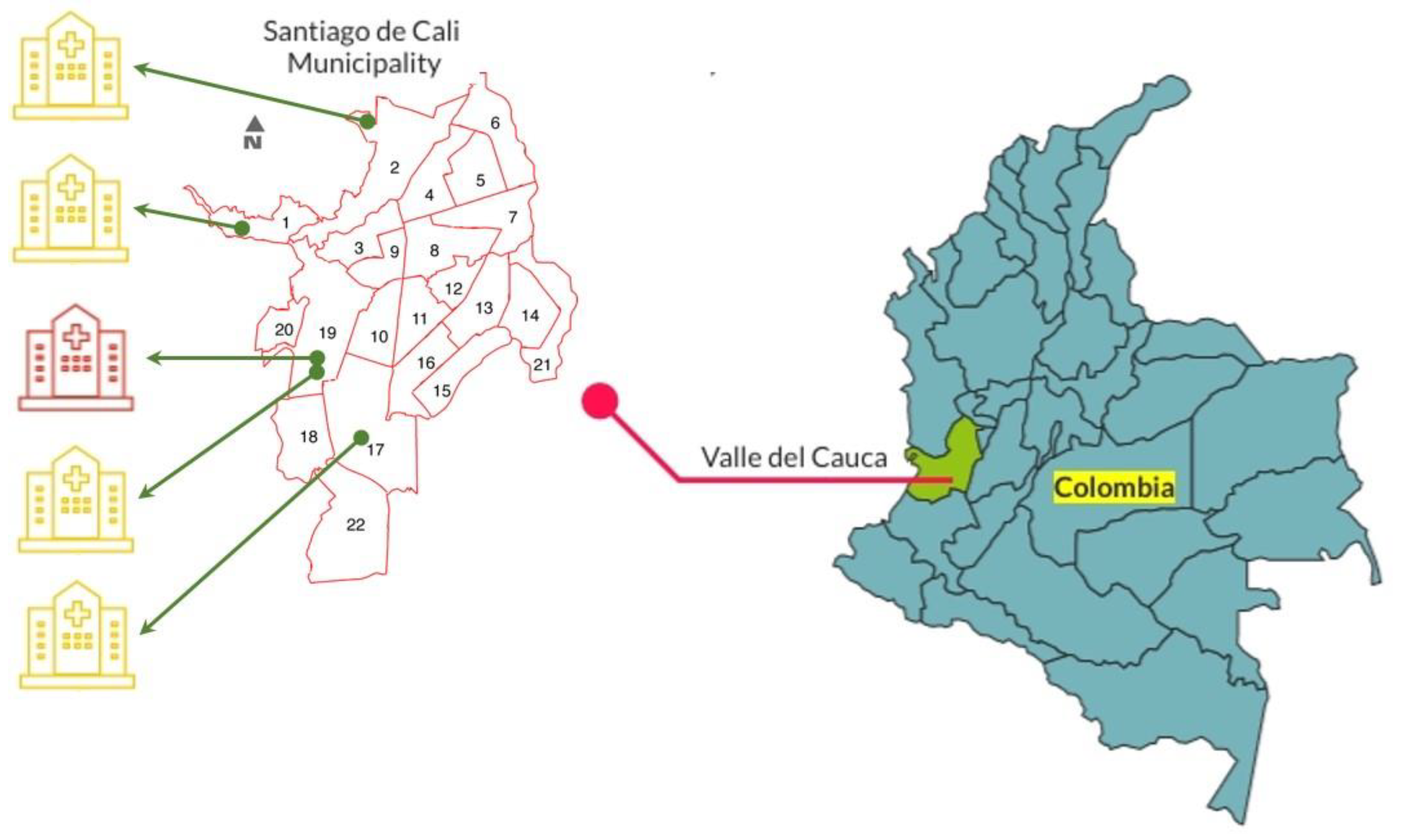
2.1. Collection of E. cloacae Complex Clinical Isolates
2.2. Detection of Genes Encoding Beta-Lactamase Enzymes
2.3. Clinical and Demographic Characteristics of the E. cloacae Complex Isolates Carrying the blaKPC-2 Gene
2.4. Molecular Genotyping of KPC-Producing E. cloacae Complex Isolates
3. Discussion
4. Materials and Methods
4.1. Selection Criteria for E. cloacae Complex Isolates and Antibiotic Sensitivity Tests
4.2. Detection of Genes Encoding Beta-Lactamase Enzymes
4.3. Molecular Genotyping of the KPC-Producing E. cloacae Complex Isolates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mustafa, A.; Ibrahim, M.; Rasheed, M.A.; Kanwal, S.; Hussain, A.; Sami, A.; Ahmed, R.; Bo, Z. Genome-wide Analysis of Four Enterobacter cloacae complex type strains: Insights into Virulence and Niche Adaptation. Sci. Rep. 2020, 10, 8150. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019, 32, 1–32. [Google Scholar] [CrossRef]
- Yang, X.; Guo, R.; Xie, B.; Lai, Q.; Xu, J.; Hu, N.; Wan, L.; Dai, M.; Zhang, B. Drug resistance of pathogens causing nosocomial infection in orthopedics from 2012 to 2017: A 6-year retrospective study. J. Orthop. Surg. Res. 2021, 16, 100. [Google Scholar] [CrossRef] [PubMed]
- Cruz-López, F.; Villarreal-Treviño, L.; Morfin-Otero, R.; Martinez-Melendez, A.; Camacho-Ortiz, A.; Rodríguez-Noriega, E.; Garza-González, E. Dynamics of colonization in patients with health care-associated infections at step-down care units from a tertiary care hospital in Mexico. Am. J. Infect. Control. 2020, 48, 1329–1335. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.-C. Multidrug-Resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S.; National Healthcare Safety Network; et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control. Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- De la Cadena, E.; Adriana, C.; Sebastian, M.J.; Rojas, L.J.; Cristhian, H.-G.; Pallares, C.; Perez, F.; Bonomo, R.A.; Villegas, M.V.; Pallares, C. Molecular characterisation of carbapenem-resistant Enterobacter cloacae complex in Colombia: Bla KPC and the “changing landscape.”. J. Glob. Antimicrob. Resist. 2018, 13, 184–189. [Google Scholar] [CrossRef]
- Salud I nacional de. Resultados Del Programa de Vigilancia Por Laboratorio De Resistencia Antimicrobiana En Infecciones Asociadas a La Atención En Salud (IAAS) 2016. Epidemiología de las Infecciones Asociadas a la Atención en Salud. 2017; 1–43. Available online: https://www.ins.gov.co/BibliotecaDigital/informe-vigilancia-por-laboratorio-resistencia-antimicrobiana-y-whonet-iaas-2016.pdf#search=Resultados%20Del%20Programa%20de%20Vigilancia%20Por%20Laboratorio%20De%20Resistencia%20Antimicrobiana%20En%20Infecciones%20Asociadas%20a%20La%20Atenci%C3%B3n%20En%20Salud%20%28IAAS%29%202016 (accessed on 8 March 2021).
- Rosa, J.F.; Rizek, C.; Marchi, A.P.; Guimaraes, T.; Miranda, L.; Carrilho, C.; Levin, A.S.; Costa, S.F. Clonality, outer-membrane proteins profile and efflux pump in KPC- producing Enterobacter sp. in Brazil. BMC Microbiol. 2017, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sacsaquispe-Contreras, R.; Bailón-Calderón, H. Original Breve Identificación De Genes De Resistencia A Carbapenémicos En Enterobacterias De Hospitales Identification of Carbapenem-Resistant Genes IN. Rev. Peru Med. Exp. Salud Publica 2018, 35, 2013–2017. [Google Scholar]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavda, K.D.; Chen, L.; Fouts, D.E.; Sutton, G.; Brinkac, L.; Jenkins, S.G.; Bonomo, R.A.; Adams, M.D.; Kreiswirth, B.N. Comprehensive Genome Analysis of Carbapenemase-Producing Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance Mechanisms. mBio 2016, 7, e02093-16. [Google Scholar] [CrossRef] [Green Version]
- Girlich, D.; Poirel, L.; Nordmann, P. Clonal distribution of multidrug-resistant Enterobacter cloacae. Diagn. Microbiol. Infect. Dis. 2015, 81, 264–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiedrowski, L.M.; Guerrero, D.M.; Perez, F.; Viau, R.A.; Rojas, L.J.; Mojica, M.F.; Rudin, S.D.; Hujer, A.M.; Marshall, S.H.; Bonomo, R.A. Carbapenem-ResistantEnterobacter cloacaeIsolates Producing KPC-3, North Dakota, USA. Emerg. Infect. Dis. 2014, 20, 1583–1585. [Google Scholar] [CrossRef] [Green Version]
- Doumith, M.; Ellington, M.J.; Livermore, D.M.; Woodford, N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 2009, 63, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A.; Urban, C.; Mariano, N.; Projan, S.J.; Rahal, J.J.; Bush, K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the foss of an outer membrane protein. Antimicrob. Agents Chemother. 1997, 41, 563–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salud I nacional de. Informe De Resultados De La Vigilancia Por Laboratorio De Resistencia Antimicrobiana En Infecciones Asociadas a La Atención En Salud (IAAS) 2017. Epidemiol las Infecc Asoc a la Atención en Salud. 2018; 1–31. Available online: https://www.ins.gov.co/buscador-eventos/Informacin%20de%20laboratorio/Informe%20Vigilancia%20por%20Laboratorio%20Resistencia%20Antimicrobiana%20y%20Whonet%20IAAS%202017.pdf#search=Informe%20de%20Resultados%20de%20la%20Vigilancia%20por%20Laboratorio%20de%20Resistencia%20antimicrobiana%20en%20Infecciones%20Asociadas%20a%20la%20Atenci%C3%B3n%20en%20Salud%20%28IAAS%29%202017 (accessed on 8 March 2021).
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of blaKPC-2 Dissemination from Non-CG258 Klebsiella pneumoniae to Other Enterobacterales via IncN Plasmids in an Area of High Endemicity. Antimicrob. Agents Chemother. 2020, 64, 64. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Parra, O.L.; Jiménez, J.N. Molecular epidemiology of carbapenem resistant gram-negative bacilli from infected pediatric population in tertiary—care hospitals in Medellín, Colombia: An increasing problem. BMC Infect. Dis. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Escandón-Vargas, K.; Reyes, S.; Gutiérrez, S.; Villegas, M.V. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti. Infect. Ther. 2016, 15, 277–297. [Google Scholar] [CrossRef]
- Pacheco, R.; Osorio, L.; Correa, A.M.; Villegas, M.V. Prevalencia de bacterias Gram negativas portadoras del gen blaKPC en hospitales de Colombia. Biomédica 2013, 34, 81. [Google Scholar] [CrossRef] [Green Version]
- Samuelsen, Ø.; Overballe-Petersen, S.; Bjørnholt, J.V.; Brisse, S.; Doumith, M.; Woodford, N.; Hopkins, K.; Aasnæs, B.; Haldorsen, B.; Sundsfjord, A. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS ONE 2017, 12, e0187832. [Google Scholar] [CrossRef] [Green Version]
- Fernández, J.F.; Montero, I.; Martinez, O.F.; Fleites, A.; Poirel, L.; Nordmann, P.; Rodicio, M.D.R.R. Dissemination of multiresistant Enterobacter cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int. J. Antimicrob. Agents 2015, 46, 469–474. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.-L.; Cheng, L.; Zhang, Z.-F.; Ning, M.-Z.; Zhou, W.-Q.; Zhang, K.; Shen, H. Survey of Clinical Extended-Spectrum Beta-Lactamase-Producing Enterobacter cloacae Isolates in a Chinese Tertiary Hospital, 2012–2014. Microb. Drug Resist. 2017, 23, 83–89. [Google Scholar] [CrossRef]
- Le-Ha, T.D.; Le, L.; Le-Vo, H.N.; Anda, M.; Motooka, D.; Nakamura, S.; Tran, L.K.; Tran, P.T.-H.; Lida, T.; Cao, V. Characterization of a carbapenem- and colistin-resistant Enterobacter cloacae carrying Tn6901 in blaNDM-1 genomic context. Infect. Drug Resist. 2019, 12, 733–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Rosa, G.; León, A.L.; Jaimes, F. Epidemiología y pronóstico de pacientes con infección del torrente sanguíneo en 10 hospitales de Colombia. Rev. Chil Infectol. 2016, 33, 141–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortés, J.A.; Leal, A.L.; Montañez, A.M.; Buitrago, G.; Castillo, J.S.; Guzman, L. Frequency of microorganisms isolated in patients with bacteremia in intensive care units in Colombia and their resistance profiles. Braz. J. Infect. Dis. 2013, 17, 346–352. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Miranda, R.; Gastélum-Acosta, M.; Guerrero-Estrada, P.; Ayala-Figueroa, R.I.; Osuna-Álvarez, L.E. Actividad antimicrobiana de ceftolozano-tazobactam y ceftazidima-avibactam contra bacilos gramnegativos clínicamente relevantes aislados en México. Gac Med. Mex 2020, 156, 592–597. [Google Scholar] [CrossRef]
- Shields, R.K.; Nguyen, M.H.; Chen, L.; Press, E.G.; Kreiswirth, B.N.; Clancy, C.J. Pneumonia and Renal Replacement Therapy Are Risk Factors for Ceftazidime-Avibactam Treatment Failures and Resistance among Patients with Carbapenem-Resistant Enterobacteriaceae Infections. Antimicrob. Agents Chemother. 2018, 62, e02497-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shields, R.K.; Iovleva, A.; Kline, E.G.; Kawai, A.; McElheny, C.L.; Doi, Y. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in enterobacter cloacae complex following exposure to cefepime. Clin. Infect. Dis. 2020, 71, 2713–2716. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Q.; Liu, F.; Xiong, H.; Yang, J. Enterobacter cloacae infection of the shoulder in a 52-year-old woman without apparent predisposing risk factor: A case report and literature review. BMC Infect. Dis. 2021, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Le, N.T.; Donadu, M.G.; Ho, D.V.; Doan, T.Q.; Le, A.T.; Raal, A.; Usai, D.; Sanna, G.; Marchetti, M.; Usai, M.; et al. Biological activities of essential oil extracted from leaves of Atalantia sessiflora Guillauminin Vietnam. J. Infect. Dev. Ctries 2020, 14, 1054–1064. [Google Scholar] [CrossRef]
- Benameur, Q.; Gervasi, T.; Pellizzeri, V.; Pľuchtová, M.; Tali-Maama, H.; Assaous, F.; Guettou, B.; Rahal, K.; Gruľová, D.; Dugo, G.; et al. Antibacterial activity of Thymus vulgaris essential oil alone and in combination with cefotaxime against blaESBL producing multidrug resistant Enterobacteriaceae isolates. Nat. Prod. Res. 2018, 33, 2647–2654. [Google Scholar] [CrossRef]
- Crespo, M.P.; Woodford, N.; Sinclair, A.; Kaufmann, M.E.; Turton, J.; Glover, J.; Velez, J.D.; Castaneda, C.R.; Recalde, M.; Livermore, D.M. Outbreak of Carbapenem-Resistant Pseudomonas aeruginosa Producing VIM-8, a Novel Metallo- -Lactamase, in a Tertiary Care Center in Cali, Colombia. J. Clin. Microbiol. 2004, 42, 5094–5101. [Google Scholar] [CrossRef] [Green Version]
- Villegas, M.V.; Lolans, K.; Olivera, M.D.R.; Suarez, C.J.; Correa, A.; Queenan, A.M.; Quinn, J.P. First Detection of Metallo-β-Lactamase VIM-2 in Pseudomonas aeruginosa Isolates from Colombia. Antimicrob. Agents Chemother. 2006, 50, 226–229. [Google Scholar] [CrossRef] [Green Version]
- Montealegre, M.C.; Correa, A.; Briceño, D.F.; Rosas, N.C.; De La Cadena, E.; Ruiz, S.J.; Mojica, M.F.; Camargo, R.D.; Zuluaga, I.; Marin, A.; et al. Novel VIM Metallo-β-Lactamase Variant, VIM-24, from a Klebsiella pneumoniae Isolate from Colombia. Antimicrob. Agents Chemother. 2011, 55, 2428–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, T.; Bustos-Cruz, R.H.; Abril, D.; Arias, S.; Uribe, L.; Rincón, J.; García, J.-C.; Escobar-Perez, J. Pseudomonas aeruginosa Coharboring BlaKPC-2 and BlaVIM-2 Carbapenemase Genes. Antibiotics 2019, 8, 98. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.A.E.; Escobar, N.M.O.; Castro-Cardozo, B.; Márquez, I.A.V.; Aguilar, M.I.G.; De La Barrera, L.M.; Barreto, E.R.B.; Marquez-Ortiz, R.A.; Guayazán, M.V.M.; Gómez, N.V. Outbreak of NDM-1-Producing Klebsiella pneumoniae in a Neonatal Unit in Colombia. Antimicrob. Agents Chemother. 2013, 57, 1957–1960. [Google Scholar] [CrossRef] [Green Version]
- Ministerio de Salud de Colombia. Informe Quincenal Epidemiológico Nacional. Inst Nac Salud Colomb 2017, 22, 65–77. Available online: https://www.ins.gov.co/buscador-eventos/IQEN/IQEN (accessed on 16 March 2021).
- Saavedra-Rojas, S.-Y.; Duarte-Valderrama, C.; González-De-Arias, M.-N.; Ovalle-Guerro, M.V. Emergencia de Providencia rettgeri NDM-1 en dos departamentos de Colombia, 2012–2013. Enferm. Infecc. y Microbiol. Clínica 2017, 35, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Rojas, L.J.; Wright, M.S.; De La Cadena, E.; Motoa, G.; Hujer, K.M.; Villegas, M.V.; Adams, M.D.; Bonomo, R.A. Initial assessment of the molecular epidemiology of blaNDM-1 in Colombia. Antimicrob. Agents Chemother. 2016, 60, 4346–4350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanegas, J.M.; Ospina, W.P.; Higuita-Gutiérrez, L.F.; Jiménez, J.N.; Higuita, L.F. First reported case of an OXA-48-producing isolate from a Colombian patient. J. Glob. Antimicrob. Resist. 2016, 6, 67–68. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 28th Edition Informational Supplement M100-S27; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Lu, J.J.; Perng, C.L.; Lee, S.Y.; Wan, C.C. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol. 2000, 38, 2076–2080. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Doménech-Sánchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, W.; Wang, J.; Pan, J.; Sun, S.; Yu, Y.; Zhao, B.; Ma, Y.; Zhang, T.; Qi, J.; et al. Identification and Characterization of the First Escherichia coli Strain Carrying NDM-1 Gene in China. PLoS ONE 2013, 8, e66666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toleman, M.A.; Simm, A.M.; Murphy, T.A.; Gales, A.C.; Biedenbach, D.J.; Jones, R.N.; Walsh, T.R. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: Report from the SENTRY antimicrobial surveillance programme. J. Antimicrob Chemother. 2002, 50, 673–679. [Google Scholar] [CrossRef]
- Poirel, L.; Castanheira, M.; Carrër, A.; Rodriguez, C.P.; Jones, R.N.; Smayevsky, J.; Nordmann, P. OXA-163, an OXA-48-Related Class D β-Lactamase with Extended Activity Toward Expanded-Spectrum Cephalosporins. Antimicrob. Agents Chemother. 2011, 55, 2546–2551. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi-Akiyama, T.; Hayakawa, K.; Ohmagari, N.; Shimojima, M.; Kirikae, T. Multilocus Sequence Typing (MLST) for Characterization of Enterobacter cloacae. PLoS ONE 2013, 8, e66358. [Google Scholar] [CrossRef] [Green Version]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009, 10, 152. [Google Scholar] [CrossRef] [Green Version]


| Key | Year Isolate | Ceftazidime | Cefepime | Ceftriaxone | Ertapenem | Meropenem | Origin | Infection | Age (Years) | Gender |
|---|---|---|---|---|---|---|---|---|---|---|
| 1416 | 2011 | >64 | 4 | >64 | 4 | <1 | Clinic 1 | Urine | 81 | F |
| 1666 | 2012 | 16 | 8 | >64 | >8 | 1 | Clinic 1 | Bloodstream | 9 | M |
| 2072 | 2012 | 16 | 2 | >64 | 4 | >16 | Clinic 1 | Urine | 41 | M |
| 1364 | 2013 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Bloodstream | 49 | M |
| 2205 | 2013 | >64 | 2 | >64 | 4 | 2 | Clinic 1 | Urine | 6 | M |
| 2249 | 2013 | 16 | 2 | 4 | 2 | 1 | Clinic 1 | Urine | 84 | F |
| 1359 | 2014 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Skin | 93 | F |
| 4290 | 2014 | 32 | 32 | >64 | >8 | >16 | Clinic 1 | Bloodstream | 23 | M |
| 4347 | 2014 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Urine | 67 | M |
| 4730 | 2015 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Ulcer | 20 | F |
| 5047 | 2016 | 16 | >64 | >64 | >8 | >16 | Clinic 1 | Urine | 17 | M |
| 5227 | 2017 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Wound | 58 | M |
| 382 | 2017 | >64 | 2 | >64 | 4 | <1 | Public | Bloodstream | 76 | F |
| 474 | 2017 | 32 | 16 | 32 | 4 | 2 | Public | Peritoneal fluid | 22 | M |
| 381 | 2017 | 16 | 16 | >32 | 4 | 1 | Public | Bloodstream | 74 | M |
| 713 | 2017 | 16 | 4 | >4 | >1 | 8 | Public | Peritoneal fluid | 54 | M |
| 332 | 2017 | 16 | 2 | >64 | 2 | 2 | Clinic 2 | Bloodstream | 21 days | M |
| 331 | 2017 | >64 | >64 | >64 | 2 | <1 | Clinic 2 | Bloodstream | 30 | M |
| 333 | 2017 | >64 | >64 | >64 | >8 | >16 | Clinic 2 | Bloodstream | 47 days | M |
| 330 | 2017 | >64 | >64 | >64 | >8 | >16 | Clinic 2 | Bloodstream | 12 days | M |
| 305 | 2017 | >64 | >64 | >64 | >8 | >16 | Clinic 3 | Sore | 30 | M |
| 31 | 2017 | >64 | 2 | >64 | 4 | 2 | Clinic 3 | Wound | 20 | F |
| 2 | 2018 | >64 | >64 | >64 | >8 | >16 | Clinic 4 | Urine culture | 70 | M |
| 1,510,006 | 2018 | >64 | >64 | >64 | >8 | >16 | Public | Catheter | 62 | M |
| 6 | 2018 | >64 | >64 | >64 | >8 | >16 | Public | Bloodstream | 62 | M |
| 5385 | 2018 | >64 | >64 | >64 | >8 | >16 | Clinic 1 | Sore | 55 | M |
| 5438 | 2018 | 16 | 2 | ≥64 | 4 | 1 | Clinic 1 | Urine | 18 | M |
| 5521-12 | 2018 | 16 | 2 | ≥64 | 4 | 1 | Clinic 1 | Urine | 1 | F |
| Key | Allele | ST | dnaA | fusA | gyrB | leuS | pyrG | rplB | rpoB |
|---|---|---|---|---|---|---|---|---|---|
| 1364 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| 1359 | blaKPC-2 | 1483 | 376 | 21 | 9 | 44 | 45 | 4 | 33 |
| 4290 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| 4730 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| 5227 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| 474 | blaKPC-2 | 513 | 171 | 1 | 190 | 168 | 1 | 22 | 113 |
| 333 | blaKPC-2 | 45 | 4 | 4 | 14 | 6 | 39 | 4 | 6 |
| 330 | blaKPC-2 | 45 | 4 | 4 | 14 | 6 | 39 | 4 | 6 |
| 305 | blaKPC-2 | 456 | 149 | 44 | 61 | 180 | 152 | 1 | 1 |
| 2 | blaKPC-2 | 456 | 149 | 44 | 61 | 180 | 152 | 1 | 1 |
| 1,510,006 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| 6 | blaKPC-2 | 510 | 4 | 4 | 4 | 209 | 171 | 4 | 115 |
| Characteristic | % of Isolates |
|---|---|
| Gender | |
| Female | 17% (2/12) |
| Male | 83% (10/12) |
| Age | |
| Newborn (0–30 days) | 8% (1/12) |
| Infant (1–12 months) | 8% (1/12) |
| Teenagers (12–20 years) | 8% (1/12) |
| Young adult (21–40 years) | 16% (2/12) |
| Middle adult (41–60 years) | 24% (3/12) |
| Elderly (>60 years) | 32% (4/12) |
| Infection site | |
| Bloodstream | 42% (5/12) |
| Skin | 8.3% (1/12) |
| Ulcer | 8.3% (1/12) |
| Wound | 8.3% (1/12) |
| Peritoneal fluid | 8.3% (1/12) |
| Sore | 8.3% (1/12) |
| Urine culture | 8.3% (1/12) |
| Catheter | 8.3% (1/12) |
| Primer Name | Primer Sequence | Product Size (bp) | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| KPC F | 5-TGTCACTGTATCGCCGTC-3 | 894 | 53 | Yigit y col., 2001 [45] |
| KPC R | 5-CTCAGTGCTCTACAGAAAACC-3 | |||
| NDM F | 5-ATGGAATTGCCCAATATTATGC-3 | 813 | 52 | Liu y col., 2013 [46] |
| NDM R | 5-TCAGCGCAGCTTGTCGGCCAT-3 | |||
| VIM F | 5-GTCTATTTGACCGCGTC-3 | 775 | 52 | Toleman y col., 2002 [47] |
| VIM R | 5-CTACTCAACGACTGAGCG-3 | |||
| OXA-48 F | 5-TATATTGCATTAAGCAAGGG-3 | 848 | 56 | Poirel y col., 2011 [48] |
| OXA-48 R | 5-CACACAAATACGCGCTAACC-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falco, A.; Guerrero, D.; García, I.; Correa, A.; Rivera, S.; Olaya, M.B.; Aranaga, C. Molecular Characterization of KPC-2-Producing Enterobacter cloacae Complex Isolates from Cali, Colombia. Antibiotics 2021, 10, 694. https://doi.org/10.3390/antibiotics10060694
Falco A, Guerrero D, García I, Correa A, Rivera S, Olaya MB, Aranaga C. Molecular Characterization of KPC-2-Producing Enterobacter cloacae Complex Isolates from Cali, Colombia. Antibiotics. 2021; 10(6):694. https://doi.org/10.3390/antibiotics10060694
Chicago/Turabian StyleFalco, Aura, Daniela Guerrero, Isabella García, Adriana Correa, Sandra Rivera, María Beatriz Olaya, and Carlos Aranaga. 2021. "Molecular Characterization of KPC-2-Producing Enterobacter cloacae Complex Isolates from Cali, Colombia" Antibiotics 10, no. 6: 694. https://doi.org/10.3390/antibiotics10060694






