Evolution of the Gram-Negative Antibiotic Resistance Spiral over Time: A Time-Series Analysis
Abstract
:1. Introduction
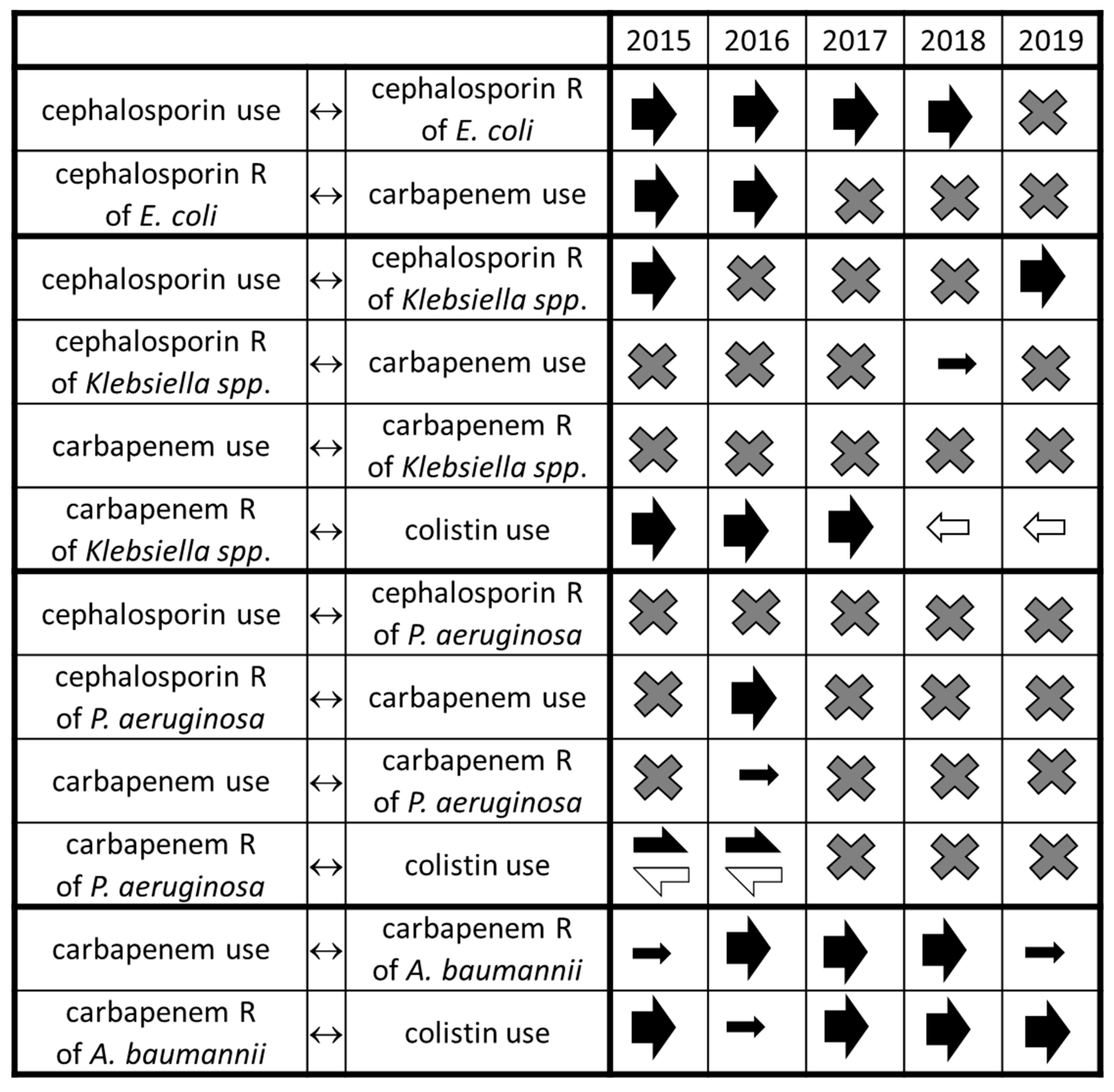
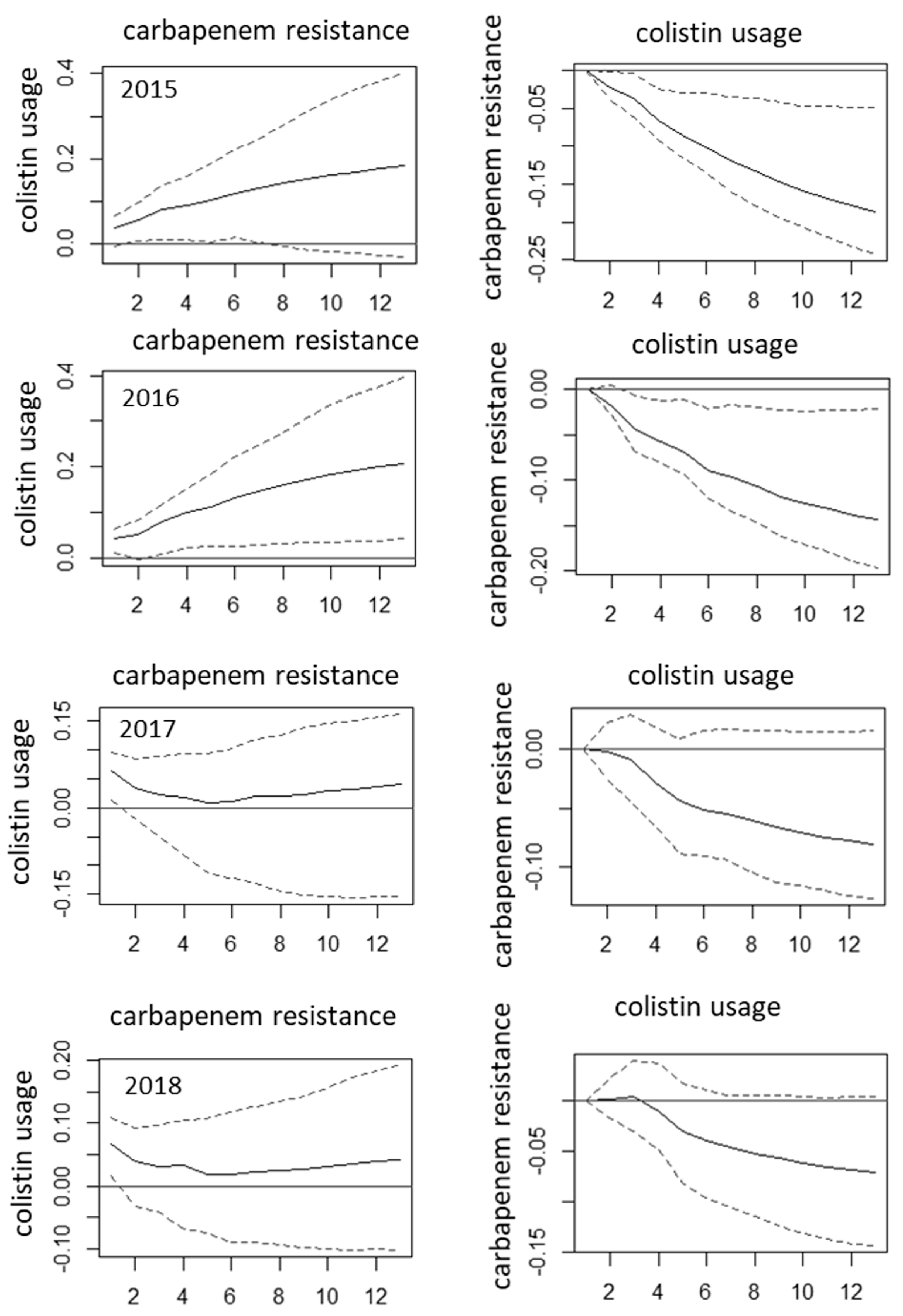
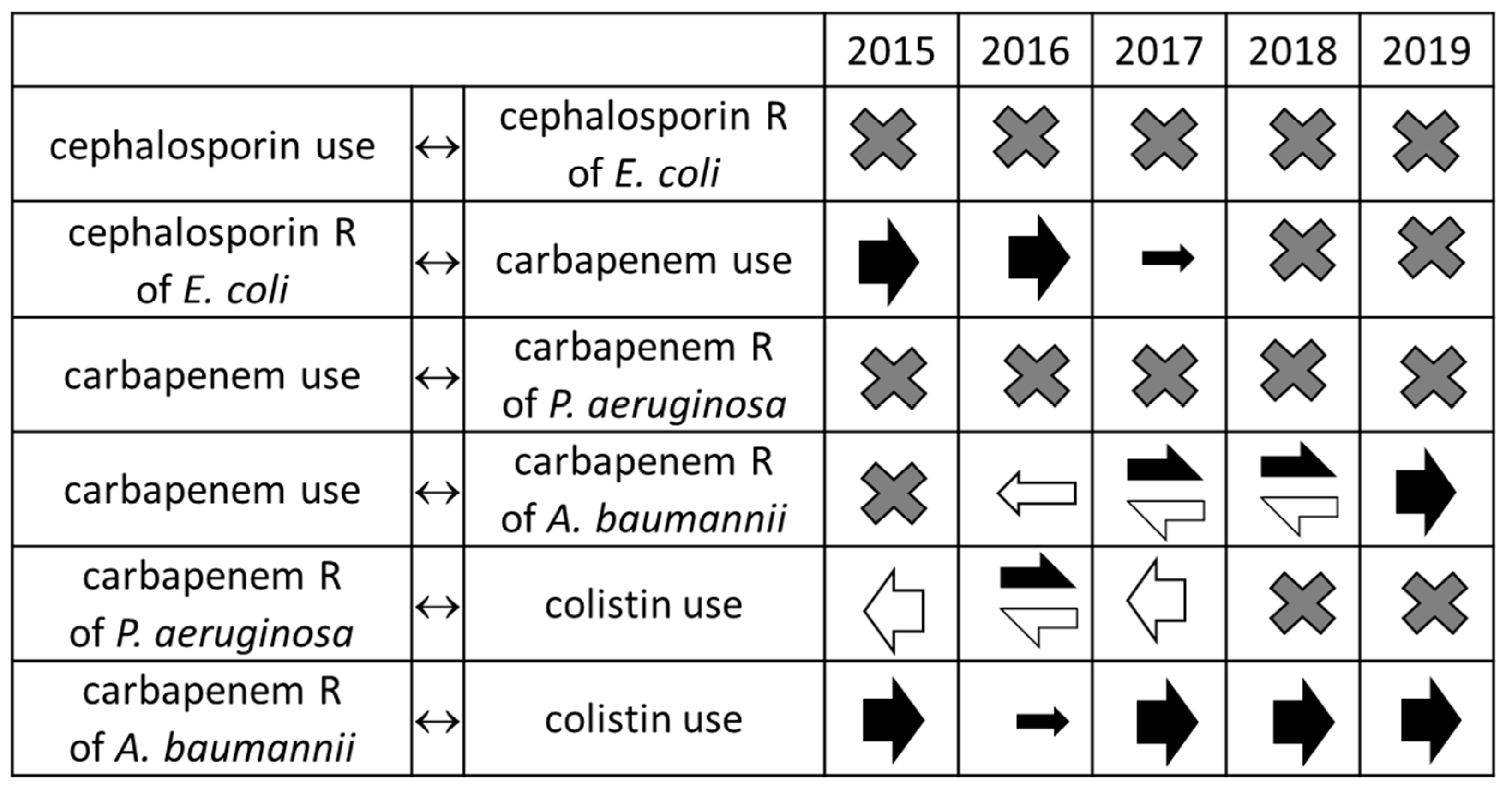
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, B.G.; Schellevis, F.; Stobberingh, E.; Goossens, H.; Pringle, M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 2014, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Carlet, J.; Ben Ali, A.; Chalfine, A. Epidemiology and control of antibiotic resistance in the intensive care unit. Curr. Opin. Infect. Dis. 2004, 17, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Perona, F.; Sotillo, A.; Troyano-Hernáez, P.; Gómez-Gil, R.; de la Vega-Bueno, Á.; Mingorance, J. Genomic path to pandrug resistance in a clinical isolate of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2018, 52, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H.; MagicBullet Working Group WP4. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Forde, B.M.; Alfaresi, M.; Alzarouni, A.; Farahat, Y.; Chong, T.-M.; Yin, W.-F.; Chan, K.G.; Li, J.; Schembri, M.; et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci. Rep. 2015, 5, 15082. [Google Scholar] [CrossRef] [PubMed]
- Tóth, H.; Fésűs, A.; Kungler-Gorácz, O.; Balázs, B.; Majoros, L.; Szarka, K.; Kardos, G. Utilization of Vector Autoregressive and Linear Transfer Models to Follow Up the Antibiotic Resistance Spiral in Gram-negative Bacteria From Cephalosporin Consumption to Colistin Resistance. Clin. Infect. Dis. 2018, 69, 1410–1421. [Google Scholar] [CrossRef]
- Baquero, F.; Alvarez-Ortega, C.; Martinez, J.L. Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 2009, 1, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Zachariasen, C.; Christiansen, L.E.; Græsbøll, K.; Toft, N.; Matthews, L.; Nielsen, S.S.; Olsen, J.E. Modeling the growth dynamics of multiple Escherichia coli strains in the pig intestine following intramuscular ampicillin treatment. BMC Microbiol. 2016, 16, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The Population Structure of Acinetobacter baumannii: Expanding Multiresistant Clones from an Ancestral Susceptible Genetic Pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef] [Green Version]
- Duong, J.; Booth, S.; McCartney, N.K.; Rabin, H.R.; Parkins, M.D.; Storey, D.G. Phenotypic and Genotypic Comparison of Epidemic and Non-Epidemic Strains of Pseudomonas aeruginosa from Individuals with Cystic Fibrosis. PLoS ONE 2015, 10, e0143466. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.K.; Jung, S.-I.; Park, K.-H.; Kim, D.H.; Choi, J.Y.; Kim, S.H.; Ko, K.S. Changes in antimicrobial susceptibility and major clones of Acinetobacter calcoaceticus–baumannii complex isolates from a single hospital in Korea over 7 years. J. Med. Microbiol. 2012, 61, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Lawes, T.; Lopez-Lozano, J.-M.; Nebot, C.; Macartney, G.; Subbarao-Sharma, R.; Dare, C.; Edwards, G.F.S.; Gould, I.M. Turning the tide or riding the waves? Impacts of antibiotic stewardship and infection control on MRSA strain dynamics in a Scottish region over 16 years: Non-linear time series analysis. BMJ Open 2015, 5, e006596. [Google Scholar] [CrossRef]
- Wang, J.; Foxman, B.; Mody, L.; Snitkin, E.S. Network of microbial and antibiotic interactions drive colonization and infection with multidrug-resistant organisms. Proc. Natl. Acad. Sci. USA 2017, 114, 10467–10472. [Google Scholar] [CrossRef] [Green Version]
- Tenover, F.C.; McGowan, J.E. Reasons for the Emergence of Antibiotic Resistance. Am. J. Med. Sci. 1996, 311, 9–16. [Google Scholar] [CrossRef]
- Tacconelli, E.; De Angelis, G.; Cataldo, M.A.; Mantengoli, E.; Spanu, T.; Pan, A.; Corti, G.; Radice, A.; Stolzuoli, L.; Antinori, S.; et al. Antibiotic Usage and Risk of Colonization and Infection with Antibiotic-Resistant Bacteria: A Hospital Population-Based Study. Antimicrob. Agents Chemother. 2009, 53, 4264–4269. [Google Scholar] [CrossRef] [Green Version]
- Kaki, R.; Elligsen, M.; Walker, S.; Simor, A.; Palmay, L.; Daneman, N. Impact of antimicrobial stewardship in critical care: A systematic review. J. Antimicrob. Chemother. 2011, 66, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Ntagiopoulos, P.G.; Paramythiotou, E.; Antoniadou, A.; Giamarellou, H.; Karabinis, A. Impact of an antibiotic restriction policy on the antibiotic resistance patterns of Gram-negative microorganisms in an Intensive Care Unit in Greece. Int. J. Antimicrob. Agents 2007, 30, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L. Squeezing the antibiotic balloon: The impact of antimicrobial classes on emerging resistance. Clin. Microbiol. Infect. 2005, 11, 4–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahal, J.J.; Urban, C.; Horn, D.; Freeman, K.; Segal-Maurer, S.; Maurer, J.; Mariano, N.; Marks, S.; Burns, J.M.; Dominick, D.; et al. Class Restriction of Cephalosporin Use to Control Total Cephalosporin Resistance in Nosocomial Klebsiella. JAMA 1998, 280, 1233–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mózes, J.; Ebrahimi, F.; Gorácz, O.; Miszti, C.; Kardos, G. Effect of carbapenem consumption patterns on the molecular epidemiology and carbapenem resistance of Acinetobacter baumannii. J. Med. Microbiol. 2014, 63, 1654–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balázs, B.; Tóth, Z.; Nagy, F.; Kovács, R.; Tóth, H.; Nagy, J.; Tóth, Á.; Szarka, K.; Majoros, L.; Kardos, G. The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement. Antibiotics 2021, 10, 127. [Google Scholar] [CrossRef]
- Agodi, A.; Voulgari, E.; Barchitta, M.; Quattrocchi, A.; Bellocchi, P.; Poulou, A.; Santangelo, C.; Castiglione, G.; Giaquinta, L.; Romeo, M.; et al. Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two Sicilian hospitals. J. Hosp. Infect. 2014, 86, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Ben-Chetrit, E.; Wiener-Well, Y.; Lesho, E.; Kopuit, P.; Broyer, C.; Bier, L.; Assous, M.V.; Benenson, S.; Cohen, M.J.; McGann, P.T.; et al. An intervention to control an ICU outbreak of carbapenem-resistant Acinetobacter baumannii: Long-term impact for the ICU and hospital. Crit. Care 2018, 22, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, G.R.; Dizbay, M.; Güven, T.; Pullukçu, H.; Taşbakan, M.; Guzel, O.T.; Tekce, Y.T.; Ozden, M.; Turhan, O.; Güner, R.; et al. Risk factors for infection with colistin-resistant gram-negative microorganisms: A multicenter study. Ann. Saudi Med. 2016, 36, 216–222. [Google Scholar] [CrossRef] [Green Version]
- Santos, I.; de Andrade, N.F.P.; Neto, O.C.D.C.; da Costa, B.S.; Marques, E.D.A.; Rocha-De-Souza, C.M.; Asensi, M.D.; Carvalho-Assef, A.P.D. Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de janeiro-Brazil: Importance of mutational mechanisms over the years (1995–2015). Infect. Genet. Evol. 2019, 73, 411–415. [Google Scholar] [CrossRef]
- McCarthy, K.L.; Kidd, T.; Paterson, D. Molecular epidemiology of Pseudomonas aeruginosa bloodstream infection isolates in a non-outbreak setting. J. Med. Microbiol. 2017, 66, 154–159. [Google Scholar] [CrossRef]
- Mózes, J.; Szűcs, I.; Molnar, D.; Jakab, P.; Fatemeh, E.; Szilasi, M.; Majoros, L.; Orosi, P.; Kardos, G. A potential role of aminoglycoside resistance in endemic occurrence of Pseudomonas aeruginosa strains in lower airways of mechanically ventilated patients. Diagn. Microbiol. Infect. Dis. 2014, 78, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, F.; Mózes, J.; Monostori, J.; Gorácz, O.; Fésűs, A.; Majoros, L.; Szarka, K.; Kardos, G. Comparison of the faecal colonization rates with extended-spectrum beta-lactamase producing enterobacteria among patients in different wards, outpatients and screened medical students. Microbiol. Immunol. 2016, 60, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Livorsi, D.; Comer, A.R.; Matthias, M.S.; Perencevich, E.; Bair, M.J. Factors Influencing Antibiotic-Prescribing Decisions Among Inpatient Physicians: A Qualitative Investigation. Infect. Control. Hosp. Epidemiol. 2015, 36, 1065–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juhász, E.; Iván, M.; Pintér, E.; Pongrácz, J.; Kristóf, K. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J. Glob. Antimicrob. Resist. 2017, 11, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Freeman, R.; Moore, L.S.P.; Charlett, A.; Donaldson, H.; Holmes, A.H. Exploring the epidemiology of carbapenem-resistant Gram-negative bacteria in west London and the utility of routinely collected hospital microbiology data. J. Antimicrob. Chemother. 2014, 70, 1212–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goel, N.; Wattal, C.; Oberoi, J.K.; Raveendran, R.; Datta, S.; Prasad, K.J. Trend analysis of antimicrobial consumption and development of resistance in non-fermenters in a tertiary care hospital in Delhi, India. J. Antimicrob. Chemother. 2011, 66, 1625–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe–Annual Report of the European Antimicrobial Resistance Surveillance Network [EARS-Net] 2017; ECDC: Stockholm, Sweden, 2018. [Google Scholar]
- Jean, S.-S.; Hsueh, P.-R. Distribution of ESBLs, AmpC ?-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 2016, 72, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, R.M.; Carrique-Mas, J.; Limmathurotsakul, D.; Day, N.P.J.; E Thwaites, G.; Baker, S.; Ashley, E.; De Balogh, K.; Baird, K.; Basnyat, B.; et al. A current perspective on antimicrobial resistance in Southeast Asia. J. Antimicrob. Chemother. 2017, 72, 2963–2972. [Google Scholar] [CrossRef] [Green Version]
- Monnet, D.L. ABC Calc—Antibiotic Consumption Calculator [Microsoft Excel Application]. Version 3.0; Statens Serum Institut: Copenhagen, Denmark, 2005. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tóth, H.; Buchholcz, G.; Fésüs, A.; Balázs, B.; Nagy, J.B.; Majoros, L.; Szarka, K.; Kardos, G. Evolution of the Gram-Negative Antibiotic Resistance Spiral over Time: A Time-Series Analysis. Antibiotics 2021, 10, 734. https://doi.org/10.3390/antibiotics10060734
Tóth H, Buchholcz G, Fésüs A, Balázs B, Nagy JB, Majoros L, Szarka K, Kardos G. Evolution of the Gram-Negative Antibiotic Resistance Spiral over Time: A Time-Series Analysis. Antibiotics. 2021; 10(6):734. https://doi.org/10.3390/antibiotics10060734
Chicago/Turabian StyleTóth, Hajnalka, Gyula Buchholcz, Adina Fésüs, Bence Balázs, József Bálint Nagy, László Majoros, Krisztina Szarka, and Gábor Kardos. 2021. "Evolution of the Gram-Negative Antibiotic Resistance Spiral over Time: A Time-Series Analysis" Antibiotics 10, no. 6: 734. https://doi.org/10.3390/antibiotics10060734
APA StyleTóth, H., Buchholcz, G., Fésüs, A., Balázs, B., Nagy, J. B., Majoros, L., Szarka, K., & Kardos, G. (2021). Evolution of the Gram-Negative Antibiotic Resistance Spiral over Time: A Time-Series Analysis. Antibiotics, 10(6), 734. https://doi.org/10.3390/antibiotics10060734





