Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World
Abstract
:1. Introduction
2. Results
2.1. Main Protease (MPro)
2.2. Papain-Like Protease
2.3. RNA-Dependent RNA Polymerase (RdRp)
2.4. Spike Glycoprotein
2.5. Helicase (Nsp13) Protein
2.6. E-Channel (Envelop Small Membrane Protein)
2.7. Non-Specific Interactions of Selected Ligands against Human Blood Proteins
2.8. ADMET Properties of Selected Ligands
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Molecular Docking
5.2. Molecular Dynamic Simulations
5.3. Pharmacokinetics and Drug-Likeness
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, Y.; Peng, F.; Wang, R.; Guan, K.; Jiang, T.; Xu, G.; Sun, J.; Chang, C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020, 109, 102434. [Google Scholar] [CrossRef]
- Rehman, M.F.; Fariha, C.; Anwar, A.; Shahzad, N.; Ahmad, M.; Mukhtar, S.; Haque, M.F.U. Novel coronavirus disease (COVID-19) pandemic: A recent mini review. Comput. Struct. Biotechnol. J. 2020, 19, 612–623. [Google Scholar] [CrossRef]
- Coronavirus-Worldwide-Graphs. Available online: https://www.worldometers.info (accessed on 12 August 2021).
- Dinleyici, E.C.; Borrow, R.; Safadi, M.A.P.; Van Damme, P.; Munoz, F.M. Vaccines and routine immunization strategies during the COVID-19 pandemic. Hum. Vaccines Immunother. 2021, 17, 400–407. [Google Scholar] [CrossRef]
- Mazzola, A.; Todesco, E.; Drouin, S.; Hazan, F.; Marot, S.; Thabut, D.; Varnous, S.; Soulié, C.; Barrou, B.; Marcelin, A.-G.; et al. Poor Antibody Response After Two Doses of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccine in Transplant Recipients. Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Mohammed, F.S.; Akgul, H.; Sevindik, M.; Khaled, B.M.T. Phenolic content and biological activities of Rhus coriaria var. zebaria. Fresenius Environ. Bull. 2018, 27, 5694–5702. [Google Scholar]
- Sevindik, M.; Akgul, H.; Selamoglu, Z.; Braidy, N. Antioxidant and antigenotoxic potential of infundibulicybe geotropa mushroom collected from Northwestern Turkey. Oxidative Med. Cell. Longev. 2020, 2020, 5620484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevindik, M.; Akgul, H.; Bal, C.; Selamoglu, Z. Phenolic contents, oxidant/antioxidant potential and heavy metal levels in Cyclocybe cylindracea. Indian J. Pharm. Educ. Res. 2018, 52, 437–441. [Google Scholar] [CrossRef] [Green Version]
- Mani, J.S.; Johnson, J.B.; Steel, J.C.; Broszczak, D.A.; Neilsen, P.M.; Walsh, K.B.; Naiker, M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020, 284, 197989. [Google Scholar] [CrossRef]
- Li, S.-y.; Chen, C.; Zhang, H.-q.; Guo, H.-y.; Wang, H.; Wang, L.; Zhang, X.; Hua, S.-n.; Yu, J.; Xiao, P.-g. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005, 67, 18–23. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, Y.I.; Park, S.J.; Kim, I.K.; Choi, Y.K.; Kim, S.-H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents 2018, 52, 730. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Mao, R.; Qiu, Y.; He, J.-S.; Tan, J.-Y.; Li, X.-H.; Liang, J.; Shen, J.; Zhu, L.-R.; Chen, Y.; Iacucci, M. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- Van Norman, G.A. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC: Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; De Clercq, E. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV); Nature Publishing Group: Berlin, Germany, 2020. [Google Scholar]
- Savarino, A.; Di Trani, L.; Donatelli, I.; Cauda, R.; Cassone, A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006, 6, 67–69. [Google Scholar] [CrossRef]
- Yan, Y.; Zou, Z.; Sun, Y.; Li, X.; Xu, K.-F.; Wei, Y.; Jin, N.; Jiang, C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013, 23, 300–302. [Google Scholar] [CrossRef] [Green Version]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases. Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Mizui, T.; Yamashina, S.; Tanida, I.; Takei, Y.; Ueno, T.; Sakamoto, N.; Ikejima, K.; Kitamura, T.; Enomoto, N.; Sakai, T. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J. Gastroenterol. 2010, 45, 195–203. [Google Scholar] [CrossRef]
- Farias, K.J.S.; Machado, P.R.L.; De Almeida Junior, R.F.; De Aquino, A.A.; Da Fonseca, B.A.L. Chloroquine interferes with dengue-2 virus replication in U937 cells. Microbiol. Immunol. 2014, 58, 318–326. [Google Scholar] [CrossRef]
- Delvecchio, R.; Higa, L.M.; Pezzuto, P.; Valadão, A.L.; Garcez, P.P.; Monteiro, F.L.; Loiola, E.C.; Dias, A.A.; Silva, F.J.; Aliota, M.T. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses 2016, 8, 322. [Google Scholar] [CrossRef] [Green Version]
- Dowall, S.D.; Bosworth, A.; Watson, R.; Bewley, K.; Taylor, I.; Rayner, E.; Hunter, L.; Pearson, G.; Easterbrook, L.; Pitman, J. Chloroquine inhibited Ebola virus replication in vitro but failed to protect against infection and disease in the in vivo guinea pig model. J. Gen. Virol. 2015, 96, 3484. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef] [Green Version]
- Fiolet, T.; Guihur, A.; Rebeaud, M.; Mulot, M.; Peiffer-Smadja, N.; Mahamat-Saleh, Y. Effect of hydroxychloroquine with or without azithromycin on the mortality of COVID-19 patients: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2020, 27, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Roustit, M.; Guilhaumou, R.; Molimard, M.; Drici, M.D.; Laporte, S.; Montastruc, J.L. Chloroquine and hydroxychloroquine in the management of COVID-19: Much kerfuffle but little evidence. Therapies 2020, 75, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E.; et al. Hydroxychloroquine in patients mainly with mild to moderate COVID-19: An open-label, randomized, controlled trial. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Borba, M.; De Almeida Val, F.; Sampaio, V.S.; Alexandre, M.A.; Melo, G.C.; Brito, M.; Mourao, M.; Sousa, J.D.B.; Guerra, M.V.F.; Hajjar, L.; et al. Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with severe respiratory syndrome in the context of coronavirus (SARS-CoV-2) infection: Preliminary safety results of a randomized, double-blinded, phase IIb clinical trial (CloroCovid-19 Study). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Guaraldi, G.; Meschiari, M.; Cozzi-Lepri, A.; Milic, J.; Tonelli, R.; Menozzi, M.; Franceschini, E.; Cuomo, G.; Orlando, G.; Borghi, V.; et al. Tocilizumab in patients with severe COVID-19: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e474–e484. [Google Scholar] [CrossRef]
- Cortegiani, A.; Ippolito, M.; Greco, M.; Granone, V.; Protti, A.; Gregoretti, C.; Giarratano, A.; Einav, S.; Cecconi, M. Rationale and evidence on the use of tocilizumab in COVID-19: A systematic review. Pulmonology 2020, 27, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Della-Torre, E.; Campochiaro, C.; Cavalli, G.; De Luca, G.; Napolitano, A.; La Marca, S.; Boffini, N.; Da Prat, V.; Di Terlizzi, G.; Lanzillotta, M.; et al. Interleukin-6 blockade with sarilumab in severe COVID-19 pneumonia with systemic hyperinflammation: An open-label cohort study. Ann. Rheum. Dis. 2020, 79, 1277–1285. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Bloch, E.M.; Shoham, S.; Casadevall, A.; Sachais, B.S.; Shaz, B.; Winters, J.L.; Van Buskirk, C.; Grossman, B.J.; Joyner, M.; Henderson, J.P. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020, 130, 2757–2765. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob. Agents Chemother. 2020, 64, e00754-20. [Google Scholar] [CrossRef] [Green Version]
- Shalhoub, S. Interferon beta-1b for COVID-19. Lancet 2020, 395, 1670–1671. [Google Scholar] [CrossRef]
- Group, T.R.C. Dexamethasone in Hospitalized Patients with Covid-19—Preliminary Report. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S. Remdesivir for the treatment of Covid-19—Preliminary report. N. Engl. J. Med. 2020, 383, 1813–1836. [Google Scholar] [CrossRef]
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL M(pro) active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Sztain, T.; Amaro, R.; McCammon, J.A. Elucidation of Cryptic and Allosteric Pockets within the SARS-CoV-2 Main Protease. J. Chem. Inf. Modeling 2021, 61, 3495–3501. [Google Scholar] [CrossRef]
- Dubanevics, I.; McLeish, T.C. Computational analysis of dynamic allostery and control in the SARS-CoV-2 main protease. J. R. Soc. Interface 2021, 18, 20200591. [Google Scholar] [CrossRef]
- Goyal, B.; Goyal, D. Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy. ACS Comb. Sci. 2020, 22, 297–305. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Bekes, M.; Huang, T.T.; Drag, M. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Lampi, G.; Deidda, D.; Pinza, M.; Pompei, R. Enhancement of anti-herpetic activity of glycyrrhizic acid by physiological proteins. Antivir. Chem. Chemother. 2001, 12, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.C. Mechanism of action of glycyrrhizic acid in inhibition of Epstein-Barr virus replication in vitro. Antivir. Res. 2003, 59, 41–47. [Google Scholar] [CrossRef]
- Crance, J.M.; Scaramozzino, N.; Jouan, A.; Garin, D. Interferon, ribavirin, 6-azauridine and glycyrrhizin: Antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 2003, 58, 73–79. [Google Scholar] [CrossRef]
- Sasaki, H.; Takei, M.; Kobayashi, M.; Pollard, R.B.; Suzuki, F. Effect of glycyrrhizin, an active component of licorice roots, on HIV replication in cultures of peripheral blood mononuclear cells from HIV-seropositive patients. Pathobiology 2002, 70, 229–236. [Google Scholar] [CrossRef]
- Miyake, K.; Tango, T.; Ota, Y.; Mitamura, K.; Yoshiba, M.; Kako, M.; Hayashi, S.; Ikeda, Y.; Hayashida, N.; Iwabuchi, S.; et al. Efficacy of Stronger Neo-Minophagen C compared between two doses administered three times a week on patients with chronic viral hepatitis. J. Gastroenterol. Hepatol. 2002, 17, 1198–1204. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Ogura, M.; Fujimoto, E.; Shono, S.; Okuda, E. Effects and cost of glycyrrhizin in the treatment of upper respiratory tract infections in members of the Japanese maritime self-defense force: Preliminary report of a prospective, randomized, double-blind, controlled, parallel-group, alternate-day treatment assignment clinical trial. Curr. Res. Clin. Exp. 2004, 65, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.T.; Lu, H.T.; Fan, K.W.; Cheng, V.C.C.; Tsui, W.H.W.; Hung, I.F.N.; Lee, T.S.W. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hoever, G.; Baltina, L.; Michaelis, M.; Kondratenko, R.; Baltina, L.; Tolstikov, G.A.; Doerr, H.W.; Cinatl, J. Antiviral activity of glycyrrhizic acid derivatives against SARS−coronavirus. J. Med. Chem. 2005, 48, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef]
- Kandeil, A.; Mostafa, A.; Kutkat, O.; Moatasim, Y.; Al-Karmalawy, A.A.; Rashad, A.A.; Kayed, A.E.; Kayed, A.E.; El-Shesheny, R.; Kayali, G.; et al. Bioactive Polyphenolic Compounds Showing Strong Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus 2. Pathogens 2021, 10, 758. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Suarez, D.; Diaz, N. SARS-CoV-2 Main Protease: A Molecular Dynamics Study. J. Chem. Inf. Model. 2020, 60, 5815–5831. [Google Scholar] [CrossRef]
- Das, S.; Sarmah, S.; Lyndem, S.; Singha Roy, A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020, 39, 3347–3357. [Google Scholar] [CrossRef]
- Srivastava, V.; Yadav, A.; Sarkar, P. Molecular docking and ADMET study of bioactive compounds of Glycyrrhiza glabra against main protease of SARS-CoV2. Mater. Today: Proc. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chandel, V.; Raj, S.; Rathi, B.; Kumar, D. In Silico Identification of Potent COVID-19 Main Protease Inhibitors from FDA Approved Antiviral Compounds and Active Phytochemicals through Molecular Docking: A Drug Repurposing Approach. Chem. Biol. Lett. 2020. [Google Scholar] [CrossRef]
- Singh, A.; Mishra, A. Leucoefdin a potential inhibitor against SARS CoV-2 Mpro. J. Biomol. Struct. Dyn. 2020, 39, 4427–4432. [Google Scholar] [CrossRef] [PubMed]
- Mittal, L.; Kumari, A.; Srivastava, M.; Singh, M.; Asthana, S. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J. Biomol. Struct. Dyn. 2020, 39, 3662–3680. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, B.; Grande, F.; Conforti, F.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Ortega-Alarcon, D.; Vega, S.; Reyburn, H.T.; Abian, O.; Velazquez-Campoy, A. Rutin Is a Low Micromolar Inhibitor of SARS-CoV-2 Main Protease 3CLpro: Implications for Drug Design of Quercetin Analogs. Biomedicines 2021, 9, 375. [Google Scholar] [CrossRef]
- Ngo, S.T.; Quynh Anh Pham, N.; Thi Le, L.; Pham, D.-H.; Vu, V.V. Computational determination of potential inhibitors of SARS-CoV-2 main protease. J. Chem. Inf. Modeling 2020. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Van de Sand, L.; Bormann, M.; Alt, M.; Schipper, L.; Heilingloh, C.S.; Steinmann, E.; Todt, D.; Dittmer, U.; Elsner, C.; Witzke, O. Glycyrrhizin Effectively Inhibits SARS-CoV-2 Replication by Inhibiting the Viral Main Protease. Viruses 2021, 13, 609. [Google Scholar] [CrossRef]
- Yuce, M.; Cicek, E.; Inan, T.; Dag, A.B.; Kurkcuoglu, O.; Sungur, F.A. Repurposing of FDA-approved drugs against active site and potential allosteric drug-binding sites of COVID-19 main protease. Proteins: Struct. Funct. Bioinform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Du, X.; Duan, Y.; Pan, X.; Sun, Y.; You, T.; Han, L.; Jin, Z.; Shang, W.; Yu, J.; et al. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Parmar, P.; Rao, P.; Sharma, A.; Shukla, A.; Rawal, R.M.; Saraf, M.; Patel, B.V.; Goswami, D. Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS-CoV2 papain-like protease (PLpro) using rigorous computational workflow. Mol. Divers. 2021. [Google Scholar] [CrossRef]
- Li, D.; Luan, J.; Zhang, L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochem. Biophys. Res. Commun. 2021, 538, 72–79. [Google Scholar] [CrossRef]
- Mitra, D.; Verma, D.; Mahakur, B.; Kamboj, A.; Srivastava, R.; Gupta, S.; Pandey, A.; Arora, B.; Pant, K.; Panneerselvam, P.; et al. Molecular docking and simulation studies of natural compounds of Vitex negundo L. against papain-like protease (PLpro) of SARS CoV-2 (coronavirus) to conquer the pandemic situation in the world. J. Biomol. Struct. Dyn. 2021. [Google Scholar] [CrossRef] [PubMed]
- Jamalan, M.; Barzegari, E.; Gholami-Borujeni, F. Structure-Based Screening to Discover New Inhibitors for Papain-like Proteinase of SARS-CoV-2: An In Silico Study. J. Proteome Res. 2021, 20, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Delre, P.; Caporuscio, F.; Saviano, M.; Mangiatordi, G.F. Repurposing Known Drugs as Covalent and Non-covalent Inhibitors of the SARS-CoV-2 Papain-Like Protease. Front. Chem. 2020, 8, 594009. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Singh, H. In silico identification and docking-based drug repurposing against the main protease of SARS-CoV-2, causative agent of COVID-19. Biol. Med. Chem. 2020, 13, 1210–1223. [Google Scholar] [CrossRef]
- Surti, M.; Patel, M.; Adnan, M.; Moin, A.; Ashraf, S.A.; Siddiqui, A.J.; Snoussi, M.; Deshpande, S.; Reddy, M.N. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: Designing, docking and molecular dynamics simulation study. RSC Adv. 2020, 10, 37707–37720. [Google Scholar] [CrossRef]
- Laskar, M.A.; Choudhury, M.D. Search for therapeutics against COVID 19 targeting SARS-CoV-2 papain-like protease: An in silico study. Res. Square 2020, Preprint. [Google Scholar] [CrossRef]
- Moghaddam, E.; Teoh, B.-T.; Sam, S.-S.; Lani, R.; Hassandarvish, P.; Chik, Z.; Yueh, A.; Abubakar, S.; Zandi, K. Baicalin, a metabolite of baicalein with antiviral activity against dengue virus. Sci. Rep. 2014, 4, 5452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayak, M.K.; Agrawal, A.S.; Bose, S.; Naskar, S.; Bhowmick, R.; Chakrabarti, S.; Sarkar, S.; Chawla-Sarkar, M. Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 2014, 69, 1298–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oo, A.; Teoh, B.T.; Sam, S.S.; Bakar, S.A.; Zandi, K. Baicalein and baicalin as Zika virus inhibitors. Arch. Virol. 2019, 164, 585–593. [Google Scholar] [CrossRef]
- Oo, A.; Rausalu, K.; Merits, A.; Higgs, S.; Vanlandingham, D.; Bakar, S.A.; Zandi, K. Deciphering the potential of baicalin as an antiviral agent for Chikungunya virus infection. Antivir. Res. 2018, 150, 101–111. [Google Scholar] [CrossRef]
- Kitamura, K.; Honda, M.; Yoshizaki, H.; Yamamoto, S.; Nakane, H.; Fukushima, M.; Ono, K.; Tokunaga, T. Baicalin, an inhibitor of HIV-1 production in vitro. Antivir. Res. 1998, 37, 131–140. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Rameshkumar, M.R.; Indu, P.; Arunagirinathan, N.; Venkatadri, B.; El-Serehy, H.A.; Ahmad, A. Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins: A molecular docking study. Saudi J. Biol. Sci. 2021, 28, 448–458. [Google Scholar] [CrossRef]
- Singh, S.; Sk, M.F.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Mishra, A.; Rathore, A.S. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J. Biomol. Struct. Dyn. 2021. [Google Scholar] [CrossRef]
- Arba, M.; Wahyudi, S.T.; Brunt, D.J.; Paradis, N.; Wu, C. Mechanistic insight on the remdesivir binding to RNA-Dependent RNA polymerase (RdRp) of SARS-cov-2. Comput. Biol. Med. 2021, 129, 104156. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.S.A.; Karim, M.A.; Hasan, M.; Jaman, J.; Karim, Z.; Tahsin, T.; Hasan, M.N.; Hosen, M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 2020, 163, 1787–1797. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.S.; Sushma, P.; Dharmashekar, C.; Beelagi, M.S.; Prasad, S.K.; Shivamallu, C.; Prasad, A.; Syed, A.; Marraiki, N.; Prasad, K.S. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J. Biol. Sci. 2021, 28, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhu, Y.; Xu, J.; Yao, G.; Zhang, P.; Wang, M.; Zhao, Y.; Lin, G.; Chen, H.; Chen, L.; et al. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine 2021, 85, 153364. [Google Scholar] [CrossRef]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L. Emergence of SARS-CoV-2 b. 1.1. 7 lineage—United States, December 29, 2020–January 12, 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 95. [Google Scholar] [CrossRef]
- Paiva, M.H.S.; Guedes, D.R.D.; Docena, C.; Bezerra, M.F.; Dezordi, F.Z.; Machado, L.C.; Krokovsky, L.; Helvecio, E.; Da Silva, A.F.; Vasconcelos, L.R.S. Multiple Introductions Followed by Ongoing Community Spread of SARS-CoV-2 at One of the Largest Metropolitan Areas of Northeast Brazil. Viruses 2020, 12, 1414. [Google Scholar] [CrossRef] [PubMed]
- Mwenda, M.; Saasa, N.; Sinyange, N.; Busby, G.; Chipimo, P.J.; Hendry, J.; Kapona, O.; Yingst, S.; Hines, J.Z.; Minchella, P.; et al. Detection of B.1.351 SARS-CoV-2 Variant Strain—Zambia, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Mujwar, S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput. Biol. Med. 2021, 136, 104748. [Google Scholar] [CrossRef] [PubMed]
- Mhatre, S.; Gurav, N.; Shah, M.; Patravale, V. Entry-inhibitory role of catechins against SARS-CoV-2 and its UK variant. Comput. Biol. Med. 2021, 135, 104560. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.U.; Froeyen, M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharm. Anal. 2020, 10, 320–328. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef] [Green Version]
- Rowaiye, A.; Onuh, O.; Asala, T.; Ogu, A.; Bur, D.; Nwankwo, E.; Orji, U.; Ibrahim, Z.; Hamza, J.; Ugorji, A. In silico identification of potential allosteric inhibitors of the SARS-CoV-2 Helicase. ChemRxiv. Prepr. 2020. [Google Scholar] [CrossRef]
- Vivek-Ananth, R.P.; Krishnaswamy, S.; Samal, A. Potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13: A molecular docking and dynamic simulation study. Mol. Divers. 2021, 1–14. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, S.; Wu, Y.; Cheng, X.; Zhang, L.-k.; Shen, X.-r.; Li, S.-q.; Xu, J.-r.; Shang, W.-j.; Gao, Z.-b.; et al. Discovery of SARS-CoV-2-E channel inhibitors as antiviral candidates. Acta Pharmacol. Sin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [Green Version]
- Bilal, S.; Hassan, M.M.; Ur Rehman, M.F.; Nasir, M.; Sami, A.J.; Hayat, A. An insect acetylcholinesterase biosensor utilizing WO3/g-C3N4 nanocomposite modified pencil graphite electrode for phosmet detection in stored grains. Food Chem. 2021, 346, 128894. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem Infor. Modeling. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Fernandez-Poza, S.; Padros, A.; Thompson, R.; Butler, L.; Islam, M.; Mosely, J.; Scrivens, J.H.; Rehman, M.F.; Akram, M.S. Tailor-made recombinant prokaryotic lectins for characterisation of glycoproteins. Anal. Chim. Acta 2021, 1155, 338352. [Google Scholar] [CrossRef]
- Motulsky, H. Prism 4 Statistics Guide—Statistical Analyses for Laboratory and Clinical Researchers; GraphPad Softw. Inc.: San Diego, CA, USA, 2003; pp. 122–126. [Google Scholar]
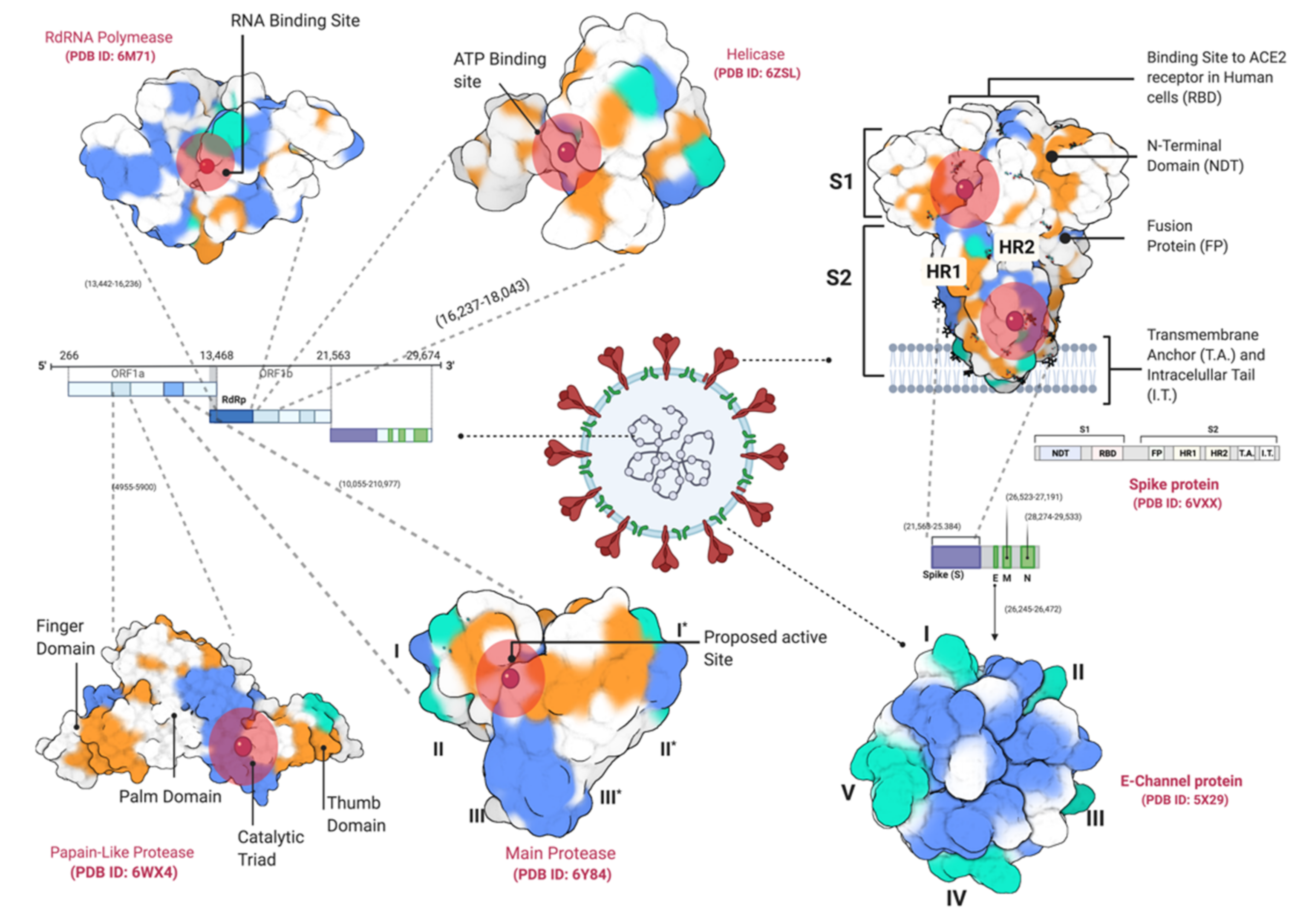












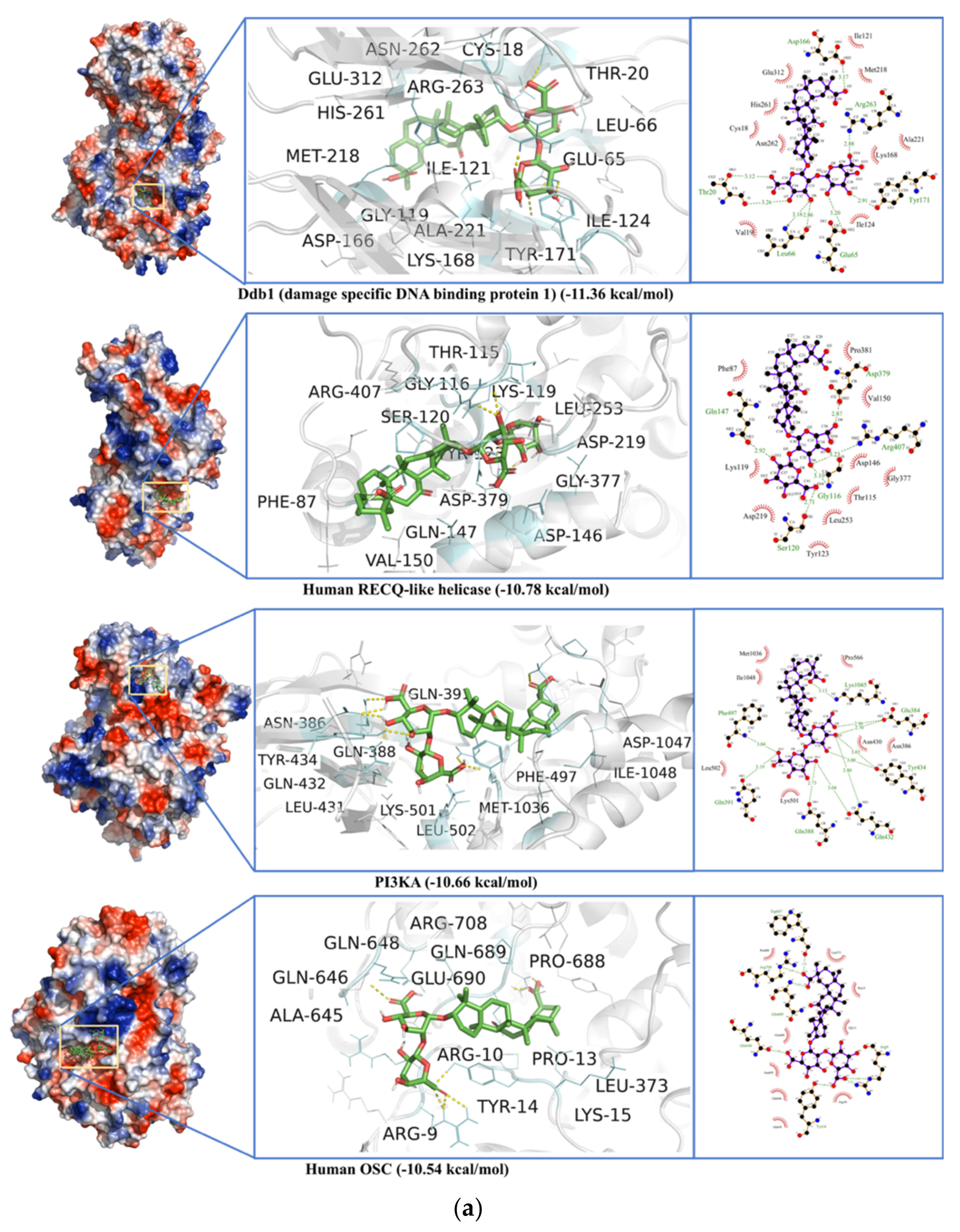




| Target | Phytochemicals | Binding Energy (kcal/mol) | Dissociation Constant (µM) | Active Site Residue |
|---|---|---|---|---|
| Main Protease (MPro) | Glycyrrhizin | −9.57 | 0.11 | Thr24, Thr25, Thr26, Leu27, Gly29, His41, Cys44, Ser46, Met49, Tyr118, Asn119, Asn142, Gly143, Cys145, His163, His164, Met165, Glu166 |
| −9.46 | 0.76 | Arg131, Asn133, Thr135, Val137, Thr169, Val171, Ala194, Gly195, Thr196, Asp197, Thr198, Thr199, Tyr237, Asn238, Tyr239, Leu272, Leu286, Leu287, Asp289 | ||
| 18,β-Glycyrrhetinic acid | −9.19 | 0.35 | Lys5, Arg131, Lys137, Asp197, Thr199, Tyr237, Tyr239, Leu272, Leu286, Leu287, Glu288, Asp289, Glu290 | |
| Rhodiolin | −9.05 | 0.23 | Thr25, His41, Cys44, Thr45, Ser46, Met49, Leu141, Asn142, Gly143, Ser144, Cys145, His163, Met165, Glu166, Leu167, Pro168, Arg188, Gln189, Thr190, Gln192 | |
| Baicalin | −8.85 | 0.33 | Val104, Ile106, Gln110, Thr111, Asn151, Ile152, Asp153, Tyr154, Pro252, Thr292, Phe294, Asp295, Val297, Arg298, Val303 | |
| Silymarin | −8.71 | 0.41 | Thr24, Thr25, Thr26, Leu27, His41, Cys44, Thr45, Ser46, Met49, Gly143, Cys145, His164, Met165, Glu166, Leu167, Pro168, Arg188, Gln189, Thr190, Gln192 | |
| Papain like protease (PLPro) | Baicalin | −10.82 | 0.01 | Cys155, Asn156, Lys157, Glu161, Leu162, Gly163, Asp164, Val165, Arg166, Glu167, Tyr171, Val202, Met206, Met208, Pro248, Tyr264, Tyr268, Gln269, Tyr273 |
| Hesperidin | −10.61 | 0.02 | Lys157, Thr158, Glu161, Leu162, Gly163, Asp164, Val165, Glu167, Leu199, Glu203, Tyr207, Met208, Lys232, Pro248, Tyr264, Tyr268, Gln269, Cys270, Tyr273, Thr301 | |
| Naringen | −10.17 | 0.04 | Cys155, Asn156, Glu161, Leu162, Gly163, Asp164, Arg166, Gln167, Ser170, Leu185, Leu199, Val202, Glu203, Met206, Tyr207, Met208, Ile222, Pro223, Lys232, Tyr268 | |
| Flemiflavanone D | −10.07 | 0.04 | Lys157, Glu161, Leu162, Gly163, Asp164, Arg166, Glu167, Ser170, Val202, Glu203, Met206, Tyr207, Met208, Tyr264, Tyr268, Gln269, Tyr273 | |
| Euchrestaflavanone A | −9.95 | 0.05 | Lys157, Glu161, Leu162, Gly163, Asp164, Arg166, Glu167, Ser170, Val202, Glu203, Met206, Tyr207, Met208, Tyr268 | |
| RNA-dependent RNA polymerase (RdRP) | Glycyrrhizin | −10.52 | 0.03 | Asp452, Tyr455, Lys551, Arg553, Ala554, Arg555, Thr556, Trp617, Asp618, Tyr619, Pro620, Lys 621, Cys622, Asp623, Arg624, Ser759, Asp760, Asp761, Ala762, Lys798, Cys799, Trp800, Glu811, Phe812, Cys813 Ser814 |
| −9.96 | 0.05 | Asp452, Tyr455, Arg553, Ala554, Arg555, Trp617, Asp618, Lys621, Cys622, Asp623, Arg624, Ser759, Asp760, Asp761, Lys798, Trp800, Glu811, Cys813, Ser814 | ||
| Hesperidin | −9.53 | 0.1 | Val166, Tyr456, Met542, Arg553, Ala554, Arg555, Thr556, Val557, Ala558, Asp618, Tyr619, Pro620, Lys621, Cys622, Asp623, Arg624, Lys676, Thr680, Ser681, Ser682, Phe793, Ser795, Lys798 | |
| Baicalin | −9.01 | 0.25 | Val31, Tyr32, Lys47, Tyr129, Ala130, His133, Phe134, Asp135, Asn138, Cys139, Thr141, Asn705, Ala706, Ser709, Thr710, Lys780, Asn781, Ser784 | |
| Naringen | −8.54 | 0.55 | Tyr32, Lys47, Tyr129, His133, Phe134, Asp135, Asn138, Ser709, Thr710, Asp711, Lys714, Ala771, Ser772, Gln773, Gly774, Ser778, Lys780, Asn781, Ser784 | |
| Oleuropein | −8.31 | 0.81 | Tyr32, Lys47, Phe48, Tyr129, Ala130, His133, Phe134, Asp135, Asn138, Cys139, Asp140, Thr141, Leu142, Thr710, Asp711, Lys714, Ser778, Lys780, Asn781, Ser784 | |
| Spike protein (S) | Glycyrrhizin | −9.49 | 0.11 | Tyr741, Ile742, Cys743, Gly744, Asp745, Phe855, Asn856, Val963, Lys964, Leu966, Ser967, Ser975, Val976, Leu977, Asn978, Arg1000 |
| −9.29 | 0.16 | Val47, His49, Lys304, Met740, Tyr741, Ile742, Cys743, Gly744, Asp745, Phe855, Asn856, Val963, Lys964, Leu966, Ser967, Ser975, Val976, Leu977, Asn978, Arg1000 | ||
| Rhodiolin | −8.68 | 0.43 | Arg102, Gly103, Trp104, Ile119, Asn121, Val126, Ile128, Phe168, Tyr170, Ser172, Arg190, Phe192, Ile203, His207, Leu226, Val227, Asp228, Leu229 | |
| Hesperidin | −8.53 | 0.56 | Tyr38, Asp40, Lys41, Val42, Phe43, Arg44, Lys206, Phe220, Ser221, Ala222, Glu224, Pro225, Leu226, Tyr279, Gly283, Thr284 | |
| −8.18 | 1.01 | Asn710, Thr1076, Ser1097, Gly1099, Thr1100, His1101, Trp1102, Ile1114, Ile1115, Val1133, Asn1134, Asn1135, Thr1136, Tyr1138 | ||
| Umbelliprenin | −8.12 | 1.11 | Trp104, Ile119, Asn121, Val126, Ile128, Phe168, Tyr170, Ser172, Arg190, Phe192, Phe194, Ile203, Ser205, Lys206, His207, Leu226, Val227, Leu229 | |
| 18,β-Glycyrrhetinic acid | −8.08 | 1.2 | Tyr38, Asp40, Lys41, Val42, Phe43, Arg44, Glu224, Pro225, Tyr279, Gly283 | |
| −7.99 | 1.38 | His1101, Trp1102, Phe1103, Ile1114, Ile1115, Asn1135, Thr1136, Val1137, Tyr1138, Asp1139, Gln1142, Pro1143 | ||
| Silyhermin | −8.05 | 1.25 | Arg355, Tyr396, Pro426, Asp428, Phe429, Thr430, Lys462, Pro463, Phe464, Ser514, Phe515, Glu516, Leu517, Leu518 | |
| Ellagic acid | −8.1 | 1.16 | Ser730, Met731, Thr732, Lys733, Gln774, Thr778, Phe823, Val860, Leu861, Pro862, Pro863, Asp867, Ile870, Ala1056, Pro1057, His1058, Gly1059 | |
| Helicase | Glycyrrhizin | −11.57 | 0.003 | Pro175, Leu176, Asn177, Lys202, Leu405, Pro406, Ala407, Pro408, Arg409, Leu412, Thr413, Gly415, Thr416, Leu417, Phe422, Ser485, Ser486, Pro514, Tyr515, Asn516, Asn519, Thr532, Val533, Asp534, His554, Asn557, Asn559, Arg560 |
| 18,β-Glycyrrhetinic acid | −9.91 | 0.054 | Ala4, Val6, Arg15, Arg21, Arg22, Pro23, Phe24, Arg129, Leu132, Phe133, Glu136, Pro234, Leu235, Ser236 | |
| Solophenol A | −9.4 | 0.13 | Pro175, Leu176, Asn177, Arg178, Thr199, Phe200, Glu201, Lys202, Gly203, Asp204, Val210, Tyr211, Arg212, Val484, Ser486, Asn516, Ser517, Asn519, Ala520, Thr530, Thr532, Asp534 | |
| Hesperidin | −8.93 | 0.283 | Lys139, Glu142, Glu143, Lys146, Arg178, Asn179, Val181, Glu197, Thr228, His230, Cys309, Arg337, Arg339, Met378, Ala379, Thr380, Tyr382, Asp383, Ala407, Pro408, Thr410 | |
| Baicalin | −8.9 | 0.29 | Asn177, Arg178, Thr199, Phe200, Glu201, Lys202, Arg212, Asn516, Asn519, Ala520, Thr530, Gln531, Thr532, Asp534 | |
| E-channel protein | Glycyrrhizin | −10.07 | 0.04 | Arg61, Asn64, leu28, val29, leu31, ala32, ile33, ala36, arg38, Leu27, Thr30, Leu31, Leu34, Leu37, Leu39, Tyr42, Cys43, Ile46, Val47, Val49, Ser50, Leu51, Pro54, Tyr57 |
| 18,β-Glycyrrhetinic acid | −9.72 | 0.07 | Arg61, Leu28, Val29, Leu31, Ala32, Thr35, Leu27, Thr30, Leu31, Ile46, Val47, Leu51, Pro54, Tyr57 | |
| Umbelliprenin | −8.90 | 0.3 | Ala22, Val25, Phe26, Leu28, Val29, Leu19, Phe20, Ala22, Phe23, Val24, Phe26, Leu27, Asn64, Leu65 | |
| Euchrestaflavanone A | −8.90 | 0.3 | Arg61, Asn64dval25dphe26dleu28dval29, Ala22, Phe23, Phe26, Leu27, Thr30, Leu34, Cys43, Ile46, Val47, Val49, Ser50, Leu51 | |
| Baicalin | −8.89 | 0.3 | Ala22, Val25, Phe26, Val29, Phe20, Ala22, Phe23, Val24, Phe26, Leu27, Thr30, Arg61, Asn64, Leu65 | |
| Silibinin A | −8.13 | 1.1 | Leu28, Leu31, Ala32, Thr35, Cys40, Ile46, Ser50, Leu51, Lys53, Pro54, Phe56, Tyr57, Tyr59, Ser60 |
| Compounds | Molecular Formula | Molecular Weight | HBA | HBD | TPSA | Log P−0.7–5 | Log S 0–6 | GI Absorption | BBB Permeant | CYPP1A2 Inhibitor | CYP2D6 Inhibitor | Log Kp (Skin Permeation), cm/s | Lipinski Violations | Ghose Violations | Veber Violations | Bioavailability Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glycyrrhizin | C42H62O16 | 822.93 | 16 | 8 | 267.04 | 1.49 | −6.24 | Low | No | No | No | −9.33 | 3 MW > 500, NorO > 10, NHorOH > 5 | 3 MW > 480, MR > 130, atoms > 70 | 1 TPSA > 140 | 0.11 |
| 18,β Glycyrrhetinic acid | C30H46O5 | 470.68 | 4 | 2 | 74.6 | 5.13 | −6.15 | High | No | No | No | −4.27 | 1 MLOGP.4.16 | 3 WLOGP > 5.6, MR > 130. | Yes | 1.56 |
| Rhodiolin | C25H20O10 | 480.42 | 10 | 5 | 159.05 | 2.3 | −4.99 | Low | No | No | No | −7.02 | 0 | 1 MW > 480 | 1 TPSA > 140 | 0.55 |
| Baicalin | C21H18O11 | 446.36 | 11 | 6 | 187.12 | 0.25 | −3.41 | Low | No | No | No | −8.23 | 3 NorO.10, NHorOH > 5 | Yes | 1 TPSA > 140 | 0.11 |
| Hesperidin | C28H34O15 | 610.56 | 15 | 8 | 234.29 | −1.06 | −3.28 | Low | No | No | No | −10.12 | 3 MW > 500, NorO > 10, NHorOH > 5 | 4 MW > 480, WLOGP < 0.4, MR > 130, #atoms > 70 | 1 TPSA > 140 | 0.17 |
| Solophenol A | C30H36O6 | 492.6 | 6 | 4 | 107.22 | 5.69 | −7.5 | Low | No | No | No | −3.8 | 0 | 4 MW > 480, WLOGP < 0.4, MR > 130, #atoms > 71 | 1 TPSA > 140 | 0.55 |
| Naringin | C26H30O14 | 566.51 | 14 | 8 | 225.06 | −0.99 | −2.68 | Low | No | No | No | −10.39 | 3 MW > 500, NorO > 10, NHorOH > 5 | 3MW > 480, WLOGP < 0.4 MR > 130, | 1 TPSA > 140 | 0.17 |
| Lopinavir | C37H48N4O5 | 628.8 | 5 | 4 | 120 | 4.37 | −6.64 | High | No | No | No | −5.93 | 1 MW > 500 | 3 MW > 480, MR > 130, #atoms > 70 | 1 Rotors > 10 | 0.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, M.F.u.; Akhter, S.; Batool, A.I.; Selamoglu, Z.; Sevindik, M.; Eman, R.; Mustaqeem, M.; Akram, M.S.; Kanwal, F.; Lu, C.; et al. Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics 2021, 10, 1011. https://doi.org/10.3390/antibiotics10081011
Rehman MFu, Akhter S, Batool AI, Selamoglu Z, Sevindik M, Eman R, Mustaqeem M, Akram MS, Kanwal F, Lu C, et al. Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics. 2021; 10(8):1011. https://doi.org/10.3390/antibiotics10081011
Chicago/Turabian StyleRehman, Muhammad Fayyaz ur, Shahzaib Akhter, Aima Iram Batool, Zeliha Selamoglu, Mustafa Sevindik, Rida Eman, Muhammad Mustaqeem, Muhammad Safwan Akram, Fariha Kanwal, Changrui Lu, and et al. 2021. "Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World" Antibiotics 10, no. 8: 1011. https://doi.org/10.3390/antibiotics10081011
APA StyleRehman, M. F. u., Akhter, S., Batool, A. I., Selamoglu, Z., Sevindik, M., Eman, R., Mustaqeem, M., Akram, M. S., Kanwal, F., Lu, C., & Aslam, M. (2021). Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics, 10(8), 1011. https://doi.org/10.3390/antibiotics10081011










