Abstract
Many antibiotic resistance genes present in human pathogenic bacteria are believed to originate from environmental bacteria. Conjugation of antibiotic resistance conferring plasmids is considered to be one of the major reasons for the increasing prevalence of antibiotic resistances. A hotspot for plasmid-based horizontal gene transfer is the phyllosphere, i.e., the surfaces of aboveground plant parts. Bacteria in the phyllosphere might serve as intermediate hosts with transfer capability to human pathogenic bacteria. In this study, the exchange of mobilisable and self-transmissible plasmids via conjugation was evaluated. The conjugation from the laboratory strain Escherichia coli S17-1, the model phyllosphere coloniser Pantoea eucalypti 299R, and the model pathogen E. coli O157:H7 to the recipient strain E. coli O157:H7::MRE103 (EcO157:H7red) in the phyllosphere of Arabidopsis thaliana was determined. The results suggest that short-term occurrence of a competent donor is sufficient to fix plasmids in a recipient population of E. coli O157:H7red. The spread of self-transmissible plasmids was limited after initial steep increases of transconjugants that contributed up to 10% of the total recipient population. The here-presented data of plasmid transfer will be important for future modelling approaches to estimate environmental spread of antibiotic resistance in agricultural production environments.
1. Introduction
With the introduction of penicillin in the 1940s, mankind entered the era of antibiotics (AB), which revolutionised therapeutic medicine [1,2]. For the first time, physicians were able to cure their patients of deadly bacterial diseases and saved millions of lives [3]. Less than a century later, bacterial diseases have yet again become a major threat to human welfare as infectious bacteria acquired antibiotic resistances (ABR) that are able to overcome every antibiotic currently available [3,4]. ABR per se is a natural phenomenon in bacteria [5] and its main function is likely a countermeasure against antibiotic-producing microorganisms that compete for the same resources [6]. It is the use of AB in anthropogenic applications such as medical treatment, animal husbandry, and agricultural practice that spreads ABR in environmental and infectious bacteria whilst pushing the selection pressure on a level beyond the natural evolutionary clock [7].
Many ABR genes present in human pathogenic bacteria are believed to originate from environmental bacteria [8,9,10,11]. This implies that, for an ABR gene to reach a human pathogenic bacterium, there needs to be an exchange of genetic material from environmental bacteria towards pathogens. Transfer of genetic material can be achieved by uptake of environmental DNA due to natural competence, phage-mediated transduction, integrative and conjugative elements, or conjugation of plasmids [12,13]. The latter is considered to be a major cause for the increased prevalence of ABR [8,9]. The ability of conjugative plasmids to move genes from one bacterium to another, not necessarily related to each other, is responsible for the rapid spread and accumulation of resistances [9,14,15,16]. A hotspot for plasmid-based horizontal gene transfer is the phyllosphere [17,18,19,20,21], representing the surface of all above-ground organs of land plants [22], thereby including the fresh plant products that are considered an important part of a healthy diet.
In today’s intensive agricultural production, fertilisers are needed to replenish soil nutrients, such as nitrogen and phosphorus. They are essential for crop growth and increased crop yield. Animal manure is an excellent source for such nutrients, but it often originates from intensive animal husbandry farms, where the widespread use of AB to preventively treat animals is the rule rather than the exception [23]. This leads not only to a relative increase of ABR bacteria in faecal waste, but also to an accumulation of ABR-conferring genetic elements, such as plasmids [23,24,25]. Bacteria that constitute the normal phyllosphere microbiota are generally not considered harmful [26,27], but they might serve as intermediate hosts for ABR-conferring plasmids with transfer capability to human pathogenic bacteria. Little is known about the number of transfer steps involved in the propagation of resistance genes and the efficacy of the mechanism participating in the exchange of genetic material in the environment. However, information about plasmid transfer and plasmid persistence will be important for future modelling and risk assessment approaches to estimate environmental spread of antibiotic resistance in agricultural production environments.
In the present study, a laboratory-scale model system was established that mimics the shortest possible route for ABR-carrying plasmids into pathogenic Escherichia coli O157:H7::MRE103 Δstx (EcO157:H7red) recipients on Arabidopsis thaliana rosettes. The exchange of the mobilisable plasmid pUC18T-mini-Tn7T-Gm-eYFP (pUC18) and self-transmissible ABR-carrying plasmids pKJK5::Plac:gfp (pKJK5) and RP4::Plac:gfp (RP4) via conjugation by filter mating and in the phyllosphere of A. thaliana was evaluated. Donors are either the model phyllosphere colonising strain Pantoea eucalypti 299R (Pe299R), the non-pathogenic laboratory strain E. coli S17-1 (EcS17-1), or E. coli O157:H7 Δstx (EcO157:H7) (Figure 1). The assay takes into account that plants can carry human pathogenic bacteria as contaminations, originating from animal manure or the digestates from biogas plants, which are used as organic fertiliser [21,28,29]. Organic fertilisers are also sources for ABR-conferring genetic elements, such as plasmids [24,25]. To mimic natural conditions, we conducted in planta experiments in the absence of antibiotic pressure.
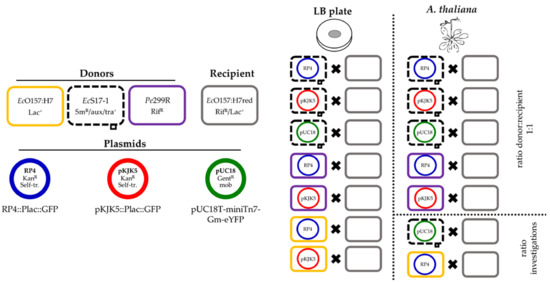
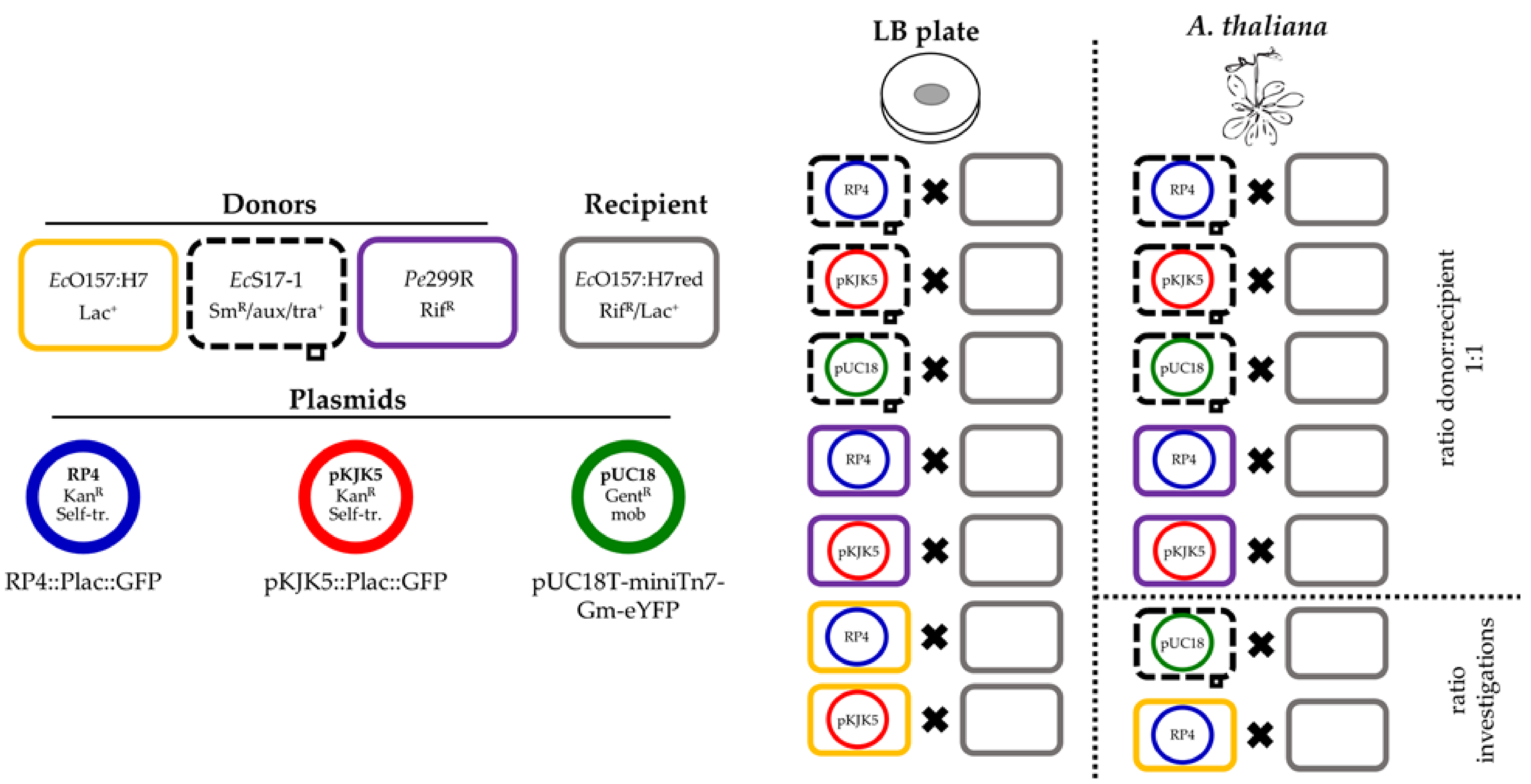
Figure 1.
Overview of performed experiments. Three donors, EcO157:H7, EcS17-1, and Pe299R, were combined with up to three different plasmids, RP4, pKJK5, or pUC18. The ability of the hosts to transfer the plasmids to the recipient EcO157:H7red was assessed on membrane filters and gnotobiotic A. thaliana. Additionally, the effect of different donor/recipient ratios onto conjugation frequency was assessed for two exemplary donor and plasmid combinations.
2. Results
2.1. Transconjugant Frequencies after Filter Mating
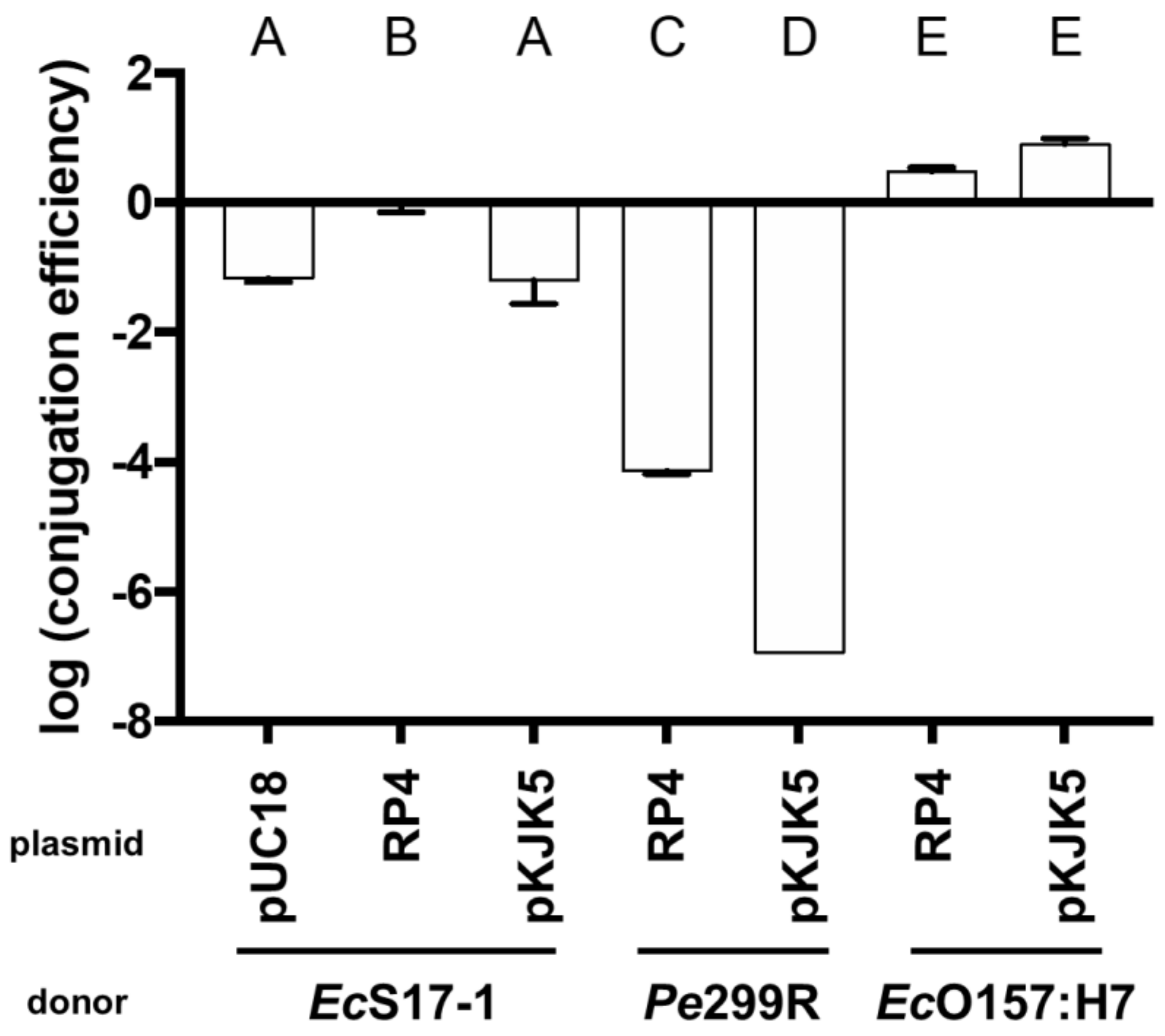
Matings on nitrocellulose filters were performed to determine transconjugant frequencies of the recipient EcO157:H7red (Figure 2). Besides Pe299R (pKJK5), all donors were able to transfer plasmids to EcO157:H7red. In the case of EcS17-1 being the donor, all plasmids were transferred at high rates, and the transconjugant frequency was between 100 and 6.31 × 10−2 per recipient cell depending on the transmitted plasmid (transconjugant frequencies pKJK5 < pUC18 < RP4). When Pe299R was donor of RP4 or pKJK5, transconjugants were on average detected at frequencies of 6.82 × 10−5 or below the limit of detection, which was 9.62 × 10−6 per recipient cell, respectively. EcO157:H7 donors transferred the plasmids pKJK5 and RP4 with the highest efficiency to EcO157:H7red with transconjugants being detected at frequencies of 3.16 × 101 and 7.94 × 101 (transconjugant frequencies RP4 < pKJK5). Since, in contrast to EcS17-1, Pe299R and EcO157:H7 are not able to mobilise plasmids, they were not tested as donors of pUC18.
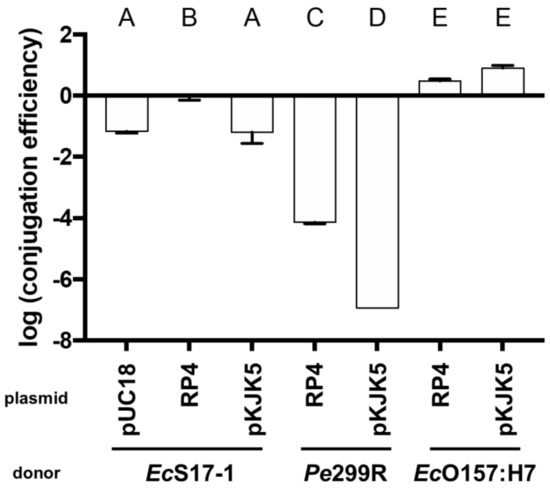
Figure 2.
Transconjugant frequencies in the recipient population after mating of the recipient EcO157:H7red with EcS17-1, Pe299R, and EcO157:H7 donors carrying plasmids pUC18, pKJK5, or RP4 on nitrocellulose filters. Error bars represent the standard deviation of the mean. Significant differences between treatments are marked with different letters (p-value < 0.01, one-way ANOVA, Tukey’s multiple comparison test).
2.2. Growth Dynamics of Individual or Co-Inoculated Bacterial Strains in Planta
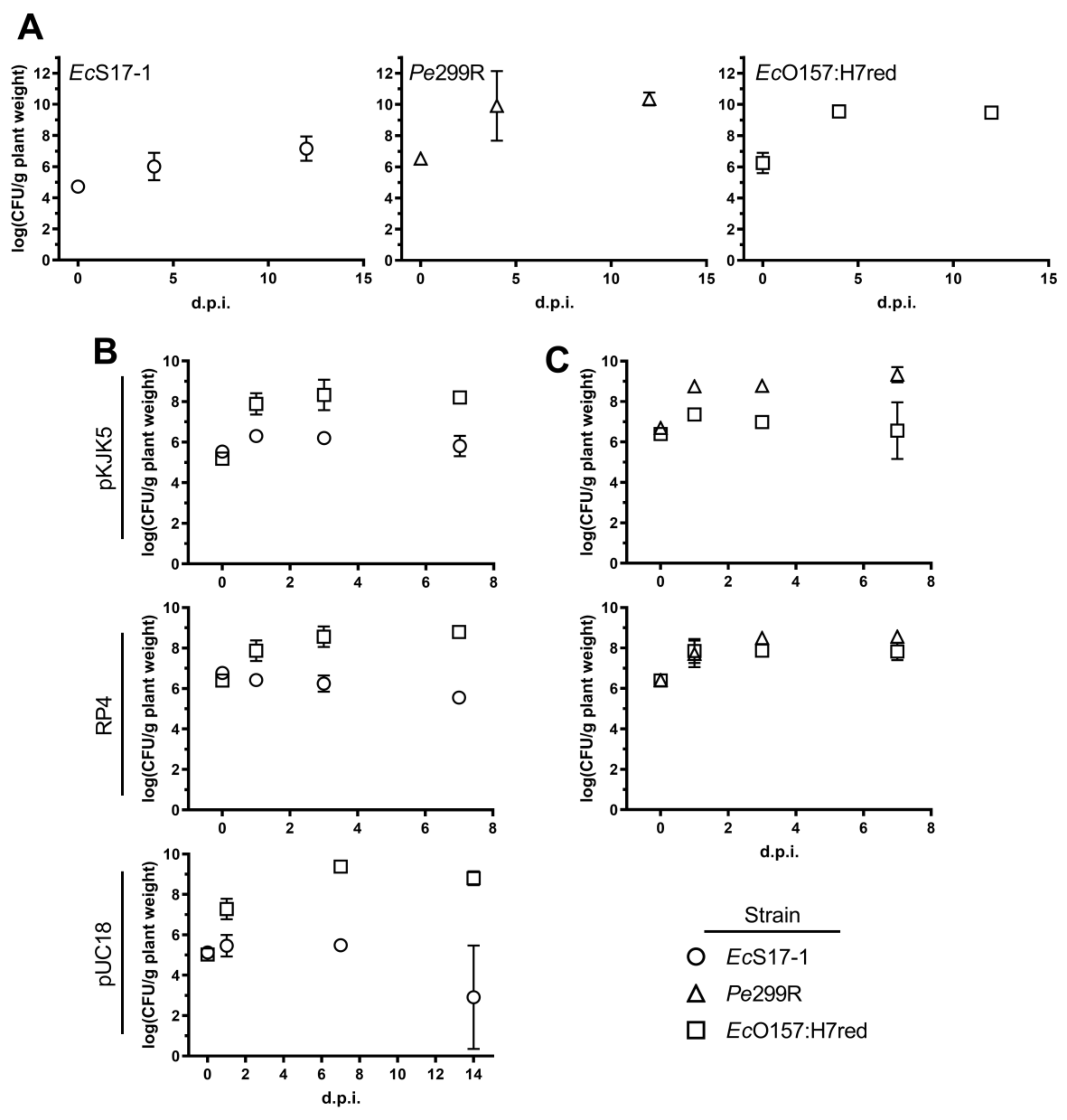
To determine the ability of the different strains to colonise A. thaliana, we inoculated EcS17-1, Pe299R, and EcO157:H7red (representative also for donor EcO157:H7) onto axenic plants. When grown individually, all bacterial strains, including the auxotrophic laboratory strain EcS17-1, were able to grow to high densities on A. thaliana, reaching CFU counts of 108−1010 bacteria per gram plant material after 4 and 12 days post-inoculation (d.p.i.) (Figure 3A); similar densities of bacterial colonisation have been reported before in gnotobiotic and environmental studies [30,31]. When EcS17-1 or Pe299R carrying either self-transmissible plasmids RP4 or pKJK5 were co-inoculated with EcO157:H7red, population development of individual strains behaved differently (Figure 3B,C). When co-cultured with EcS17-1 (pKJK5), the EcO157:H7red population reached similar densities as grown on A. thaliana only, i.e., EcO157:H7red multiplied to densities of >108 CFU g−1 and maintained those densities until the end of the experiment. In contrast, EcS17-1 (pKJK5) increased to a maximum of >106 CFU g−1 after 24 h, but then the population steadily declined below 106 CFU g−1 until 7 d.p.i. (Figure 3B, top). When EcO157:H7red was co-cultured with EcS17-1 (RP4), the initial population size of EcS17-1 (RP4) was 107 CFU g−1 and the population did not further increase and steadily declined during the experiment (Figure 3B, middle). The EcO157:H7red population increased by two magnitudes to 5 × 108 CFU g−1 and remained stable. When in co-culture with EcS17-1 (pUC18), EcO157:H7red showed a strong initial increase and reached a maximum density of >109 CFU g−1, 7 days after inoculation (Figure 3B, bottom). EcS17-1 (pUC18) reached a maximum density of ≈105 CFU g−1, 1 day after inoculation. Afterwards, the density remained stable and started declining 7 days after inoculation.
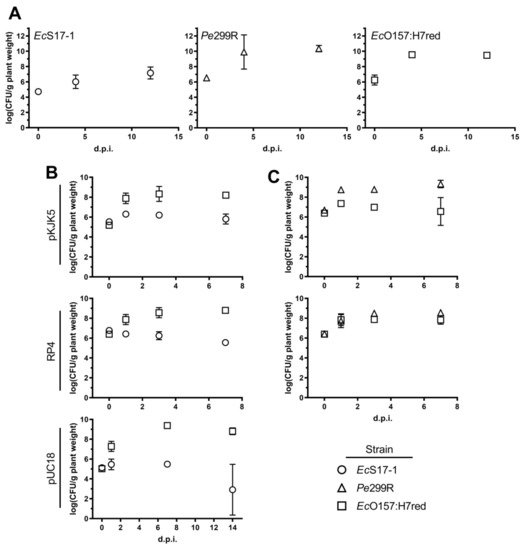
Figure 3.
Bacterial population development on gnotobiotic A. thaliana. (A) Populations after monoinoculation of EcS17-1 (circles), Pe299R (triangles), or EcO157:H7red (squares) at 4 and 12 days post-inoculation (d.p.i.). (B) Population developments after co-inoculation of EcS17-1 (circles) harbouring pKJK5 (top), RP4 (middle), and pUC18 (bottom) with the recipient EcO157:H7red (squares) onto gnotobiotic A. thaliana after 1, 3, and 7 d.p.i. (C) Population development after co-inoculation of Pe299R (triangles) harbouring pKJK5 (top) and RP4 (bottom) with the recipient EcO157:H7red (squares) onto gnotobiotic A. thaliana after 1, 3, and 7 d.p.i. Error bars represent the standard deviation of the mean.
When in competition with Pe299R (pKJK5), the EcO157:H7 population increased by one magnitude to 107 CFU g−1 and never reached densities of above 108 CFU g−1, while Pe299R (pKJK5) increased from 106 CFU g−1 to 109 and remained stable for the duration of the experiment (Figure 3C, top). In competition with Pe299R (RP4), both EcO157:H7red and Pe299R (RP4) reached densities of approximately 108 CFU g−1 and remained stable thereafter (Figure 3C, bottom).
When EcO157:H7 represented the donor, the combined EcO157:H7 population (sum of donor and recipient CFU) reached cell numbers above 108 CFU g−1 (Figure S1).
2.3. Conjugation Dynamics of Self-Transmissible Plasmids in Planta
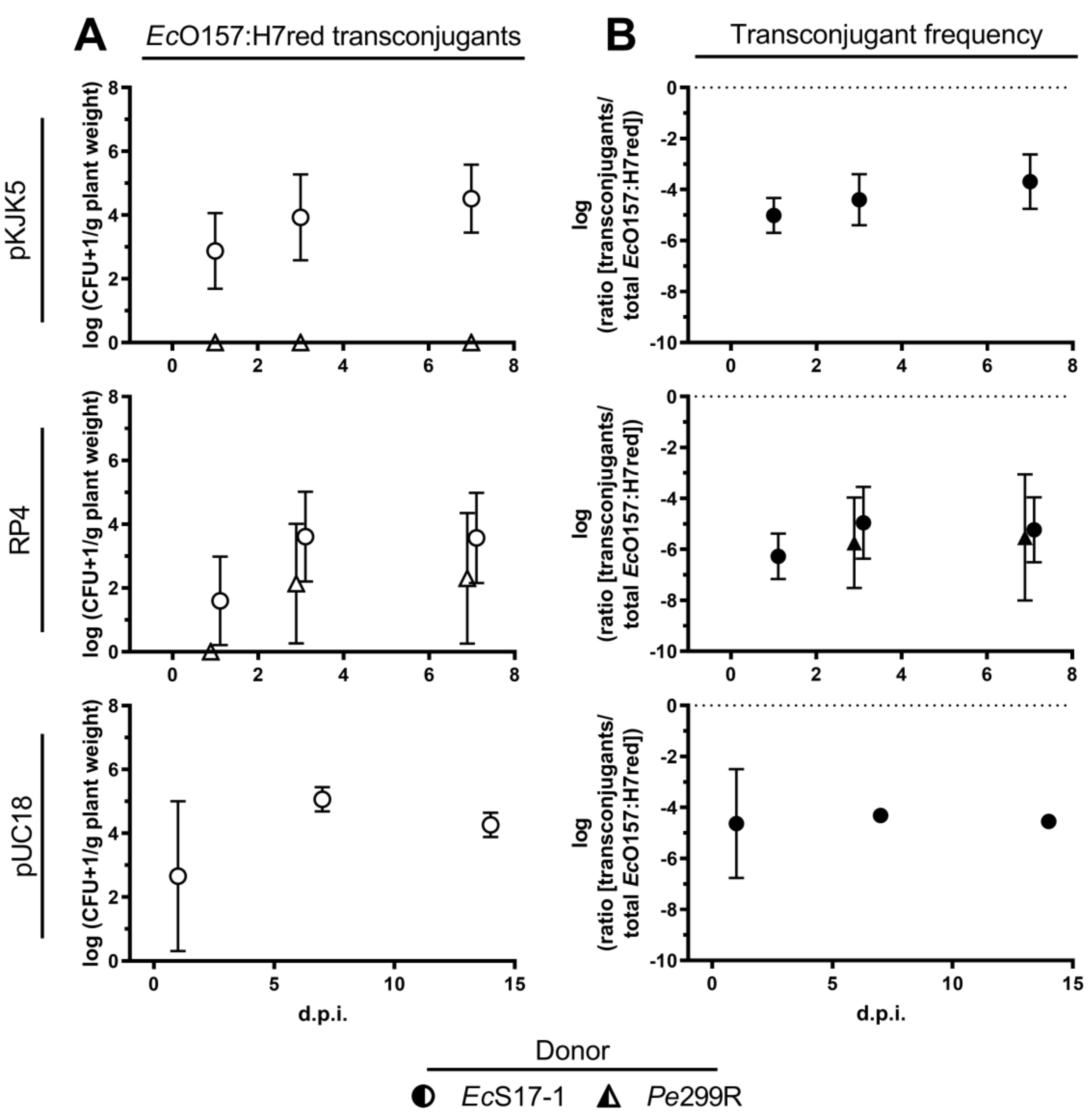
EcS17-1 was able to transfer self-transmissible pKJK5 and RP4 to EcO157:H7red on A. thaliana. When co-inoculated for 24 h with EcS17-1 (pKJK5) or EcS17-1 (RP4), on average, more than 103 EcO157:H7red (pKJK5) and ≈102 EcO157:H7red (RP4) transconjugants g−1 plant were detected (Figure 4A, top and middle). The average ratio of EcO157:H7red transconjugants in the recipient population slowly increased over time, but not significantly (Figure 4B, top and middle).
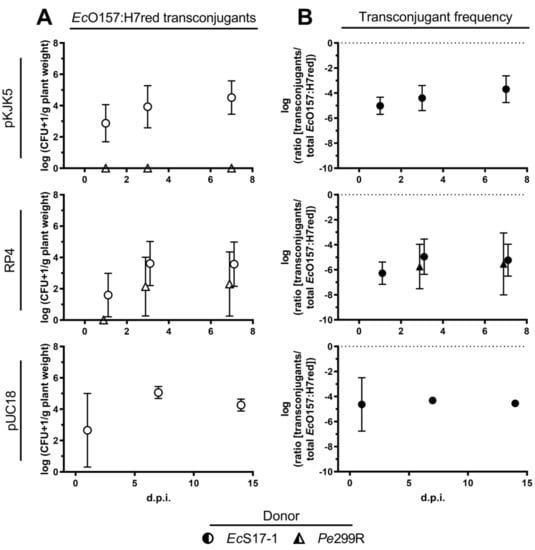
Figure 4.
Conjugation dynamics of transconjugant EcO157:H7red on gnotobiotic A. thaliana. (A) Population development of transconjugant EcO157:H7red after co-inoculation with donors EcS17-1 (circles) and Pe299R (triangles) harbouring pKJK5 (top), RP4 (middle), and pUC18 (bottom) onto gnotobiotic A. thaliana after 1, 3, and 7 d.p.i. (B) Frequency of EcO157:H7red transconjugants in the recipient population after co-inoculation with donors EcS17-1 (circles) and Pe299R (triangles) harbouring pKJK5 (top), RP4 (middle), and pUC18 (bottom) onto gnotobiotic A. thaliana after 1, 3, and 7 d.p.i. The conjugation with Pe299R (pKJK5) did not yield transconjugants above the limit of detection. Error bars represent the standard deviation of the mean.
No transconjugants could be detected after co-inoculation of Pe299R (pKJK5) and EcO157:H7red. After co-inoculation of Pe299R (RP4) and EcO157:H7red, 5 × 102 EcO157:H7red transconjugants were detected 7 days after inoculation (Figure 4A, middle). Between replicates, the occurrence of transconjugants was highly fluctuating, resulting in large standard deviations. The frequency of transconjugants remained stable during the period of 7 days (Figure 4B, middle).
2.4. Effect of Non-Self-Transmissible but Mobilisable Plasmids on Transconjugant Frequencies in Planta
EcS17-1 was able to transfer the mobilisable plasmid pUC18 to EcO157:H7red on A. thaliana. When co-inoculated for 24 h with EcS17-1 (pUC18), on average, 103 EcO157:H7red (pUC18) transconjugants g−1 plant were detected, reaching a maximum of ≈105 transconjugants g−1 plant 7 d.p.i. (Figure 4A, bottom). The average frequency of EcO157:H7red (pUC18) transconjugants reached 10−4 per recipient cell after 7 days and did not decrease within 14 days, despite the lack of selective agents (i.e., antibiotics) and a potential fitness cost of the plasmid (Figure 4B, bottom).
2.5. Effect of Ratio Differences between Donor Cells to Recipients on Transconjugant Frequencies in Planta
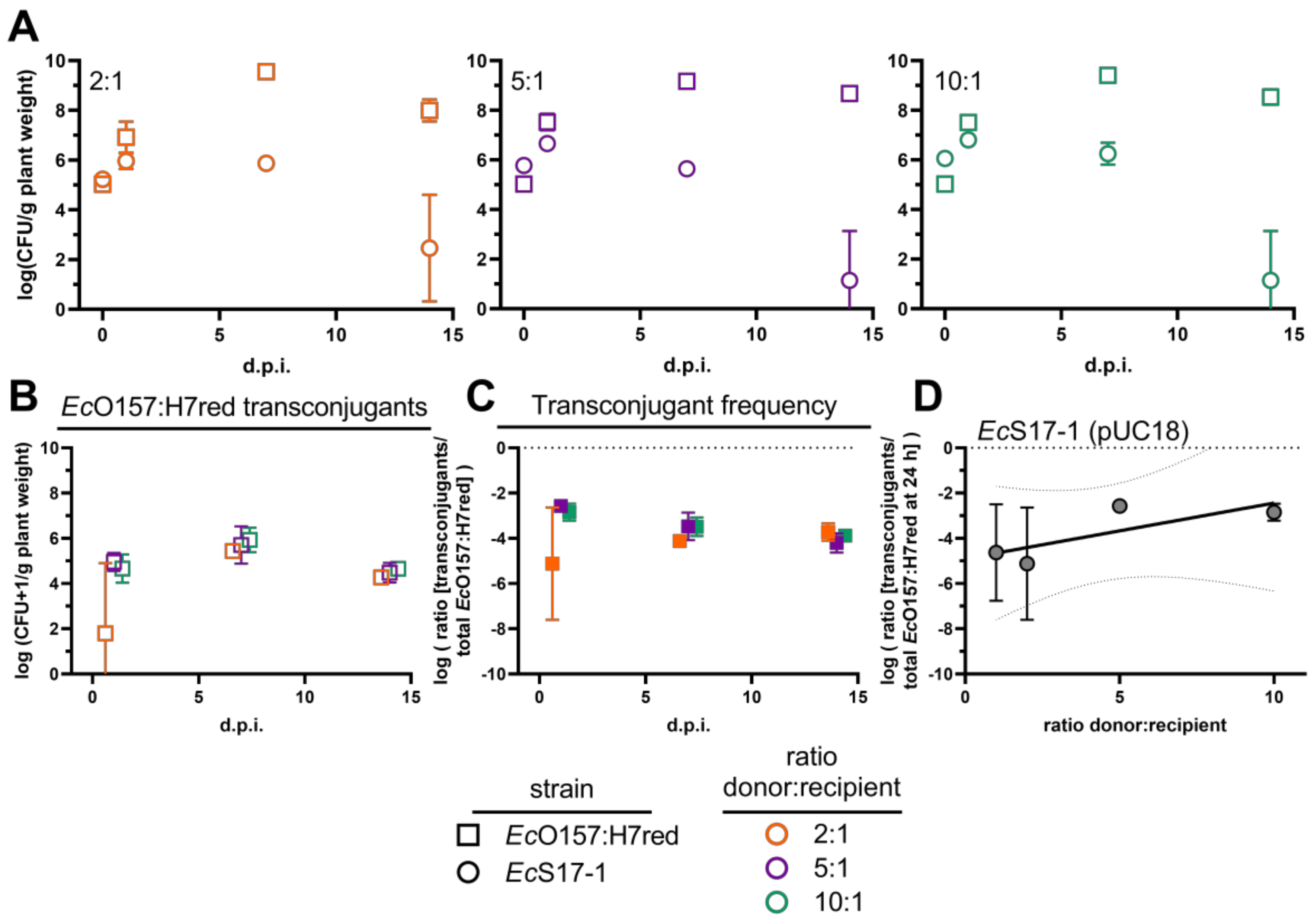
To separate the effect of secondary horizontal transfer of plasmids from primary conjugations, i.e., from a freshly conjugated cell to another recipient vs. from an original donor to a recipient cell, we tested three different initial densities of donor EcS17-1 carrying the mobilisable, but non-self-transmissible, plasmid pUC18 and EcO157:H7red as recipient. Donor and recipient were mixed in ratios of 1:1 (see Section 2.4), 2:1, 5:1, and 10:1. Similar to the same starting densities of donor and recipient, EcO157:H7red showed a strong initial increase and reached a maximum density of >109 CFU g−1, 7 days after inoculation for all donor/recipient ratios (Figure 5A). EcS17-1 (pUC18) reached a maximum density at day 1 after inoculation and then started declining. The donor was outcompeted by the recipient during the experiment (Figure 5A). Additionally, a strong initial increase of EcO157:H7red transconjugants occurred within the first 24 h for all of the different donor/recipient ratios and remained stable until the end of the experiment (Figure 5B). Interestingly, while not statistically significant, all analysed timepoints showed a trend of higher transconjugant numbers in the presence of increased donor densities (Figure 5B). As a consequence of the competition between donor cells and recipient cells, the probability over time for recipients to encounter donor cells decreased, which was supported by a slight decline of transconjugant frequencies over the course of time (Figure 5C).
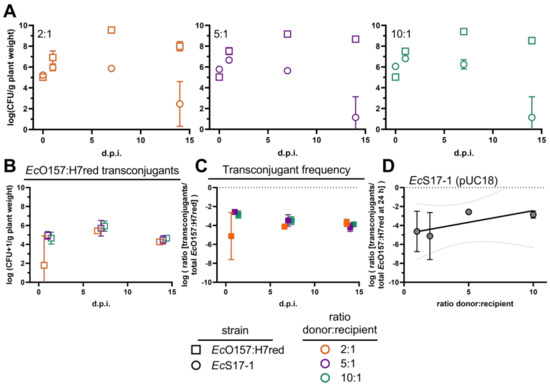
Figure 5.
Conjugation dynamics of the mobilisable plasmid pUC18 after inoculating different densities of donors. (A) Population developments after co-inoculation of different ratios of the donor EcS17-1 (circles) harbouring pUC18 with the recipient EcO157:H7red (squares) onto gnotobiotic A. thaliana after 1, 7, and 14 d.p.i. The initial recipient inoculation density remained constant, whilst EcS17-1 donors were inoculated at different densities, resulting in donor/recipient ratios of 2:1 (left, orange), 5:1 (middle, purple), and 10:1 (right, green). (B) Population development of transconjugant EcO157:H7red after co-inoculation with different densities of donor EcS17-1 (pUC18) onto gnotobiotic A. thaliana after 1, 7, and 14 d.p.i. (C) Frequency of EcO157:H7red transconjugants in the recipient population after co-inoculation with different densities of donor EcS17-1 (pUC18) onto gnotobiotic A. thaliana after 1, 7, and 14 d.p.i. (D) Transconjugant frequencies of different donor/recipient ratios after 24 h. No significant differences in the conjugation efficiency after treatments with different EcS17-1 donor concentrations could be detected, however, the variation within treatments was lower at high donor densities. A linear regression was fitted (r2 = 0.61, broken lines 95% confidence intervals of the linear regression). Error bars represent the standard deviation of the mean.
When comparing the transconjugant frequencies after treatment with different donor densities 24 h after inoculation, we found no significant difference between the different donor and recipient ratios. However, we found a positive trend between donor density and transconjugant frequencies after 24 h (Figure 5D).
To investigate the effect of donor to recipient ratios onto the ability of self-transmissible plasmids to invade a population of E. coli O157:H7, EcO157:H7 (RP4) donor, and EcO157:H7red, we inoculated recipient ratios 1:10, 1:5, 1:2, 2:1, 5:1, and 10:1 onto A. thaliana plants. All ratio mixtures yielded >104 transconjugants per gram of plant after 24 h, and the transconjugant population levelled off between 106 and 107 transconjugants per gram of plant (Figure S1A). This translated to transconjugant frequencies of 2.5 × 10−2 to 9 × 10−4 for the different ratio mixtures 24 h after inoculation (Figure S1B).
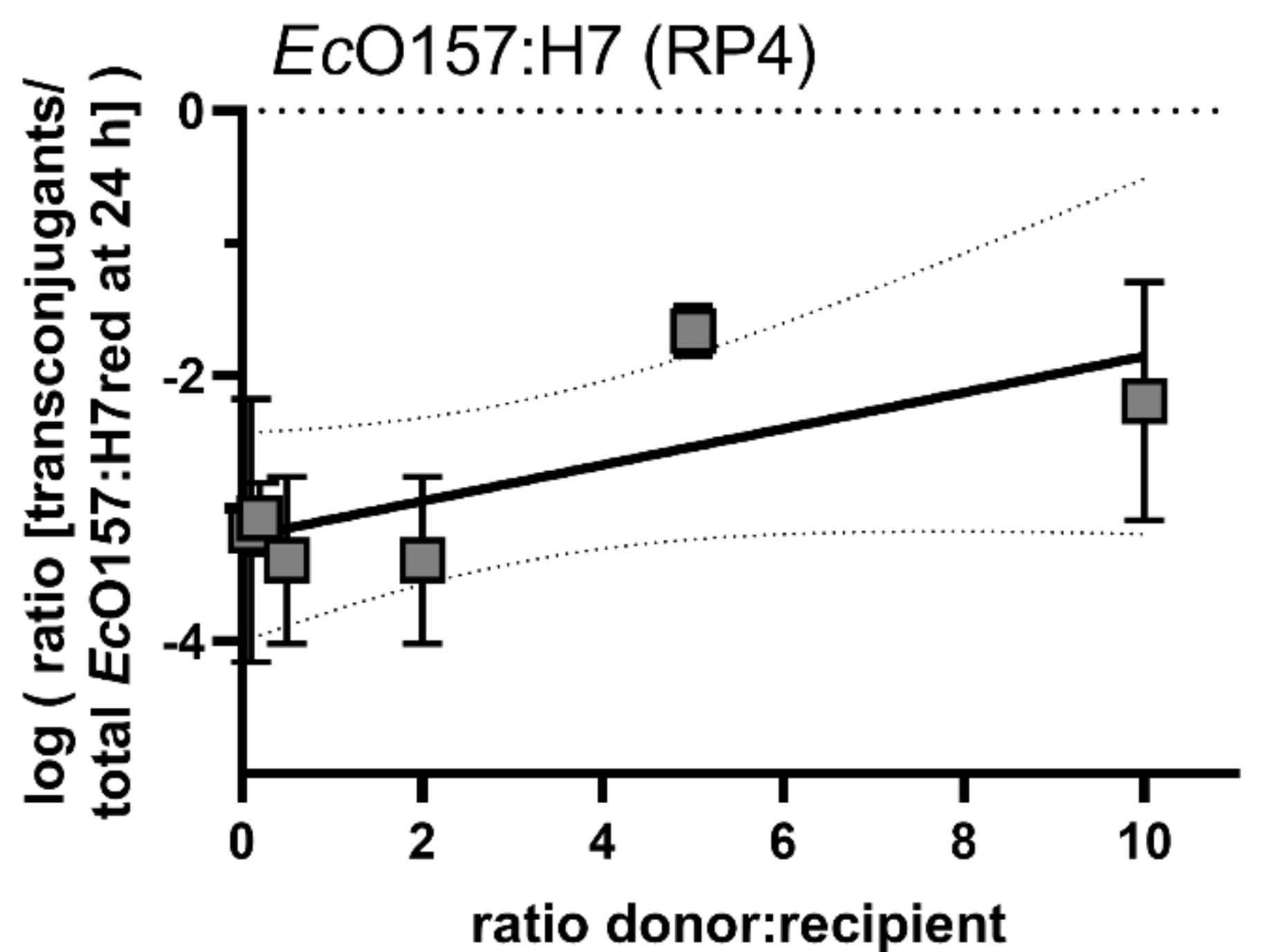
Similar to the results with mobilisable plasmid pUC18, transconjugant frequency data suggest a low correlation between the donor/recipient ratio and the transconjugant frequency after 24 h (Figure 6).
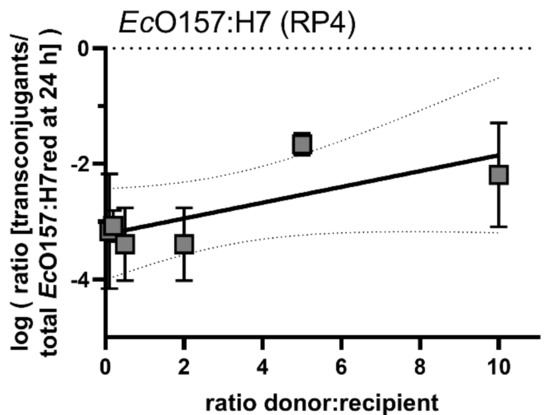
Figure 6.
Conjugation dynamics of the self-transmissible plasmid RP4 after inoculating different densities of donor EcO157:H7 with the recipient EcO157:H7red. Shown are transconjugant frequencies of different donor/recipient ratios after 24 h. No significant differences in the conjugation efficiency after treatments with different EcO157:H7 donor concentrations could be detected. A linear regression was fitted (r2 = 0.55, broken lines 95% confidence intervals of the linear regression). Error bars represent the standard deviation of the mean.
3. Discussion
To study the probability of horizontal gene transfer towards pathogenic bacteria on plant leaf surfaces, we established a model system for the exchange of self-transmissible- and non-self-transmissible but mobilisable plasmids. The model system provided insights into the conjugation between Enterobacteriaceae in the phyllosphere of A. thaliana. Besides the phyllosphere colonising strain Pe299R (pKJK5), all donor strains tested were able to transfer plasmids in measurable rates to the model human pathogenic EcO157:H7red after being co-inoculated onto nitrocellulose filters, although Pe299R did so at a much lower frequency compared to EcS17-1. The reason for this transfer barrier [12,32] is currently unknown, given that Pe299R was a competent recipient of the self-transmissible plasmid, that EcO157:H7red had no issue with receiving the same plasmids from EcS17-1, and that both donor and recipient are members of the family Enterobacteriaceae.
In planta, EcO157:H7red outgrew EcS17-1 and thereby outcompeted it. This is not unexpected, since both strains of E. coli should have a close to identical resource demand and EcS17-1 is an auxotroph, lab-adapted strain [33], which is therefore prone to be less competitive. When co-inoculated with the phyllosphere-competent strain Pe299R carrying plasmid pKJK5, EcO157:H7red did not reach the same high densities as in a monoculture, and the cell density was decreased to less than 107 CFU on average. Potentially, Pe299R is outcompeting EcO157:H7red due to nutritional competition [34]. There is no indication that Pe299R produces antibiotics which inhibit the growth of EcO157:H7red [35] as no antibiotic production genes are annotated in the Pe299R genome [36] and no growth halos were formed around Pe299R colonies on agar plates indicative for growth inhibition of EcO157:H7red (data not shown).
After co-inoculation of EcS17-1 containing different self-transmissible and mobilisable plasmids with EcO157:H7red as recipient, transconjugants could be detected after 24 h (Figure 4) at high rates, underlining the donor’s ability to transfer plasmids on plant leaves. These rates are comparable to previously reported conjugation rates in planta [18,19,37].
The physicochemical nature of plant leaf surfaces presents a spatially segregated, heterogeneous environment [38] that promotes clonal cluster formation and limits movement, thereby limiting the potential spread of invasive plasmids [39,40]. This might explain why self-transmissible plasmids did not further invade the recipient population and the relative contribution of plasmid-bearing transconjugants did not over-proportionally increase in time (Figure 4).
The extent to which the self-transmissible plasmid RP4 is able to invade the recipient population was tested by using EcO157:H7 as donor and EcO157:H7red as recipient. After an initial steep increase of the emerging transconjugant population within the first 24 h, the transconjugant population’s increase exhibited a slope that was slightly higher than the overall recipient’s population increase. This indicates that the plasmid was horizontally propagating to new recipients and the increase in the population not exclusively vertically during bacterial growth. Generally, after three days of growth, the increase in transconjugants levelled off and the contribution of transconjugants to the total EcO157:H7red population did not further increase. This indicates that the ability of plasmids to invade the complete population is limited and directly connected to active growth of the donor and recipient populations. Once the plant is saturated with colonisers, the transmission of the plasmid slows to a hold and can best be explained by vertical transmission rather than horizontal transmission. By using a wide range of donor vs. recipient ratios that were initially inoculated, we determined the relationship between donor and recipient ratios and transconjugant frequencies. The transconjugant frequency was correlated with the number of donors inoculated (r2 = 0.55, Figure 6). The transconjugants frequency is likely a combined result of the maximal load of local leaf habitats [39] and the probability of members of the two populations to colonise the leaf at the same site [40,41].
When the non-self-transmissible but mobilisable plasmid pUC18 is conjugated by EcS17-1, the transconjugant population is not over-proportionally increasing in comparison to self-transmissible plasmids. This lack of increase is likely depicting a stable total population of transconjugants that ceased in growth. As pUC18 does not contain the transfer machinery necessary to further conjugate itself, EcO157:H7red transconjugants are incapable of transmitting the acquired plasmid to other cells. As expected, the ability of pUC18 to invade the recipient population is limited and the transconjugant population is increasing proportionally slower than the total recipient population. As the generation of new transconjugants is limited by the presence of the donor strain and vertical transfer of the plasmid from primary transconjugants to daughter cells, this can be interpreted as a cease of growth or decrease of the donor population and a cease of growth of the primary transconjugant population. Indeed, the donor population stopped growing after 1 day and started to decrease after 7 days (Figure 3B).
In line with previous findings, we observed that conjugation efficiency of plasmids was high in the absence of antibiotic pressure [42]. Even for mobilisable plasmids, which only propagate vertically after the initial conjugation, we found that transconjugants were not lost from the system, i.e., they were not outcompeted by the non-plasmid-bearing population. This finding is concerning as it indicates that even low frequencies of plasmid transfer on plant foodstuffs might fix a plasmid bearing antibiotic resistance in a population of bacteria.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
Strains and plasmids used in this study and their abbreviations are listed in Table 1. E. coli strains and Pe299R were routinely grown on lysogeny broth agar (LB). To determine total colony forming units (CFU) of E. coli after conjugation experiments, M9 minimal medium agar containing lactose as sole carbon source (15 g L−1 agar, 100 mL 10 × M9 salts (85.1 g L−1 Na2HPO4·2H2O, 30 g L−1 KH2PO4, 5 g L−1 NaCl, and 10 g L−1 NH4Cl, pH 7), 2 mL 1 M MgSO4, 1 mL 0.1 M CaCl, 40 mL 10% w/v lactose solution) or LB supplemented with rifampicin were employed. E. coli colonies were assessed after 7 days of incubation at room temperature, Pe299R colonies on the same agar plates after additional 7 days of incubation. To select for EcO157:H7red transconjugants, we employed M9 minimal medium agar containing lactose as sole carbon source and appropriate antibiotics. EcS17-1 CFU were determined by plating on LB agar containing streptomycin. To select for EcO157:H7 (RP4) donor cells, we used LB containing kanamycin (transconjugants contributed to less than 10% of the donor population that was also kanamycin-resistant). Where appropriate, antibiotics were used in the following concentrations: kanamycin 50 µg mL−1, gentamicin 15 µg mL−1, streptomycin 100 µg mL−1, and rifampicin 100 µg mL−1.

Table 1.
Strains and plasmids used in this study and their abbreviations.
4.2. Plasmids Used in the Study
The plasmids employed in this study (Table 1) are the two self-transmissible plasmids RP4::Plac::GFP (RP4), pKJK5::Plac::GFP (pKJK5) [15], and the mobilisable plasmid pUC18T-mini-Tn7T-Gm-eYFP (pUC18) [43]. Both self-transmissible plasmids are promiscuous and have a broad host range, RP4 is a IncP-1α incompatibility group plasmid [44] and pKJK5 is an IncP-1ε incompatibility group plasmid [45]. Plasmid pUC18 is a synthetic construct replicating only in Enterobacteriaceae and is present in high copy numbers when carried by E. coli [43]. For horizontal transfer plasmid pUC18 requires the tra operon, which EcS17-1 contains in its chromosome.
4.3. Conjugation Experiments
An overview of the performed conjugation experiments is shown in Figure 1.
4.3.1. Conjugation on Nitrocellulose Filters
To determine in vitro conjugation rates, we grew donors and recipients as described above. To prepare conjugation mixtures, we harvested a loop-full of cell material from freshly grown bacterial lawns on agar plates. Each individual strain was resuspended in 1 mL 1 × PBS (8 g L−1 NaCl, 0.24 g L−1 KCl, 1.42 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4) by vortexing and pipetting, washed twice by centrifugation at 3500× g, and resuspended in 10 mL 1 × PBS. Optical density at 600 nm was determined for the cell suspensions, and 109 CFU of donors and recipients, i.e., 1 mL of OD600nm = 1 were mixed and concentrated by centrifugation. The mixtures were resuspended in 100 µL 1 × PBS, pipetted onto a nitrocellulose filter (0.22 µm pore diameter, Millipore, Burlington, MA, USA), placed on top of LB agar plates, and incubated at 30 °C. Bacteria were harvested after 24 h by placing the filter in an Eppendorf vial containing 1 mL 1 × PBS. The vial was vortexed until the complete bacterial biomass was dislodged and resuspended. From this suspension, a serial dilution was prepared up to 10−11, and 3 µL droplets were plated onto M9 lactose agar containing appropriate antibiotics to select for transconjugant EcO157:H7red. Conjugation frequency was calculated by the ratio of EcH157:H7red transconjugants to the total EcO157:H7red population. Conjugation data are known to be log-normal distributed; thus, transconjugant and donor CFU numbers were log10 transformed before mean values were calculated.
4.3.2. Plant Growth
A. thaliana Col0 seeds were surface sterilised by adding 1 mL 70% EtOH to ≈50 seeds. The seeds were incubated under constant agitation for 2 min, before they were collected by centrifugation at 1500× g for 1 min. The supernatant was discarded, and 1 mL sterilisation solution was added (1.17 mL bleach (12% NaOCl), 0.83 mL ddH2O, 20 µL 20% Triton × 100). The seeds were then incubated under constant agitation for five minutes before they were collected by centrifugation at 1500× g for 1 min. To remove residual sterilisation solution, we washed the seeds five times by adding 1 mL sterile water, centrifuging them, and then discarding the supernatant, after which we added 1 mL of sterile water. For stratification, seeds were stored at 4 °C for four days.
For plant cultivation, all wells of 24-well microtiter plates were filled with 1 mL ½ strength Murashige and Skoog (MS) agar (2.2 g L−1 MS powder including vitamins (Duchefa, Haarlem, The Netherlands), 10 g L−1 sucrose, 5.5 g L−1 plant agar (Duchefa), pH adjusted to 5.8), after which the plates were exposed to UV light in a laminar flow for 15 min [47]. Individual stratified seeds were placed into each well of the prepared microtiter plates; then, the plate was closed using Parafilm® and placed in a translucent plastic bag. Plants were then grown in a plant growth chamber (Percival, Perry, IA, USA) at long day conditions (16 h day/8 h night, 22 °C day, 18 °C night, 70% relative humidity). Plants were grown 3 to 3.5 weeks and developed between six to eight leaves before they were inoculated with bacteria.
4.3.3. Plant Inoculation with Conjugation Partners and Harvest
Bacterial strains were grown overnight on LB agar plates containing appropriate antibiotics. Freshly grown colonies of each bacterial strain were harvested using an inoculation loop, and the bacteria were resuspended in 10 mL 1 × PBS, washed twice by centrifugation at 3500× g, and resuspended in 1 × PBS. Optical density at 600 nm was determined for the cell suspensions. For single strain growth experiments, the optical density of each strain was set to OD600nm = 0.2 before 20 µL of bacterial suspension were pipetted onto the middle of individual plant rosettes. For in planta conjugation experiments, donor and recipient were mixed in 1 × PBS and 20 µL of the mixture were pipetted onto individual plants. The inoculation densities were dependent on the experiment and inoculation densities ranged from OD600nm = 0.05, 0.1, 0.25, to 0.5 of donor and recipient. For experiments described in Figure 3 and Figure 4, donors and recipients were each co-inoculated at an OD600nm of 0.05. For experiments described in Figure 5, donors and recipients were mixed in ratios 1:1 (OD600nm 0.05/0.05), 2:1 (OD600nm 0.1/0.05), 5:1 (OD600nm0.25/0.05), or 10:1 (OD600nm 0.5/0.05). For experiments described in Figure 6, donors and recipients were mixed in ratios 1:2 (OD600nm 0.05/0.1), 1:5 (OD600nm 0.05/0.25), 1:10 (OD600nm 0.05/0.5), 2:1 (OD600nm 0.1/0.05), 5:1 (OD600nm 0.25/0.05), or 10:1 (OD600nm 0.5/0.05). The inoculated plants were further incubated at standard growth conditions (16 h day/8 h night, 22 °C day, 18 °C night, 70% relative humidity). Plants were harvested at different time points, and bacteria were washed off to determine the CFU of each strain and transconjugants. To that end, 3 individual plants per treatment were individually processed. Plants were harvested using sterile forceps and the roots cut from the plants on a sterile surface with a sterile scalpel. Plants were transferred to pre-weighed 2 mL tubes and their weight was determined. To dislodge bacteria from plants, we added 1 mL 1 × PBS to a tube, vortexed it for 15 s, and after 7 min of sonication vortexed it again for 15 s. A total of 100 µL of the wash was spread on M9lactose + appropriate antibiotic to select for transconjugants when EcS17-1 or Pe299R were used as donors. When EcO157:H7 was used as a donor, transconjugants were selected on LBrif + appropriate antibiotic. To extend the range of transconjugants detection, we performed a 10-times dilution series from the leaf wash and placed 3 µL droplets on appropriate agar selective for transconjugants.
4.3.4. Statistical Analysis
Data were statistically analysed using the software Prism 7 (Graphpad Software, San Diego, CA, USA). All CFU were log-transformed before plotting or statistical tests were performed. To accommodate values below the limit of detection, we added a 1 to all values. To compare the difference of the mean between treatments, we performed a one-way ANOVA using Kruskal–Wallis test with Dunn’s correction for multiple comparisons.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10080928/s1, Figure S1: Overview of experiments.
Author Contributions
M.N.P.R.-E. conceived the study, planned the experiments, performed experiments, analysed the data, and wrote the manuscript. D.A. analysed data and wrote the manuscript. C.P. conceived the study, performed experiments, and wrote the manuscript. P.G. performed the experiments. D.D. conceived the study and provided infrastructure, project management, and input on writing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Programme “Antimicrobial Resistance” (NRP 72, grant number 407240_167068) of the Swiss National Science Foundation. The publication of this article was funded by Freie Universität Berlin.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information file).
Acknowledgments
The authors thank Jack A. Heinemann, University of Canterbury, Christchurch, New Zealand, for valuable discussion and insights about plasmid conjugation. Uli Klümper and Barth Smets, University of Copenhagen, Denmark, kindly donated plasmids RP4::Plac::GFP and pKJK5::Plac::GFP. The authors acknowledge André Imboden, ETH Zurich, for providing A. thaliana seeds, and Katharina Schneider for technical assistance. The authors thank the Agroscope research program Reduction and Dynamics of Antibiotic-resistant and Persistent Microorganisms along Food Chains (REDYMO).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kardos, N.; Demain, A.L. Penicillin: The medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol. 2011, 92, 677–687. [Google Scholar] [CrossRef]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Neu, H.C. The crisis in antibiotic resistance. Science 1992, 257, 1064–1073. [Google Scholar] [CrossRef]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Palumbi, S.R. Humans as the World’s Greatest Evolutionary Force. Science 2001, 293, 1786–1790. [Google Scholar] [CrossRef] [PubMed]
- Cantas, L.; Shah, S.Q.A.; Cavaco, L.M.; Manaia, C.; Walsh, F.; Popowska, M.; Garelick, H.; Bürgmann, H.; Sørum, H. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.F.; Pla, M.D.P.; Mayer, K.H.; Kishi, H.; Gilleece, E.; Syvanen, M.; Hopkins, J.D. Intercontinental spread of a new antibiotic resistance gene on an epidemic plasmid. Science 1985, 230, 87–88. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Tedim, A.P.; Coque, T.M. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 2013, 4, 15. [Google Scholar] [CrossRef]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.H.; Sørensen, S.J.; Smets, B.F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015, 9, 934–945. [Google Scholar] [CrossRef]
- Colombi, E.; Straub, C.; Künzel, S.; Templeton, M.D.; McCann, H.C.; Rainey, P.B. Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids. Environ. Microbiol. 2017, 19, 819–832. [Google Scholar] [PubMed]
- Powell, B.J.; Purdy, K.J.; Thompson, I.P.; Bailey, M.J. Demonstration of tra plasmid activity in bacteria indigenous to the phyllosphere of sugar beet; gene transfer to a recombinant pseudomonad. FEMS Microbiol. Ecol. 1993, 12, 195–206. [Google Scholar] [CrossRef][Green Version]
- Normander, B.; Christensen, B.B.; Molin, S.; Kroer, N. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 1998, 64, 1902–1909. [Google Scholar] [CrossRef] [PubMed]
- Björklöf, K.; Nurmiaho-Lassila, E.L.; Klinger, N.; Haahtela, K.; Romantschuk, M. Colonization strategies and conjugal gene transfer of inoculated Pseudomonas syringae on the leaf surface. J. Appl. Microbiol. 2000, 89, 423–432. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Turner, S.; Bailey, M.J. Horizontal gene transfer in the phytosphere. New Phytol. 2003, 157, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Blau, K.; Bettermann, A.; Jechalke, S.; Fornefeld, E.; Vanrobaeys, Y.; Stalder, T.; Top, E.M.; Smalla, K. The transferable resistome of produce. bioRxiv 2018, 350629. [Google Scholar] [CrossRef] [PubMed]
- Ruinen, J. The phyllosphere. Plant Soil 1961, 15, 81–109. [Google Scholar] [CrossRef]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef]
- Wolters, B.; Kyselková, M.; Krögerrecklenfort, E.; Kreuzig, R.; Smalla, K. Transferable antibiotic resistance plasmids from biogas plant digestates often belong to the IncP-1ε subgroup. Front. Microbiol. 2014, 5, 765. [Google Scholar]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H.J. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brandl, M.T. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 2006, 44, 367–392. [Google Scholar] [CrossRef]
- Heaton, J.C.; Jones, K. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: A review. J. Appl. Microbiol. 2008, 104, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Lücker, S.; Müller, D.B.; Potthoff, E.; Daims, H.; Vorholt, J.A. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ. Microbiol. 2014, 16, 2329–2340. [Google Scholar] [CrossRef]
- Heinemann, J.A. Genetics of gene transfer between species. Trends Genet. 1991, 7, 181–185. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Biotechnology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Wilson, M.; Lindow, S.E. Ecological Similarity and Coexistence of Epiphytic Ice-Nucleating (Ice+) Pseudomonas syringae Strains and a Non-Ice-Nucleating (Ice−) Biological Control Agent. Appl. Environ. Microbiol. 1994, 60, 3128–3137. [Google Scholar] [CrossRef]
- Smits, T.H.M.; Rezzonico, F.; Kamber, T.; Blom, J.; Goesmann, A.; Ishimaru, C.A.; Frey, J.E.; Stockwell, V.O.; Duffy, B. Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1. PLoS ONE 2011, 6, e22247. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Kim, E.B.; Marco, M.L.; Tecon, R.; Leveau, J.H.J. Draft Genome Sequence of the Phyllosphere Model Bacterium Pantoea agglomerans 299R. Genome Announc. 2013, 1, e00036-13. [Google Scholar] [CrossRef]
- Lilley, A.K.; Bailey, M.J.; Barr, M.; Kilshaw, K.; Timms-Wilson, T.M.; Day, M.J.; Norris, S.J.; Jones, T.H.; Godfray, H.C.J. Population dynamics and gene transfer in genetically modified bacteria in a model microcosm: Gene transfer in phytosphere bacteria. Mol. Ecol. 2003, 12, 3097–3107. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Schlechter, R.O. Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.P.; Tecon, R.; Kowalchuk, G.A.; Leveau, J.H.J. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 2012, 6, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Tecon, R.; Leveau, J.H.J. The mechanics of bacterial cluster formation on plant leaf surfaces as revealed by bioreporter technology. Environ. Microbiol. 2012, 14, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Monier, J.-M.; Lindow, S.E. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl. Environ. Microbiol. 2005, 71, 5484–5493. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lopatkin, A.J.; Huang, S.; Smith, R.P.; Srimani, J.K.; Sysoeva, T.A.; Bewick, S.; Karig, D.K.; You, L. Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 2016, 1, 16044. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Barth, P.T.; Grinter, N.J. Map of plasmid RP4 derived by insertion of transposon C. J. Mol. Biol. 1977, 113, 455–474. [Google Scholar] [CrossRef]
- Sengeløv, G.; Kristensen, K.J.; Sørensen, A.H.; Kroer, N.; Sørensen, S.J. Effect of genomic location on horizontal transfer of a recombinant gene cassette between Pseudomonas strains in the rhizosphere and spermosphere of barley seedlings. Curr. Microbiol. 2001, 42, 160–167. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Gisler, P.; Drissner, D. MiniTn7-transposon delivery vectors for inducible or constitutive fluorescent protein expression in Enterobacteriaceae. FEMS Microbiol. Lett. 2016, 363, fnw178. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Innerebner, G.; Zingg, J.; Guder, J.; Vorholt, J.A. Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp. strain Fr1 against Pseudomonas syringae DC3000. Appl. Environ. Microbiol. 2012, 78, 5529–5535. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).