Bacteriophage SRD2021 Recognizing Capsular Polysaccharide Shows Therapeutic Potential in Serotype K47 Klebsiella pneumoniae Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Bacteriophage Isolation and Purification
2.3. Phage Characterization Assay
2.3.1. Host Range Determination
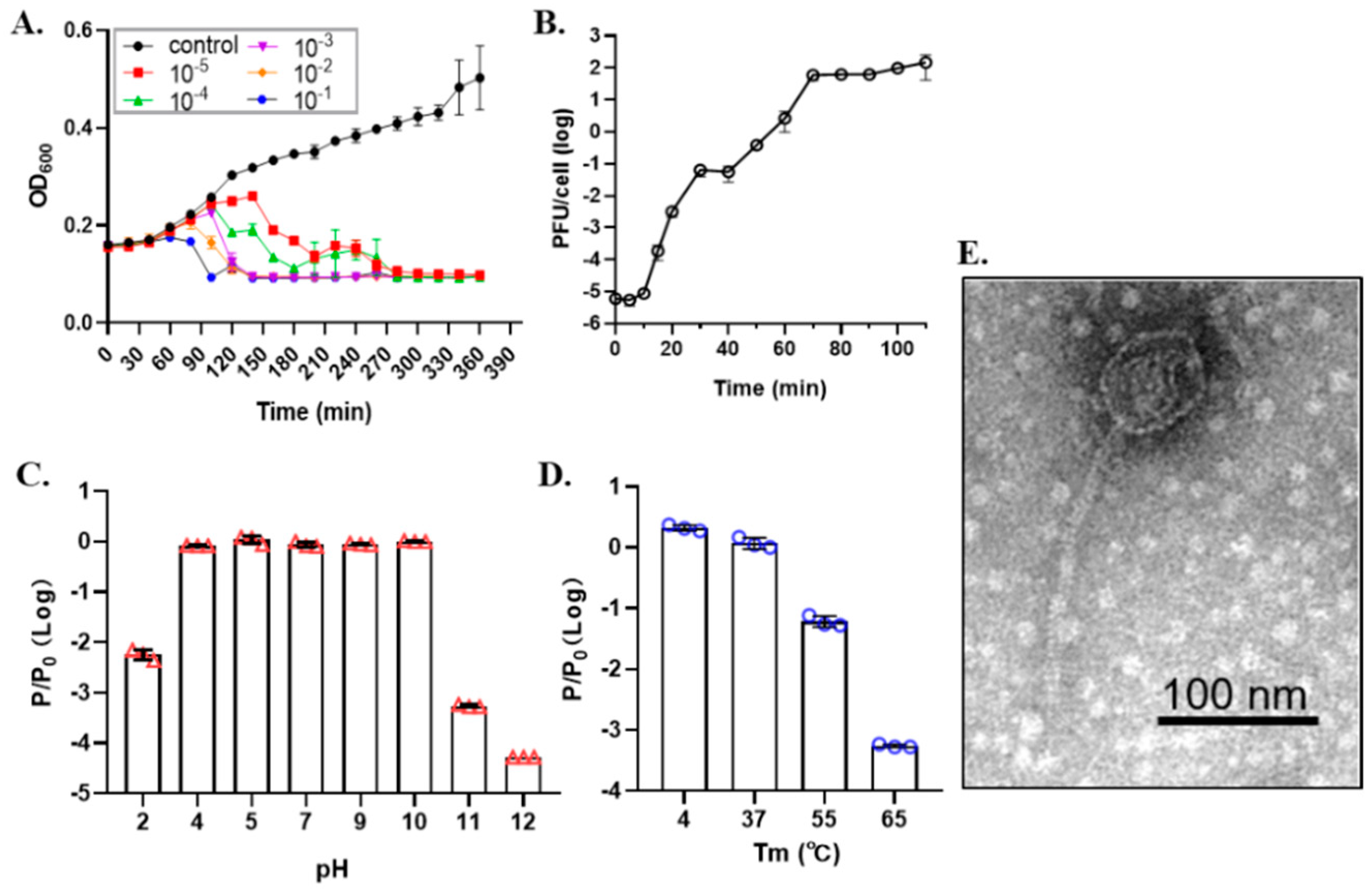
2.3.2. One-Step Growth Curve and Stability Test
2.3.3. Transmission Electron Microscopy
2.4. Bacteriophage Adsorption and Lytic Activity Assays
2.5. Phage Genomic Sequencing and Bioinformatics Analysis
2.6. Phage Resistant Mutants Screening
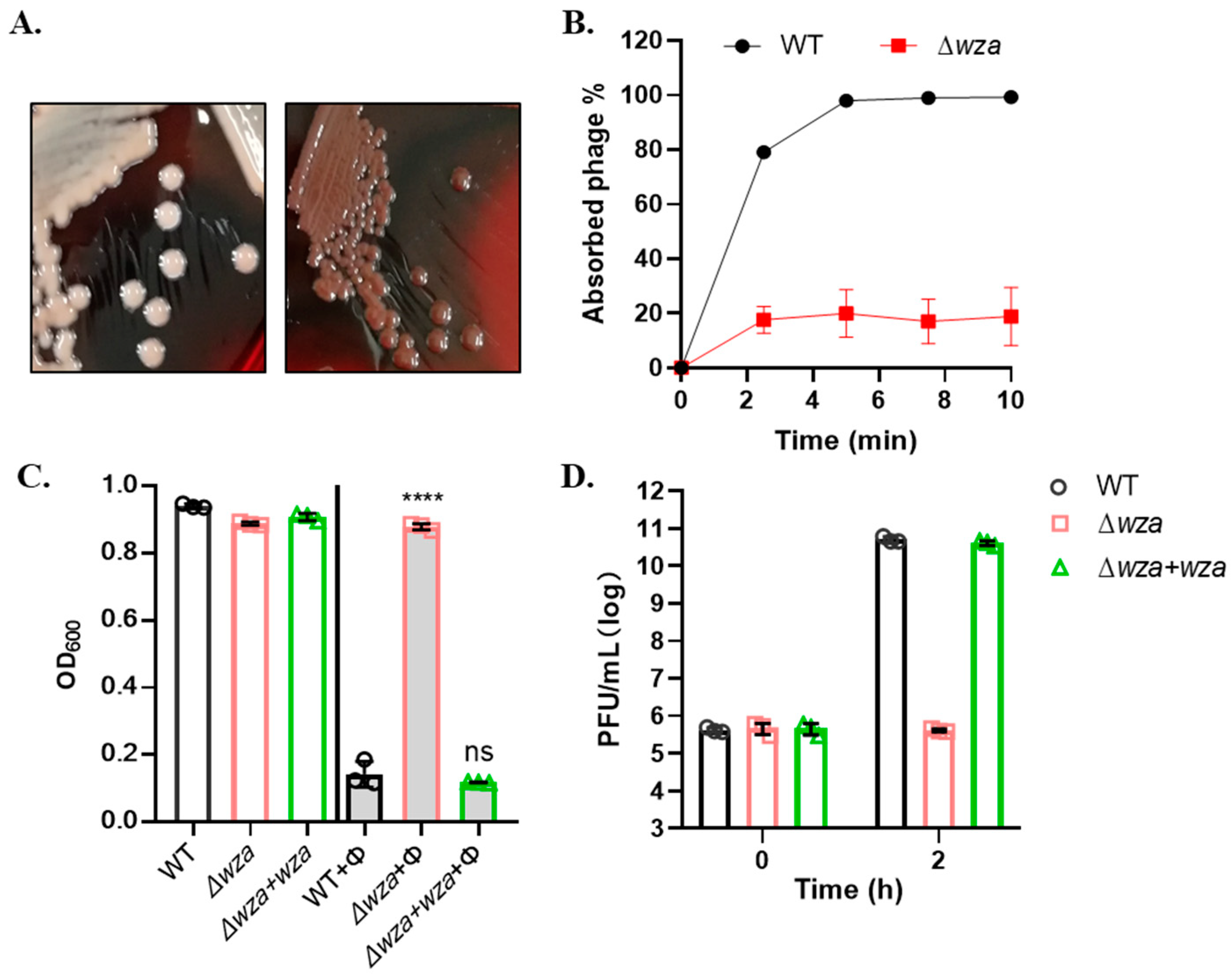
2.7. Construction of CPS Mutant
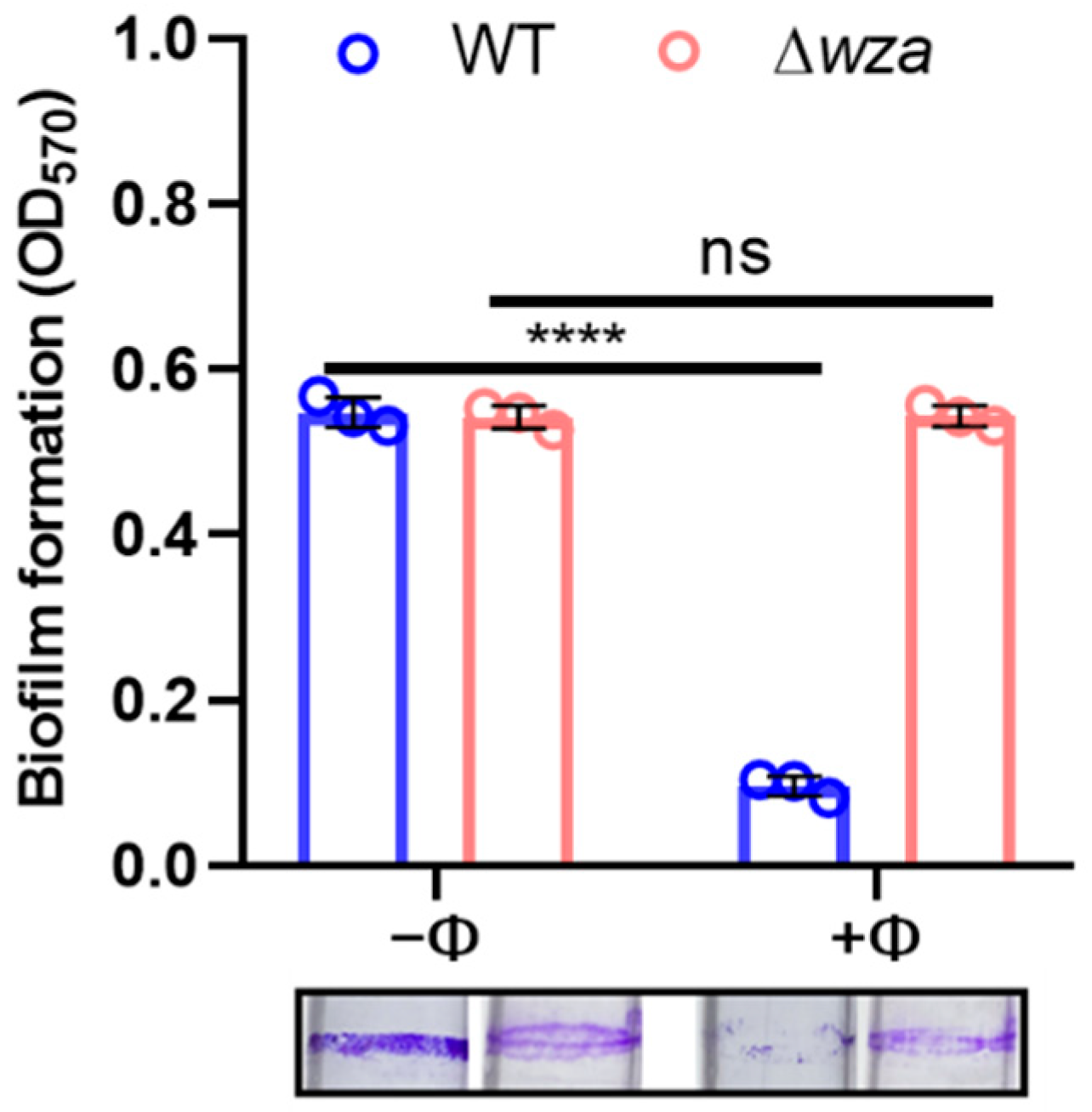
2.8. Biofilm Formation and Crystal Violet Assay
2.9. Galleria Mellonella Model
2.10. The Adult Mouse Model
3. Results
3.1. Characteristics of Phage SRD2021
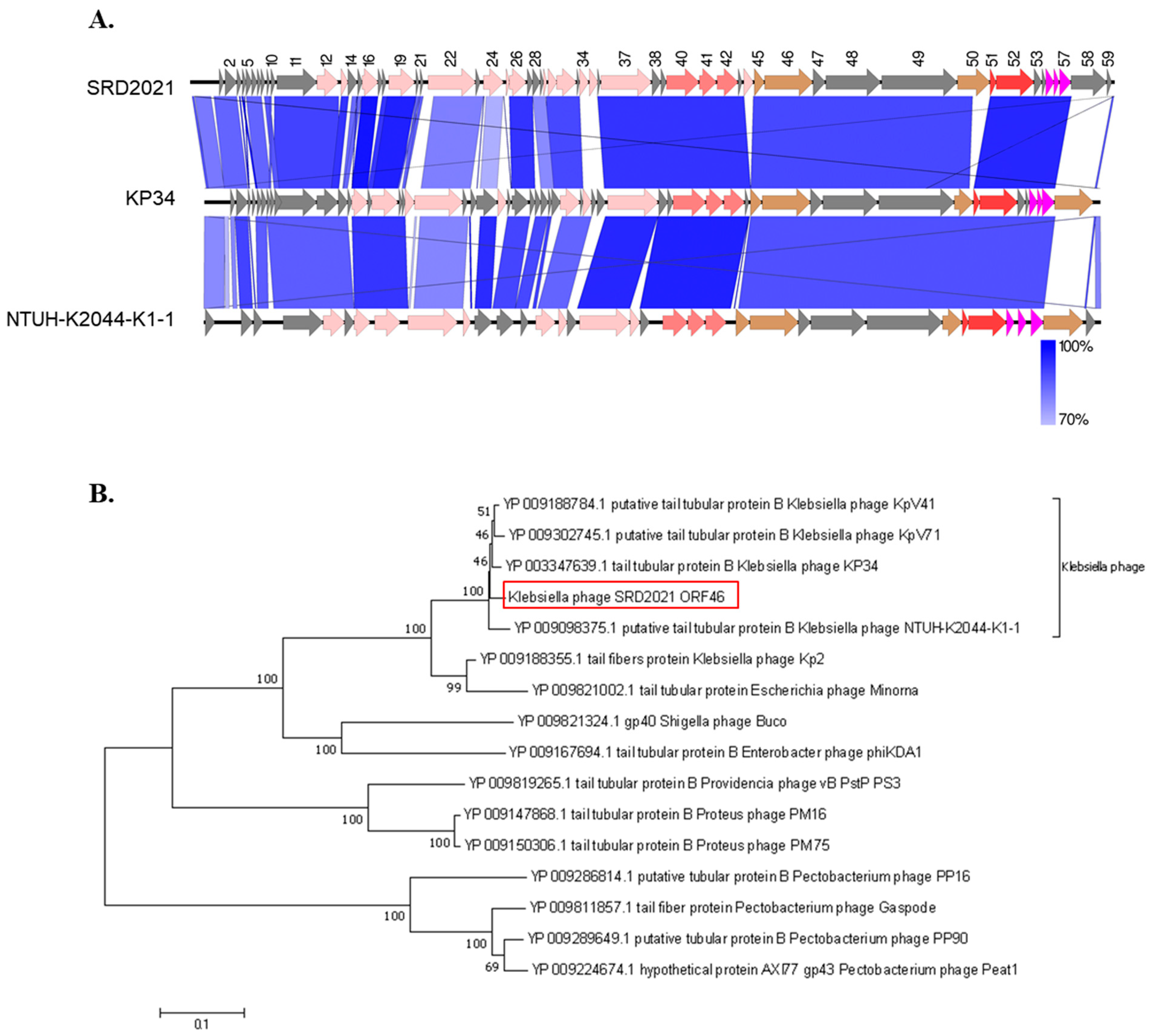
3.2. Phage Genome Analysis
3.3. Phage SRD2021 Encodes Polysaccharide Depolymerase Gene in Its Tailspike Protein
3.4. Phage SRD2021 Uses K47 Capsular Polysaccharide as a Receptor
3.5. Phage SRD2021 Lysed Mature Biofilms Efficiently
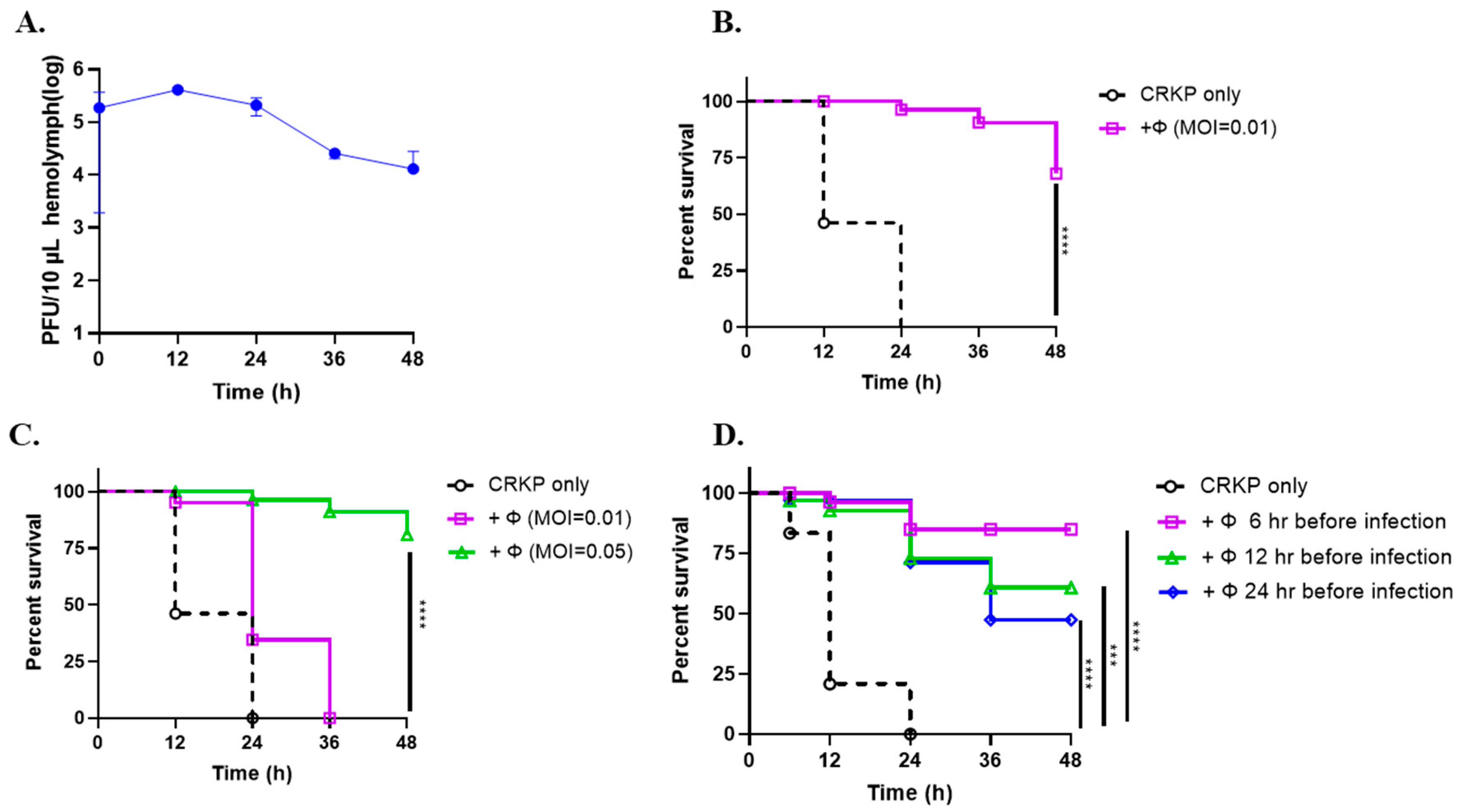
3.6. Phage SRD2021 Showed Preventive and Therapeutic Effect in G. mellonella Model
3.7. Phage SRD2021 Reduced Colonized K. pneumoniae in Mouse Intestine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arakawa, Y.; Wacharotayankun, R.; Nagatsuka, T.; Ito, H.; Kato, N.; Ohta, M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J. Bacteriol. 1995, 177, 1788–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef]
- Schembri, M.A.; Blom, J.; Krogfelt, K.A.; Klemm, P. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 2005, 73, 4626–4633. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Jia, X.; Zhou, M.; Zhang, H.; Yang, W.; Kudinha, T.; Xu, Y. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: A molecular, biological, and epidemiological study. Emerg. Microbes Infect. 2020, 9, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.-J.; Lin, T.-L.; Chen, C.-C.; Tsai, Y.-T.; Cheng, Y.-H.; Chen, Y.-Y.; Hsieh, P.-F.; Lin, Y.-T.; Wang, J.-T. Klebsiella Phage Phi ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91, e02457-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solovieva, E.V.; Myakinina, V.P.; Kislichkina, A.A.; Krasilnikova, V.M.; Verevkin, V.V.; Mochalov, V.V.; Lev, A.I.; Fursova, N.K.; Volozhantsev, N.V. Comparative genome analysis of novel Podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Res. 2018, 243, 10–18. [Google Scholar] [CrossRef]
- Bessler, W.; Freund-Mölbert, E.; Knüfermann, H.; Rudolph, C.; Thurow, H.; Stirm, S. A bacteriophage-induced depolymerase active on Klebsiella K11 capsular polysaccharide. Virology 1973, 56, 134–151. [Google Scholar] [CrossRef]
- Lin, T.-L.; Hsieh, P.-F.; Huang, Y.-T.; Lee, W.-C.; Tsai, Y.-T.; Su, P.-A.; Pan, Y.-J.; Hsu, C.-R.; Wu, M.-C.; Wang, J.-T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: Implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef] [Green Version]
- de Sousa, J.A.M.; Buffet, A.; Haudiquet, M.; Rocha, E.P.C.; Rendueles, O. Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J. 2020, 14, 2980–2996. [Google Scholar] [CrossRef]
- Hao, G.; Yuan, C.; Shu, R.; Jia, Y.; Zhao, S.; Xie, S.; Liu, M.; Zhou, H.; Sun, S.; Wang, H. O-antigen serves as a two-faced host factor for bacteriophage NJS1 infecting nonmucoid Klebsiella pneumoniae. Microb. Pathog. 2021, 155, 104897. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.L.R. Bacteriophages; Humana Press: Totowa, NJ, USA, 2009. [Google Scholar]
- Heller, K.; Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 1979, 139, 32–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallet, R.; Lenormand, T.; Wang, I.N. Phenotypic stochasticity protects lytic bacteriophage populations from extinction during the bacterial stationary phase. Evolution 2012, 66, 3485–3494. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.; Gerdes, S.; Olsen, G.J.; Olson, R.J.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, M.J.; Petty, N.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underwood, A.P.; Mulder, A.; Gharbia, S.; Green, J. Virulence Searcher: A tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 2005, 11, 770–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Larsen, R.A.; Wilson, M.M.; Guss, A.; Metcalf, W.W. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 2002, 178, 193–201. [Google Scholar] [CrossRef]
- Judson, N.; Mekalanos, J.J. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 2000, 18, 740–745. [Google Scholar] [CrossRef]
- Kalogeraki, V.S.; Winans, S.C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 1997, 188, 69–75. [Google Scholar] [CrossRef]
- Zhu, J.; Mekalanos, J.J. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 2003, 5, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Chen, A.I.; Liu, M.; Zhou, H.; Egan, M.; Yang, X.; Kan, B.; Wang, H.; Goulian, M.; Zhu, J. Colistin-resistance-mediated bacterial surface modification sensitizes phage infection. Antimicrob. Agents Chemother. 2019, 12, e01609-19. [Google Scholar] [CrossRef]
- Insua, J.L.; Llobet, E.; Moranta, D.; Pérez-Gutiérrez, C.; Tomás, A.; Garmendia, J.; Bengoechea, J.A. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect. Immun. 2013, 81, 3552–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front. Microbiol. 2019, 10, 2649. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.; Pulido-Cid, M.; Chagoyen, M.; Arranz, R.; González-García, V.A.; Doval, C.G.; Caston, J.; Valpuesta, J.; van Raaij, M.J.; Martín-Benito, J.; et al. Structural characterization of the bacteriophage T7 tail machinery. J. Biol. Chem. 2013, 288, 26290–26299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beis, K.; Collins, R.F.; Ford, R.C.; Kamis, A.B.; Whitfield, C.; Naismith, J.H. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J. Biol. Chem. 2004, 279, 28227–28232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyckoff, T.J.O.; Thomas, B.; Hassett, D.J.; Wozniak, D.J. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 2002, 148, 3423–3430. [Google Scholar] [CrossRef] [Green Version]
- Limoli, D.H.; Whitfield, G.B.; Kitao, T.; Ivey, M.L.; Davis, M.R.; Grahl, N.; Hogan, D.A.; Rahme, L.G.; Howell, P.L.; O’Toole, G.A.; et al. Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. mBio 2017, 8, e00186-17. [Google Scholar] [CrossRef] [Green Version]
- Chhibber, S.; Aggarwal, S.; Yadav, V. Contribution of capsular and lipopolysaccharide antigens to the pathogenesis of Klebsiella pneumoniae respiratory tract infection. Folia Microbiol. 2003, 48, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadha, P.; Katare, O.P.; Chhibber, S. In vivo efficacy of single phage versus phage cocktail in resolving burn wound infection in BALB/c mice. Microb. Pathog. 2016, 99, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.; Taylor, N.M.I.; Buth, S.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef]
- Rakhuba, D.; Kolomiets, E.; Dey, E.S.; Novik, G. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Taylor, K. Physical and chemical changes of Vi-polysaccharide due to Vi-phage II action. Acta Biochim. Pol. 1966, 13, 97–106. [Google Scholar]
- Lindsay, D.; von Holy, A. Bacterial biofilms within the clinical setting: What healthcare professionals should know. J. Hosp. Infect. 2006, 64, 313–325. [Google Scholar] [CrossRef]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef] [Green Version]
- Tascini, C.; Lipsky, B.A.; Iacopi, E.; Ripoli, A.; Sbrana, F.; Coppelli, A.; Goretti, C.; Piaggesi, A.; Menichetti, F. KPC-producing Klebsiella pneumoniae rectal colonization is a risk factor for mortality in patients with diabetic foot infections. Clin. Microbiol. Infect. 2015, 21, 790.e1–790.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain | MLST a | Colony Phenotype | Characteristics of Bla b | Source |
|---|---|---|---|---|
| A1806 | ST1493 | mucoid | NDM-1 | unknown |
| A1502 | ST11 | mucoid | KPC-2 | lower respiratory secretions |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, G.; Shu, R.; Ding, L.; Chen, X.; Miao, Y.; Wu, J.; Zhou, H.; Wang, H. Bacteriophage SRD2021 Recognizing Capsular Polysaccharide Shows Therapeutic Potential in Serotype K47 Klebsiella pneumoniae Infections. Antibiotics 2021, 10, 894. https://doi.org/10.3390/antibiotics10080894
Hao G, Shu R, Ding L, Chen X, Miao Y, Wu J, Zhou H, Wang H. Bacteriophage SRD2021 Recognizing Capsular Polysaccharide Shows Therapeutic Potential in Serotype K47 Klebsiella pneumoniae Infections. Antibiotics. 2021; 10(8):894. https://doi.org/10.3390/antibiotics10080894
Chicago/Turabian StyleHao, Guijuan, Rundong Shu, Liqin Ding, Xia Chen, Yonghao Miao, Jiaqi Wu, Haijian Zhou, and Hui Wang. 2021. "Bacteriophage SRD2021 Recognizing Capsular Polysaccharide Shows Therapeutic Potential in Serotype K47 Klebsiella pneumoniae Infections" Antibiotics 10, no. 8: 894. https://doi.org/10.3390/antibiotics10080894
APA StyleHao, G., Shu, R., Ding, L., Chen, X., Miao, Y., Wu, J., Zhou, H., & Wang, H. (2021). Bacteriophage SRD2021 Recognizing Capsular Polysaccharide Shows Therapeutic Potential in Serotype K47 Klebsiella pneumoniae Infections. Antibiotics, 10(8), 894. https://doi.org/10.3390/antibiotics10080894






