Detection of Multidrug-Resistant Enterobacterales—From ESBLs to Carbapenemases
Abstract
1. Intestinal Colonization by Multidrug-Resistant Bacteria and the Impact on Global Health
2. Detection and Characterization of MDR Enterobacterales
2.1. Preanalytical Considerations
2.2. Culture Based Methods
2.2.1. Screening with Selective Chromogenic and Non-Chromogenic Media
2.2.2. Susceptibility Testing of MDRE
2.2.3. Confirmation Tests
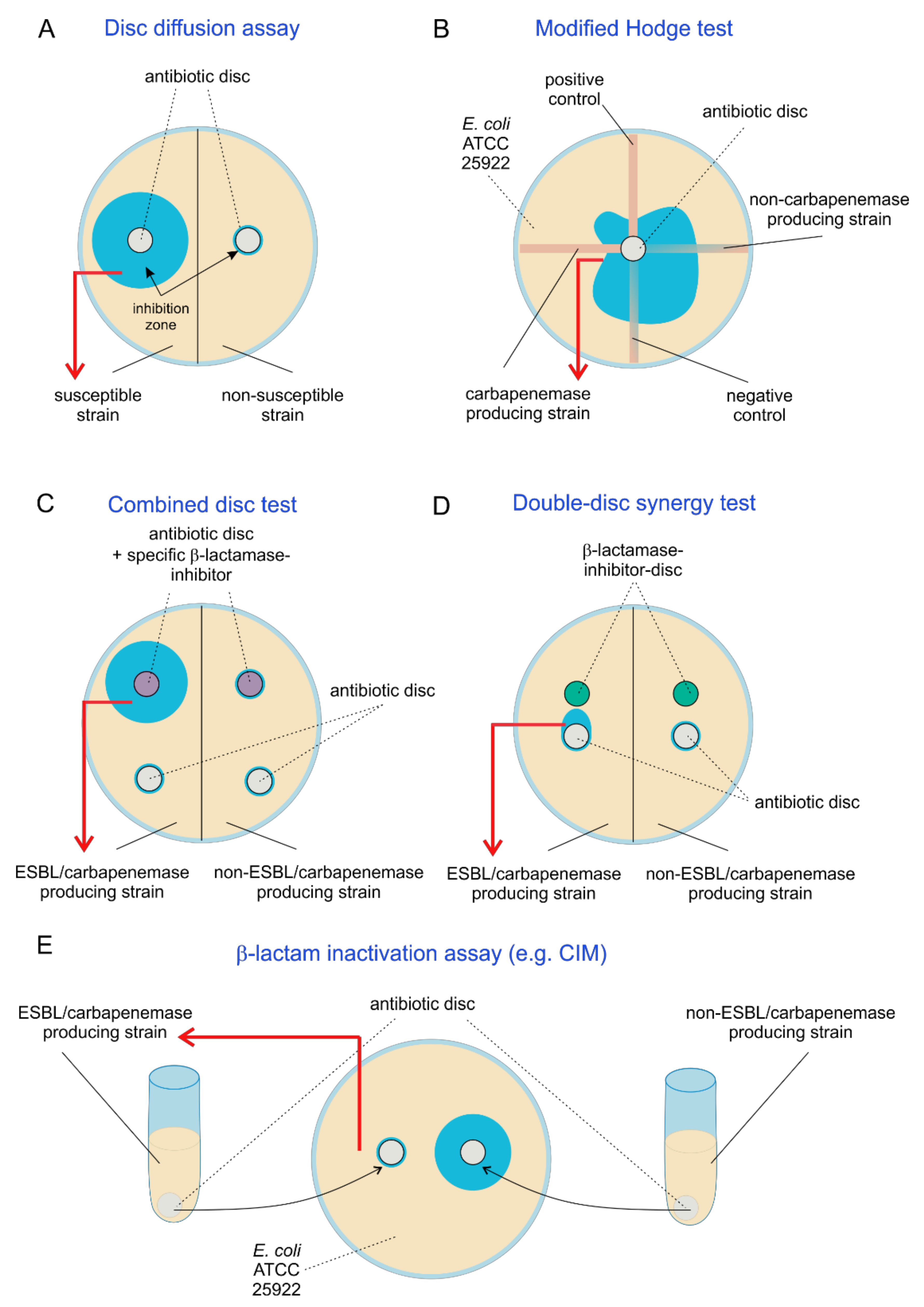
2.2.4. Disc Diffusion Assays for Detection of ESBL and Carbapenemase Production
2.2.5. Modified Hodge Test
2.2.6. β-Lactam Inactivation Assay (e.g., CIM)
2.2.7. Colorimetric Assays
2.3. Molecular Methods
2.3.1. PCR, RT-PCR and Microarray Techniques
2.3.2. Loop-Mediated Isothermal Amplification Assay (LAMP)
2.4. Further Methods
2.4.1. Immunochromatographic Test (ICT)
2.4.2. Electrochemical Assays
2.4.3. MALDI-TOF MS
3. Summary and Future Perspectives
Funding
References
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P. Carbapenemase-producing Enterobacteriaceae: Overview of a major public health challenge. Med. Mal. Infect. 2014, 44, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Vrancianu, C.O.; Dobre, E.G.; Gheorghe, I.; Barbu, I.; Cristian, R.E.; Chifiriuc, M.C. Present and Future Perspectives on Therapeutic Options for Carbapenemase-Producing Enterobacterales Infections. Microorganisms 2021, 9, 730. [Google Scholar] [CrossRef]
- Mota, R.; Pinto, M.; Palmeira, J.; Goncalves, D.; Ferreira, H. Multidrug-resistant bacteria as intestinal colonizers and evolution of intestinal colonization in healthy university students in Portugal. Access Microbiol. 2021, 3, acmi000182. [Google Scholar] [CrossRef] [PubMed]
- Ruppe, E.; Andremont, A. Causes, consequences, and perspectives in the variations of intestinal density of colonization of multidrug-resistant enterobacteria. Front. Microbiol. 2013, 4, 129. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Mozes, J.; Monostori, J.; Goracz, O.; Fesus, A.; Majoros, L.; Szarka, K.; Kardos, G. Comparison of rates of fecal colonization with extended-spectrum beta-lactamase-producing enterobacteria among patients in different wards, outpatients and medical students. Microbiol. Immunol. 2016, 60, 285–294. [Google Scholar] [CrossRef]
- Angelin, M.; Forsell, J.; Granlund, M.; Evengard, B.; Palmgren, H.; Johansson, A. Risk factors for colonization with extended-spectrum beta-lactamase producing Enterobacteriaceae in healthcare students on clinical assignment abroad: A prospective study. Travel Med. Infect. Dis. 2015, 13, 223–229. [Google Scholar] [CrossRef]
- Maharjan, A.; Bhetwal, A.; Shakya, S.; Satyal, D.; Shah, S.; Joshi, G.; Khanal, P.R.; Parajuli, N.P. Ugly bugs in healthy guts! Carriage of multidrug-resistant and ESBL-producing commensal Enterobacteriaceae in the intestine of healthy Nepalese adults. Infect. Drug Resist. 2018, 11, 547–554. [Google Scholar] [CrossRef]
- Chirindze, L.M.; Zimba, T.F.; Sekyere, J.O.; Govinden, U.; Chenia, H.Y.; Sundsfjord, A.; Essack, S.Y.; Simonsen, G.S. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infect. Dis. 2018, 18, 244. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Lai, L.C.; Chen, Y.A.; Lin, K.Y.; Chou, Y.H.; Chen, H.C.; Wang, S.S.; Wang, J.T.; Chang, S.C. Colonization with Multidrug-Resistant Organisms Among Healthy Adults in the Community Setting: Prevalence, Risk Factors, and Composition of Gut Microbiome. Front. Microbiol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Arcilla, M.S.; van Hattem, J.M.; Haverkate, M.R.; Bootsma, M.C.J.; van Genderen, P.J.J.; Goorhuis, A.; Grobusch, M.P.; Lashof, A.M.O.; Molhoek, N.; Schultsz, C.; et al. Import and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): A prospective, multicentre cohort study. Lancet Infect. Dis. 2017, 17, 78–85. [Google Scholar] [CrossRef]
- Hamprecht, A.; Rohde, A.M.; Behnke, M.; Feihl, S.; Gastmeier, P.; Gebhardt, F.; Kern, W.V.; Knobloch, J.K.; Mischnik, A.; Obermann, B.; et al. Colonization with third-generation cephalosporin-resistant Enterobacteriaceae on hospital admission: Prevalence and risk factors. J. Antimicrob. Chemother. 2016, 71, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Villodres, A.; Martin-Gandul, C.; Penalva, G.; Guisado-Gil, A.B.; Crespo-Rivas, J.C.; Pachon-Ibanez, M.E.; Lepe, J.A.; Cisneros, J.M. Prevalence and Risk Factors for Multidrug-Resistant Organisms Colonization in Long-Term Care Facilities Around the World: A Review. Antibiotics 2021, 10, 680. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Maechler, F.; Pena Diaz, L.A.; Schroder, C.; Geffers, C.; Behnke, M.; Gastmeier, P. Prevalence of carbapenem-resistant organisms and other Gram-negative MDRO in German ICUs: First results from the national nosocomial infection surveillance system (KISS). Infection 2015, 43, 163–168. [Google Scholar] [CrossRef]
- Zaha, D.C.; Kiss, R.; Hegedus, C.; Gesztelyi, R.; Bombicz, M.; Muresan, M.; Pallag, A.; Zrinyi, M.; Pall, D.; Vesa, C.M.; et al. Recent Advances in Investigation, Prevention, and Management of Healthcare-Associated Infections (HAIs): Resistant Multidrug Strain Colonization and Its Risk Factors in an Intensive Care Unit of a University Hospital. Biomed. Res. Int 2019, 2019, 2510875. [Google Scholar] [CrossRef] [PubMed]
- Arena, F.; Vannetti, F.; Di Pilato, V.; Fabbri, L.; Colavecchio, O.L.; Giani, T.; Marraccini, C.; Pupillo, R.; Macchi, C.; Converti, F.; et al. Diversity of the epidemiology of carbapenemase-producing Enterobacteriaceae in long-term acute care rehabilitation settings from an area of hyperendemicity, and evaluation of an intervention bundle. J. Hosp. Infect. 2018, 100, 29–34. [Google Scholar] [CrossRef]
- Magrini, E.T.N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- ECDC. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2020.
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum beta-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Proliferation and significance of clinically relevant beta-lactamases. Ann. N. Y. Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Front. Microbiol. 2019, 10, 1823. [Google Scholar] [CrossRef] [PubMed]
- Arnold, R.S.; Thom, K.A.; Sharma, S.; Phillips, M.; Kristie Johnson, J.; Morgan, D.J. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South. Med. J. 2011, 104, 40–45. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Nordmann, P.; Mariotte, S.; Naas, T.; Labia, R.; Nicolas, M.H. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 1993, 37, 939–946. [Google Scholar] [CrossRef]
- Rasmussen, B.A.; Bush, K.; Keeney, D.; Yang, Y.; Hare, R.; O’Gara, C.; Medeiros, A.A. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 1996, 40, 2080–2086. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]
- Arakawa, Y.; Murakami, M.; Suzuki, K.; Ito, H.; Wacharotayankun, R.; Ohsuka, S.; Kato, N.; Ohta, M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 1995, 39, 1612–1615. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Gniadkowski, M.; Giske, C.G.; Poirel, L.; Woodford, N.; Miriagou, V.; European Network on, C. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 2012, 18, 432–438. [Google Scholar] [CrossRef]
- Larson, E. Community factors in the development of antibiotic resistance. Annu Rev. Public Health 2007, 28, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Voor, A.F.; Mourik, K.; Beishuizen, B.; van der Schoor, A.S.; Verbon, A.; Vos, M.C.; Severin, J.A. Acquisition of multidrug-resistant Enterobacterales during international travel: A systematic review of clinical and microbiological characteristics and meta-analyses of risk factors. Antimicrob. Resist. Infect. Control 2020, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Cataldo, M.A.; Dancer, S.J.; De Angelis, G.; Falcone, M.; Frank, U.; Kahlmeter, G.; Pan, A.; Petrosillo, N.; Rodriguez-Bano, J.; et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 2014, 20 (Suppl. 1), 1–55. [Google Scholar] [CrossRef]
- CDC. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb. Mortal Wkly. Rep. 2009, 58, 256–260. [Google Scholar]
- Jazmati, T.; Hamprecht, A.; Jazmati, N. Comparison of stool samples and rectal swabs with and without pre-enrichment for the detection of third-generation cephalosporin-resistant Enterobacterales (3GCREB). Eur. J. Clin. Microbiol. Infect. Dis. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.D.; Rhinehart, E.; Jackson, M.; Chiarello, L.; Healthcare Infection Control Practices Advisory, C. Management of multidrug-resistant organisms in health care settings, 2006. Am. J. Infect. Control. 2007, 35, S165–S193. [Google Scholar] [CrossRef]
- Van Prehn, J.; Kaiser, A.M.; van der Werff, S.D.; van Mansfeld, R.; Vandenbroucke-Grauls, C. Colonization sites in carriers of ESBL-producing Gram-negative bacteria. Antimicrob. Resist. Infect. Control 2018, 7, 52. [Google Scholar] [CrossRef]
- Warnke, P.; Johanna Pohl, F.P.; Kundt, G.; Podbielski, A. Screening for Gram-negative bacteria: Impact of preanalytical parameters. Sci. Rep. 2016, 6, 30427. [Google Scholar] [CrossRef] [PubMed]
- Dyakova, E.; Bisnauthsing, K.N.; Querol-Rubiera, A.; Patel, A.; Ahanonu, C.; Tosas Auguet, O.; Edgeworth, J.D.; Goldenberg, S.D.; Otter, J.A. Efficacy and acceptability of rectal and perineal sampling for identifying gastrointestinal colonization with extended spectrum beta-lactamase Enterobacteriaceae. Clin. Microbiol. Infect. 2017, 23, 577.e1–577.e3. [Google Scholar] [CrossRef] [PubMed]
- Sturod, K.; Dahle, U.R.; Berg, E.S.; Steinbakk, M.; Wester, A.L. Evaluation of the ability of four ESBL-screening media to detect ESBL-producing Salmonella and Shigella. BMC Microbiol. 2014, 14, 217. [Google Scholar] [CrossRef]
- Lucena Baeza, L.; Hamprecht, A. A profile of the GenePOC Carba C assay for the detection and differentiation of gene sequences associated with carbapenem-non-susceptibility. Expert Rev. Mol. Diagn. 2020, 20, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Göttig, S.; Walker, S.V.; Saleh, A.; Koroska, F.; Sommer, J.; Stelzer, Y.; Steinmann, J.; Hamprecht, A. Comparison of nine different selective agars for the detection of carbapenemase-producing Enterobacterales (CPE). Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 923–927. [Google Scholar] [CrossRef]
- Simner, P.J.; Gilmour, M.W.; DeGagne, P.; Nichol, K.; Karlowsky, J.A. Evaluation of five chromogenic agar media and the Rosco Rapid Carb screen kit for detection and confirmation of carbapenemase production in Gram-negative bacilli. J. Clin. Microbiol. 2015, 53, 105–112. [Google Scholar] [CrossRef]
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef]
- Sattler, J.; Brunke, A.; Hamprecht, A. Systematic comparison of three commercially available combination disc tests and zCIM for carbapenemase detection in Enterobacterales isolates. J. Clin. Microbiol. 2021, 59, e0314020. [Google Scholar] [CrossRef] [PubMed]
- Pantel, A.; Souzy, D.; Sotto, A.; Lavigne, J.P. Evaluation of Two Phenotypic Screening Tests for Carbapenemase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2015, 53, 3359–3362. [Google Scholar] [CrossRef]
- Girlich, D.; Poirel, L.; Nordmann, P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 477–479. [Google Scholar] [CrossRef]
- Van der Zwaluw, K.; de Haan, A.; Pluister, G.N.; Bootsma, H.J.; de Neeling, A.J.; Schouls, L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef]
- Baeza, L.L.; Pfennigwerth, N.; Greissl, C.; Gottig, S.; Saleh, A.; Stelzer, Y.; Gatermann, S.G.; Hamprecht, A. Comparison of five methods for detection of carbapenemases in Enterobacterales with proposal of a new algorithm. Clin. Microbiol. Infect. 2019, 25, 1286.e9–1286.e15. [Google Scholar] [CrossRef]
- Meier, M.; Hamprecht, A. Systematic Comparison of Four Methods for Detection of Carbapenemase-Producing Enterobacterales Directly from Blood Cultures. J. Clin. Microbiol. 2019, 57, e00709-19. [Google Scholar] [CrossRef]
- Jing, X.; Min, X.; Zhang, X.; Gong, L.; Wu, T.; Sun, R.; Chen, L.; Liu, R.; Zeng, J. The Rapid Carbapenemase Detection Method (rCDM) for Rapid and Accurate Detection of Carbapenemase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 2019, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Brechard, L.; Poirel, L.; Nordmann, P. Rapid detection of carbapenemase-producing Enterobacteriaceae from blood cultures. Clin. Microbiol. Infect. 2014, 20, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Agathine, A.; Naas, T.; Cuzon, G.; Poirel, L.; Nordmann, P. Evaluation of the RAPIDEC(R) CARBA NP, the Rapid CARB Screen(R) and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2015, 70, 3014–3022. [Google Scholar] [CrossRef]
- Bernabeu, S.; Dortet, L.; Naas, T. Evaluation of the beta-CARBA test, a colorimetric test for the rapid detection of carbapenemase activity in Gram-negative bacilli. J. Antimicrob. Chemother. 2017, 72, 1646–1658. [Google Scholar] [CrossRef]
- Mancini, S.; Kieffer, N.; Poirel, L.; Nordmann, P. Evaluation of the RAPIDEC(R) CARBA NP and beta-CARBA(R) tests for rapid detection of Carbapenemase-producing Enterobacteriaceae. Diagn Microbiol. Infect. Dis. 2017, 88, 293–297. [Google Scholar] [CrossRef]
- Sattler, J.; Brunke, A.; Hamprecht, A. Evaluation of CARBA PAcE, a novel rapid test for detection of carbapenemase-producing Enterobacterales. J. Med. Microbiol. 2021, 70, 001290. [Google Scholar] [CrossRef] [PubMed]
- Pires, J.; Novais, A.; Peixe, L. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J. Clin. Microbiol. 2013, 51, 4281–4283. [Google Scholar] [CrossRef] [PubMed]
- Novais, A.; Brilhante, M.; Pires, J.; Peixe, L. Evaluation of the Recently Launched Rapid Carb Blue Kit for Detection of Carbapenemase-Producing Gram-Negative Bacteria. J. Clin. Microbiol. 2015, 53, 3105–3107. [Google Scholar] [CrossRef] [PubMed]
- Pasteran, F.; Tijet, N.; Melano, R.G.; Corso, A. Simplified Protocol for Carba NP Test for Enhanced Detection of Carbapenemase Producers Directly from Bacterial Cultures. J. Clin. Microbiol. 2015, 53, 3908–3911. [Google Scholar] [CrossRef]
- Ma, C.W.; Ng, K.K.; Yam, B.H.; Ho, P.L.; Kao, R.Y.; Yang, D. Rapid Broad Spectrum Detection of Carbapenemases with a Dual Fluorogenic-Colorimetric Probe. J. Am. Chem. Soc. 2021, 143, 6886–6894. [Google Scholar] [CrossRef]
- Hamprecht, A.; Vehreschild, J.J.; Seifert, H.; Saleh, A. Rapid detection of NDM, KPC and OXA-48 carbapenemases directly from positive blood cultures using a new multiplex immunochromatographic assay. PLoS ONE 2018, 13, e0204157. [Google Scholar] [CrossRef]
- Greissl, C.; Saleh, A.; Hamprecht, A. Rapid detection of OXA-48-like, KPC, NDM, and VIM carbapenemases in Enterobacterales by a new multiplex immunochromatographic test. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 331–335. [Google Scholar] [CrossRef]
- Han, R.; Guo, Y.; Peng, M.; Shi, Q.; Wu, S.; Yang, Y.; Zheng, Y.; Yin, D.; Hu, F. Evaluation of the Immunochromatographic NG-Test Carba 5, RESIST-5 O.O.K.N.V., and IMP K-SeT for Rapid Detection of KPC-, NDM-, IMP-, VIM-type, and OXA-48-like Carbapenemase Among Enterobacterales. Front. Microbiol. 2020, 11, 609856. [Google Scholar] [CrossRef]
- Takissian, J.; Bonnin, R.A.; Naas, T.; Dortet, L. NG-Test Carba 5 for Rapid Detection of Carbapenemase-Producing Enterobacterales from Positive Blood Cultures. Antimicrob. Agents Chemother. 2019, 63, e00011-19. [Google Scholar] [CrossRef]
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar] [CrossRef]
- Hopkins, K.L.; Meunier, D.; Naas, T.; Volland, H.; Woodford, N. Evaluation of the NG-Test CARBA 5 multiplex immunochromatographic assay for the detection of KPC, OXA-48-like, NDM, VIM and IMP carbapenemases. J. Antimicrob. Chemother. 2018, 73, 3523–3526. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Riazzo, C.; Miranda-Casas, C.; Rojo-Martin, M.D.; Gutierrez-Fernandez, J.; Navarro-Mari, J.M. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J. Microbiol. Methods 2014, 105, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Akyar, I.; Kaya Ayas, M.; Karatuna, O. Performance Evaluation of MALDI-TOF MS MBT STAR-BL Versus In-House Carba NP Testing for the Rapid Detection of Carbapenemase Activity in Escherichia coli and Klebsiella pneumoniae Strains. Microb. Drug Resist. 2019, 25, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Anantharajah, A.; Tossens, B.; Olive, N.; Kabamba-Mukadi, B.; Rodriguez-Villalobos, H.; Verroken, A. Performance Evaluation of the MBT STAR((R))-Carba IVD Assay for the Detection of Carbapenemases with MALDI-TOF MS. Front. Microbiol. 2019, 10, 1413. [Google Scholar] [CrossRef] [PubMed]
- Cordovana, M.; Abdalla, M.; Ambretti, S. Evaluation of the MBT STAR-Carba Assay for the Detection of Carbapenemase Production in Enterobacteriaceae and Hafniaceae with a Large Collection of Routine Isolates from Plate Cultures and Patient-Derived Positive Blood Cultures. Microb. Drug Resist. 2020, 26, 1298–1306. [Google Scholar] [CrossRef]
- Dortet, L.; Tande, D.; de Briel, D.; Bernabeu, S.; Lasserre, C.; Gregorowicz, G.; Jousset, A.B.; Naas, T. MALDI-TOF for the rapid detection of carbapenemase-producing Enterobacteriaceae: Comparison of the commercialized MBT STAR(R)-Carba IVD Kit with two in-house MALDI-TOF techniques and the RAPIDEC(R) CARBA NP. J. Antimicrob. Chemother. 2018, 73, 2352–2359. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018, 24, 738–743. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Storck, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Rapid Direct Susceptibility Testing from Positive Blood Cultures by the Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry-Based Direct-on-Target Microdroplet Growth Assay. J. Clin. Microbiol. 2018, 56, e00913-18. [Google Scholar] [CrossRef]
- Bogaerts, P.; Yunus, S.; Massart, M.; Huang, T.D.; Glupczynski, Y. Evaluation of the BYG Carba Test, a New Electrochemical Assay for Rapid Laboratory Detection of Carbapenemase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.; Boudet, A.; Lavigne, J.P.; Pantel, A. Evaluation of Two Methods for the Detection of Third Generation Cephalosporins Resistant Enterobacterales Directly from Positive Blood Cultures. Front. Cell Infect. Microbiol. 2020, 10, 491. [Google Scholar] [CrossRef]
- Garcia-Fernandez, S.; Morosini, M.I.; Marco, F.; Gijon, D.; Vergara, A.; Vila, J.; Ruiz-Garbajosa, P.; Canton, R. Evaluation of the eazyplex(R) SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J. Antimicrob. Chemother. 2015, 70, 1047–1050. [Google Scholar] [CrossRef]
- Girlich, D.; Oueslati, S.; Bernabeu, S.; Langlois, I.; Begasse, C.; Arangia, N.; Creton, E.; Cotellon, G.; Sauvadet, A.; Dortet, L.; et al. Evaluation of the BD MAX Check-Points CPO Assay for the Detection of Carbapenemase Producers Directly from Rectal Swabs. J. Mol. Diagn. 2020, 22, 294–300. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Bogaerts, P.; Glupczynski, Y.; Nordmann, P. Evaluation of a DNA microarray for the rapid detection of extended-spectrum beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J. Antimicrob. Chemother. 2012, 67, 1865–1869. [Google Scholar] [CrossRef]
- Girlich, D.; Bernabeu, S.; Fortineau, N.; Dortet, L.; Naas, T. Evaluation of the CRE and ESBL ELITe MGB(R) kits for the accurate detection of carbapenemase- or CTX-M-producing bacteria. Diagn. Microbiol. Infect. Dis. 2018, 92, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, S.; Girlich, D.; Dortet, L.; Naas, T. Evaluation of the Amplidiag CarbaR+VRE Kit for Accurate Detection of Carbapenemase-Producing Bacteria. J. Clin. Microbiol. 2018, 56, e01092-17. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Bernabeu, S.; Grosperrin, V.; Langlois, I.; Begasse, C.; Arangia, N.; Creton, E.; Cotellon, G.; Sauvadet, A.; Dortet, L.; et al. Evaluation of the Amplidiag CarbaR+MCR Kit for Accurate Detection of Carbapenemase-Producing and Colistin-Resistant Bacteria. J. Clin. Microbiol. 2019, 57, e01800-18. [Google Scholar] [CrossRef]
- Dortet, L.; Fusaro, M.; Naas, T. Improvement of the Xpert Carba-R Kit for the Detection of Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Tojo, M.; Fujita, T.; Ainoda, Y.; Nagamatsu, M.; Hayakawa, K.; Mezaki, K.; Sakurai, A.; Masui, Y.; Yazaki, H.; Takahashi, H.; et al. Evaluation of an automated rapid diagnostic assay for detection of Gram-negative bacteria and their drug-resistance genes in positive blood cultures. PLoS ONE 2014, 9, e94064. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Verroken, A.; Despas, N.; Rodriguez-Villalobos, H.; Laterre, P.F. The impact of a rapid molecular identification test on positive blood cultures from critically ill with bacteremia: A pre-post intervention study. PLoS ONE 2019, 14, e0223122. [Google Scholar] [CrossRef]
- Hopkins, T.M.; Juang, P.; Weaver, K.; Kollef, M.H.; Betthauser, K.D. Outcomes of Macrolide Deescalation in Severe Community-acquired Pneumonia. Clin. Ther. 2019, 41, 2540–2548. [Google Scholar] [CrossRef]
- Huang, T.D.; Melnik, E.; Bogaerts, P.; Evrard, S.; Glupczynski, Y. Evaluation of the ePlex Blood Culture Identification Panels for Detection of Pathogens in Bloodstream Infections. J. Clin. Microbiol. 2019, 57, e01597-18. [Google Scholar] [CrossRef]
- Burrack-Lange, S.C.; Personne, Y.; Huber, M.; Winkler, E.; Weile, J.; Knabbe, C.; Gorig, J.; Rohde, H. Multicenter assessment of the rapid Unyvero Blood Culture molecular assay. J. Med. Microbiol. 2018, 67, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Kaase, M.; Szabados, F.; Wassill, L.; Gatermann, S.G. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J. Clin. Microbiol. 2012, 50, 3115–3118. [Google Scholar] [CrossRef] [PubMed]
- Ceyssens, P.J.; Garcia-Graells, C.; Fux, F.; Botteldoorn, N.; Mattheus, W.; Wuyts, V.; De Keersmaecker, S.; Dierick, K.; Bertrand, S. Development of a Luminex xTAG(R) assay for cost-effective multiplex detection of beta-lactamases in Gram-negative bacteria. J. Antimicrob. Chemother. 2016, 71, 2479–2483. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, P.; Hamels, S.; de Mendonca, R.; Huang, T.D.; Roisin, S.; Remacle, J.; Markine-Goriaynoff, N.; de Longueville, F.; Pluster, W.; Denis, O.; et al. Analytical validation of a novel high multiplexing real-time PCR array for the identification of key pathogens causative of bacterial ventilator-associated pneumonia and their associated resistance genes. J. Antimicrob. Chemother. 2013, 68, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015, 33, 296–300. [Google Scholar] [CrossRef]
- Reglier-Poupet, H.; Naas, T.; Carrer, A.; Cady, A.; Adam, J.M.; Fortineau, N.; Poyart, C.; Nordmann, P. Performance of chromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum beta-lactamases. J. Med. Microbiol. 2008, 57, 310–315. [Google Scholar] [CrossRef]
- Huang, T.D.; Bogaerts, P.; Berhin, C.; Guisset, A.; Glupczynski, Y. Evaluation of Brilliance ESBL agar, a novel chromogenic medium for detection of extended-spectrum-beta- lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2010, 48, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Grohs, P.; Tillecovidin, B.; Caumont-Prim, A.; Carbonnelle, E.; Day, N.; Podglajen, I.; Gutmann, L. Comparison of five media for detection of extended-spectrum Beta-lactamase by use of the wasp instrument for automated specimen processing. J. Clin. Microbiol. 2013, 51, 2713–2716. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans-van den Bergh, M.F.; Verhulst, C.; Willemsen, L.E.; Verkade, E.; Bonten, M.J.; Kluytmans, J.A. Rectal Carriage of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae in Hospitalized Patients: Selective Preenrichment Increases Yield of Screening. J. Clin. Microbiol. 2015, 53, 2709–2712. [Google Scholar] [CrossRef]
- Jazmati, N.; Hein, R.; Hamprecht, A. Use of an Enrichment Broth Improves Detection of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae in Clinical Stool Samples. J. Clin. Microbiol. 2016, 54, 467–470. [Google Scholar] [CrossRef]
- Jazmati, N.; Jazmati, T.; Hamprecht, A. Importance of pre-enrichment for detection of third-generation cephalosporin-resistant Enterobacteriaceae (3GCREB) from rectal swabs. Eur J. Clin. Microbiol. Infect. Dis. 2017, 36, 1847–1851. [Google Scholar] [CrossRef] [PubMed]
- Farber, J.; Moder, K.A.; Layer, F.; Tammer, I.; Konig, W.; Konig, B. Extended-spectrum Beta-lactamase detection with different panels for automated susceptibility testing and with a chromogenic medium. J. Clin. Microbiol. 2008, 46, 3721–3727. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Geiss, H.K.; Mack, D.; Sturenburg, E.; Seifert, H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J. Clin. Microbiol. 2007, 45, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Espinar, M.J.; Rocha, R.; Ribeiro, M.; Goncalves Rodrigues, A.; Pina-Vaz, C. Extended-spectrum beta-lactamases of Escherichia coli and Klebsiella pneumoniae screened by the VITEK 2 system. J. Med. Microbiol. 2011, 60, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Gatermann, S.; Pfeifer, Y.; Reischl, U.; Gessner, A.; Jantsch, J. Evaluation of the automated BD Phoenix CPO Detect panel in combination with the beta-CARBA assay for detection and classification of carbapenemase-producing Enterobacterales. J. Microbiol. Methods 2019, 156, 29–33. [Google Scholar] [CrossRef]
- Jonas, D.; Reuter, S.; Klassen, S.; Weber, S.; Buck, M.; Giani, T.; Rossolini, G.M.; Grundmann, H. Evaluation of the BD Phoenix CPO detect panel for prediction of Ambler class carbapenemases. Sci. Rep. 2021, 11, 13150. [Google Scholar] [CrossRef]
- He, Q.; Chen, W.; Huang, L.; Lin, Q.; Zhang, J.; Liu, R.; Li, B. Performance evaluation of three automated identification systems in detecting carbapenem-resistant Enterobacteriaceae. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 40. [Google Scholar] [CrossRef]
- EUCAST. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 12 August 2021).
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 90–103. [Google Scholar] [CrossRef]
- Kaur, J.; Chopra, S.; Sheevani, G.M. Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella pneumoniae. J. Clin. Diagn Res. 2013, 7, 229–233. [Google Scholar] [CrossRef]
- Doi, Y.; Potoski, B.A.; Adams-Haduch, J.M.; Sidjabat, H.E.; Pasculle, A.W.; Paterson, D.L. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type beta-lactamase by use of a boronic acid compound. J. Clin. Microbiol. 2008, 46, 4083–4086. [Google Scholar] [CrossRef]
- Giske, C.G.; Gezelius, L.; Samuelsen, O.; Warner, M.; Sundsfjord, A.; Woodford, N. A sensitive and specific phenotypic assay for detection of metallo-beta-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 2011, 17, 552–556. [Google Scholar] [CrossRef]
- Tsakris, A.; Poulou, A.; Bogaerts, P.; Dimitroulia, E.; Pournaras, S.; Glupczynski, Y. Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J. Clin. Microbiol. 2015, 53, 1245–1251. [Google Scholar] [CrossRef]
- Huang, T.D.; Poirel, L.; Bogaerts, P.; Berhin, C.; Nordmann, P.; Glupczynski, Y. Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J. Antimicrob. Chemother. 2014, 69, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Leverstein-Van Hall, M.A.; Cohen Stuart, J.W. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Day, K.M.; Pike, R.; Winstanley, T.G.; Lanyon, C.; Cummings, S.P.; Raza, M.W.; Woodford, N.; Perry, J.D. Use of faropenem as an indicator of carbapenemase activity in the Enterobacteriaceae. J. Clin. Microbiol. 2013, 51, 1881–1886. [Google Scholar] [CrossRef]
- Koroska, F.; Gottig, S.; Kaase, M.; Steinmann, J.; Gatermann, S.; Sommer, J.; Wille, T.; Plum, G.; Hamprecht, A. Comparison of Phenotypic Tests and an Immunochromatographic Assay and Development of a New Algorithm for Detection of OXA-48-like Carbapenemases. J. Clin. Microbiol. 2017, 55, 877–883. [Google Scholar] [CrossRef]
- CLSI. The Modified Hodge Test for Suspected Carbapenemase Production in Enterobacteriaceae. Available online: https://clsi.org/media/1899/_m100_archived_methods_table.pdf (accessed on 12 August 2021).
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 12 August 2021).
- Bianco, G.; Boattini, M.; Iannaccone, M.; Fossati, L.; Cavallo, R.; Costa, C. Direct Beta-Lactam Inactivation Method: A New Low-Cost Assay for Rapid Detection of Carbapenemase- or Extended-Spectrum-beta-Lactamase-Producing Enterobacterales Directly from Positive Blood Culture Bottles. J. Clin. Microbiol. 2019, 58, e01178-19. [Google Scholar] [CrossRef]
- Camelena, F.; Cointe, A.; Mathy, V.; Hobson, C.; Doit, C.; Bercot, B.; Decre, D.; Podglajen, I.; Dortet, L.; Monjault, A.; et al. Within-a-Day Detection and Rapid Characterization of Carbapenemase by Use of a New Carbapenem Inactivation Method-Based Test, CIMplus. J. Clin. Microbiol. 2018, 56, e00137-18. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, M.M.; Hayden, J.A.; Fauntleroy, K.A.; Mazur, C.; Johnson, J.K.; Simner, P.J.; Das, S.; Satlin, M.J.; Jenkins, S.G.; Westblade, L.F. EDTA-Modified Carbapenem Inactivation Method: A Phenotypic Method for Detecting Metallo-beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, e01757-18. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Rapid detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 3016–3022. [Google Scholar] [CrossRef]
- Poirel, L.; Fernandez, J.; Nordmann, P. Comparison of Three Biochemical Tests for Rapid Detection of Extended-Spectrum-beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Gallah, S.; Decre, D.; Genel, N.; Arlet, G. The beta-Lacta test for direct detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in urine. J. Clin. Microbiol. 2014, 52, 3792–3794. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Poirel, L.; Nordmann, P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 2012, 56, 6437–6440. [Google Scholar] [CrossRef]
- Tijet, N.; Boyd, D.; Patel, S.N.; Mulvey, M.R.; Melano, R.G. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4578–4580. [Google Scholar] [CrossRef] [PubMed]
- Gazin, M.; Paasch, F.; Goossens, H.; Malhotra-Kumar, S.; Mosar, W.P.; Teams, S.W.S. Current trends in culture-based and molecular detection of extended-spectrum-beta-lactamase-harboring and carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 1140–1146. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Roschanski, N.; Fischer, J.; Guerra, B.; Roesler, U. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS ONE 2014, 9, e100956. [Google Scholar] [CrossRef]
- Chavda, K.D.; Satlin, M.J.; Chen, L.; Manca, C.; Jenkins, S.G.; Walsh, T.J.; Kreiswirth, B.N. Evaluation of a Multiplex PCR Assay to Rapidly Detect Enterobacteriaceae with a Broad Range of beta-Lactamases Directly from Perianal Swabs. Antimicrob. Agents Chemother. 2016, 60, 6957–6961. [Google Scholar] [CrossRef]
- Ogutu, J.O.; Zhang, Q.; Huang, Y.; Yan, H.; Su, L.; Gao, B.; Zhang, W.; Zhao, J.; Cai, W.; Li, W.; et al. Development of a multiplex PCR system and its application in detection of blaSHV, blaTEM, blaCTX-M-1, blaCTX-M-9 and blaOXA-1 group genes in clinical Klebsiella pneumoniae and Escherichia coli strains. J. Antibiot. 2015, 68, 725–733. [Google Scholar] [CrossRef]
- Souverein, D.; Euser, S.M.; van der Reijden, W.A.; Herpers, B.L.; Kluytmans, J.; Rossen, J.W.A.; Den Boer, J.W. Clinical sensitivity and specificity of the Check-Points Check-Direct ESBL Screen for BD MAX, a real-time PCR for direct ESBL detection from rectal swabs. J. Antimicrob. Chemother. 2017, 72, 2512–2518. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-S.; Lee, M. Verification of the performance of the BD MAX Check-Points CPO Assay on clinical isolates. J. Lab. Med. 2020, 44, 165–168. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Vasoo, S.; Patel, R. Evaluation of the Check-Points Check MDR CT103 and CT103 XL Microarray Kits by Use of Preparatory Rapid Cell Lysis. J. Clin. Microbiol. 2016, 54, 1368–1371. [Google Scholar] [CrossRef]
- Ko, Y.J.; Kim, J.; Kim, H.N.; Yoon, S.Y.; Lim, C.S.; Lee, C.K. Diagnostic performance of the Xpert Carba-R assay for active surveillance of rectal carbapenemase-producing organisms in intensive care unit patients. Antimicrob. Resist. Infect. Control 2019, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Traczewski, M.M.; Carretto, E.; Canton, R.; Moore, N.M.; Carba, R.S.T. Multicenter Evaluation of the Xpert Carba-R Assay for Detection of Carbapenemase Genes in Gram-Negative Isolates. J. Clin. Microbiol. 2018, 56, e00272-18. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.M.; Canton, R.; Carretto, E.; Peterson, L.R.; Sautter, R.L.; Traczewski, M.M.; Carba, R.S.T. Rapid Identification of Five Classes of Carbapenem Resistance Genes Directly from Rectal Swabs by Use of the Xpert Carba-R Assay. J. Clin. Microbiol. 2017, 55, 2268–2275. [Google Scholar] [CrossRef]
- Zalas-Wiecek, P.; Gospodarek-Komkowska, E.; Smalczewska, A. Rapid Detection of Genes Encoding Extended-Spectrum Beta-Lactamase and Carbapenemase in Clinical Escherichia coli Isolates with eazyplex SuperBug CRE System. Microb. Drug Resist. 2020, 26, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Sekowska, A.; Bogiel, T.; Gospodarek-Komkowska, E. Evaluation of eazyplex((R)) SuperBug CRE Test for Beta-Lactamase Genes Detection in Klebsiella spp. and P. aeruginosa Strains. Curr. Microbiol. 2020, 77, 99–103. [Google Scholar] [CrossRef]
- Findlay, J.; Hopkins, K.L.; Meunier, D.; Woodford, N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J. Antimicrob. Chemother. 2015, 70, 1338–1342. [Google Scholar] [CrossRef]
- Hinic, V.; Ziegler, J.; Straub, C.; Goldenberger, D.; Frei, R. Extended-spectrum beta-lactamase (ESBL) detection directly from urine samples with the rapid isothermal amplification-based eazyplex(R) SuperBug CRE assay: Proof of concept. J. Microbiol. Methods 2015, 119, 203–205. [Google Scholar] [CrossRef]
- Bernabeu, S.; Ratnam, K.C.; Boutal, H.; Gonzalez, C.; Vogel, A.; Devilliers, K.; Plaisance, M.; Oueslati, S.; Malhotra-Kumar, S.; Dortet, L.; et al. A Lateral Flow Immunoassay for the Rapid Identification of CTX-M-Producing Enterobacterales from Culture Plates and Positive Blood Cultures. Diagnostics 2020, 10, 764. [Google Scholar] [CrossRef]
- Cointe, A.; Bonacorsi, S.; Truong, J.; Hobson, C.; Doit, C.; Monjault, A.; Bidet, P.; Birgy, A. Detection of Carbapenemase-Producing Enterobacteriaceae in Positive Blood Culture Using an Immunochromatographic RESIST-4 O.K.N.V. Assay. Antimicrob. Agents Chemother. 2018, 62, e01828-18. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Boattini, M.; Iannaccone, M.; Bondi, A.; Ghibaudo, D.; Zanotto, E.; Peradotto, M.; Cavallo, R.; Costa, C. Carbapenemase detection testing in the era of ceftazidime/avibactam-resistant KPC-producing Enterobacterales: A 2-year experience. J. Glob. Antimicrob. Resist. 2021, 24, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Gottig, S.; Hamprecht, A.G. Multiplex Immunochromatographic Detection of OXA-48, KPC, and NDM Carbapenemases: Impact of Inoculum, Antibiotics, and Agar. J. Clin. Microbiol. 2018, 56, e00050-18. [Google Scholar] [CrossRef]
- Boattini, M.; Bianco, G.; Iannaccone, M.; Ghibaudo, D.; Almeida, A.; Cavallo, R.; Costa, C. Fast-track identification of CTX-M-extended-spectrum-beta-lactamase- and carbapenemase-producing Enterobacterales in bloodstream infections: Implications on the likelihood of deduction of antibiotic susceptibility in emergency and internal medicine departments. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1495–1501. [Google Scholar] [CrossRef]
- Bogaerts, P.; Oueslati, S.; Meunier, D.; Nonhoff, C.; Yunus, S.; Massart, M.; Denis, O.; Woodford, N.; Hopkins, K.L.; Naas, T.; et al. Multicentre evaluation of the BYG Carba v2.0 test, a simplified electrochemical assay for the rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. Sci. Rep. 2017, 7, 9937. [Google Scholar] [CrossRef]
- Rochelet, M.; Solanas, S.; Betelli, L.; Neuwirth, C.; Vienney, F.; Hartmann, A. Amperometric detection of extended-spectrum beta-lactamase activity: Application to the characterization of resistant E. coli strains. Analyst 2015, 140, 3551–3556. [Google Scholar] [CrossRef] [PubMed]
- Feucherolles, M.; Cauchie, H.M.; Penny, C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, J.; Walkova, R.; Studentova, V.; Chudackova, E.; Bergerova, T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 3222–3227. [Google Scholar] [CrossRef] [PubMed]
- Correa-Martinez, C.L.; Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid Detection of Extended-Spectrum Beta-Lactamases (ESBL) and AmpC beta-Lactamases in Enterobacterales: Development of a Screening Panel Using the MALDI-TOF MS-Based Direct-on-Target Microdroplet Growth Assay. Front. Microbiol. 2019, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Huang, Y.; Lizou, Y.; Li, J.; Zhang, R. Evaluation of Staphylococcus aureus Subtyping Module for Methicillin-Resistant Staphylococcus aureus Detection Based on Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Front. Microbiol. 2019, 10, 2504. [Google Scholar] [CrossRef]
- Cordovana, M.; Kostrzewa, M.; Soki, J.; Witt, E.; Ambretti, S.; Pranada, A.B. Bacteroides fragilis: A whole MALDI-based workflow from identification to confirmation of carbapenemase production for routine laboratories. Anaerobe 2018, 54, 246–253. [Google Scholar] [CrossRef]
- Camara, J.E.; Hays, F.A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2007, 389, 1633–1638. [Google Scholar] [CrossRef]
- Figueroa-Espinosa, R.; Costa, A.; Cejas, D.; Barrios, R.; Vay, C.; Radice, M.; Gutkind, G.; Di Conza, J. MALDI-TOF MS based procedure to detect KPC-2 directly from positive blood culture bottles and colonies. J. Microbiol. Methods 2019, 159, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Lee, E.H.; Hwang, D.H.; Lee, H.; Baek, J.H.; Jeong, S.H. Direct detection of intact Klebsiella pneumoniae carbapenemases produced by Enterobacterales using MALDI-TOF MS. J. Antimicrob. Chemother. 2020, 75, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Cordovana, M.; Kostrzewa, M.; Glandorf, J.; Bienia, M.; Ambretti, S.; Pranada, A.B. A Full MALDI-Based Approach to Detect Plasmid-Encoded KPC-Producing Klebsiella pneumoniae. Front. Microbiol. 2018, 9, 2854. [Google Scholar] [CrossRef]
- Knight, G.M.; Dyakova, E.; Mookerjee, S.; Davies, F.; Brannigan, E.T.; Otter, J.A.; Holmes, A.H. Fast and expensive (PCR) or cheap and slow (culture)? A mathematical modelling study to explore screening for carbapenem resistance in UK hospitals. BMC Med. 2018, 16, 141. [Google Scholar] [CrossRef]
- Doern, C.D. The Confounding Role of Antimicrobial Stewardship Programs in Understanding the Impact of Technology on Patient Care. J. Clin. Microbiol. 2016, 54, 2420–2423. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Bathoorn, E.; Chlebowicz, M.A.; Couto, N.; Ferdous, M.; Garcia-Cobos, S.; Kooistra-Smid, A.M.; Raangs, E.C.; Rosema, S.; Veloo, A.C.; et al. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol 2017, 243, 16–24. [Google Scholar] [CrossRef]
- Berglund, F.; Osterlund, T.; Boulund, F.; Marathe, N.P.; Larsson, D.G.J.; Kristiansson, E. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Ellington, M.J.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.T.G.; Hopkins, K.L.; et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Rossen, J.W.A.; Friedrich, A.W.; Moran-Gilad, J.; on behalf ofthe ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD). Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin. Microbiol. Infect. 2018, 24, 355–360. [Google Scholar] [CrossRef] [PubMed]
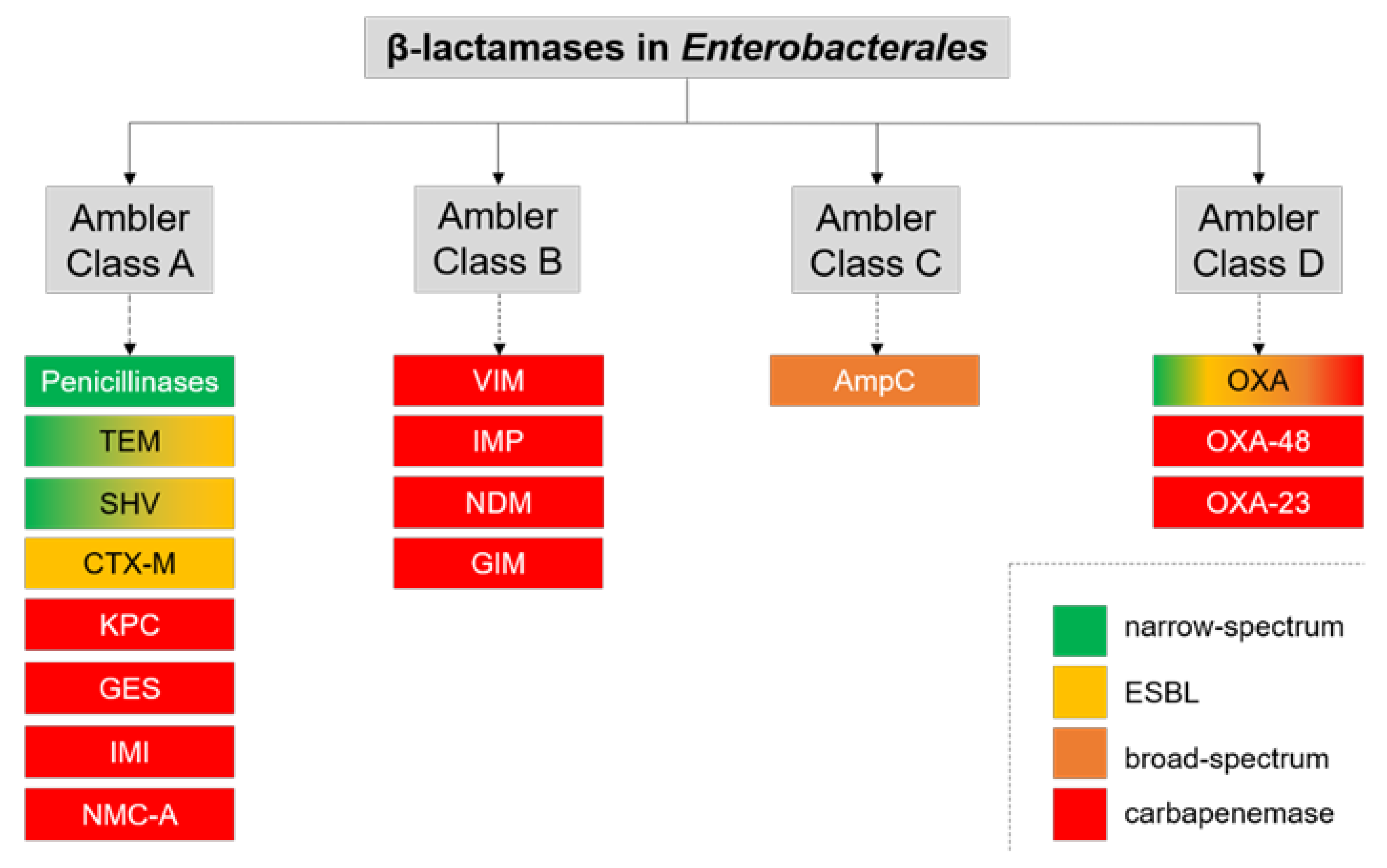



| Method | Test | Time to Result | Ease of Interpretation | Target Enzymes/ Genes | Sens. (%) | Spec. (%) | Tested on Clinical Specimens | Reference |
|---|---|---|---|---|---|---|---|---|
| Selective chromogenic/non-chromogenic agar | Brilliance CRE (Oxoid) | 18–48 h | easy | Classes A, B, D | 77.6–98.6 | 60–87.1 | bacterial colonies | [47,48] |
| Chromatic CRE (Liofilchem) | 18–48 h | easy | Classes A, B, D | 94.2 | 60 | bacterial colonies | [47] | |
| chromID CARBA (bioMérieux) | 18–48 h | easy | Classes A, B, D | 85.5–89.8 | 87.5–95 | bacterial colonies | [47,48] | |
| chromID OXA-48 (bioMérieux) | 18–48 h | easy | OXA-48 | 34.8 (all CPE) 100 (OXA-48) | 100 | bacterial colonies | [47] | |
| McConkey supplemented with ertapenem, cloxacillin, zinc-sulfate and ticarcillin | 24–48 h | easy | Classes A, B, D | 97.1 | 77.5 | bacterial colonies | [47] | |
| Disc diffusion and related assays | Mastdiscs TM Carbapenemase Detection Set (MAST GROUP) | 18–24 h | easy | Classes A, B | 78 | 93 | bacterial colonies | [49] |
| Combi Carba Plus Kit (MAST GROUP) | 18 h | easy | Classes A, B, D | 86 | 98 | bacterial colonies | [50] | |
| KPC/MBL & OXA-48 Confirm Kits (Rosco Diagnostica) | 18–24 h | subjective | Classes A, B, D | 86–98.8 | 93.1–98 | bacterial colonies | [50,51] | |
| KPC&MBL&OXA-48 disc kit (Liofilchem) | 18 h | Classes A, B, D | 96 | 87 | bacterial colonies | [50] | ||
| faropenem disc | 18 h | easy | Classes A, B, D | 99 | 81 | bacterial colonies | [50] | |
| Modified Hodge Test | 18–24 h | subjective | Classes A, B, D | 77.4 | 38.9 | bacterial colonies | [52] | |
| Carbapenem inactivation method (CIM) | 18–24 h | easy | Classes A, B, D | n.a. | n.a. | bacterial colonies | [53] | |
| mCIM | 18–24 h | subjective | Classes A, B, D | 97 | 99 | bacterial colonies | [54,55] | |
| zCIM | 18–24 | easy | Classes A, B, D | 97.4–98 | 97.7–100 | bacterial colonies | [50] | |
| bcCIM | 18–24 h | easy | Classes A, B, D | 100 | 100 | blood culture fluid | [56] | |
| rapid carbapenemase detection method (rCDM) | 5–6 h | easy | Classes A, B, D | 100 | 99.6 | bacterial colonies | [57] | |
| Colorimetric assays | Carba NP test | 5–120 min | subjective | Classes A, B, D | 97.9 | 100 | spiked blood cultures | [58] |
| bcCarba NP test | 5–120 min | subjective | Classes A, B, D | 99 | 95.1 | blood culture fluid | [56] | |
| Neo-Rapid CARB Screen (Rosco Diagnostica) | 15–120 min | subjective | Classes A, B, D | 89.5 | 70.9 | blood culture fluid, urine | [59] | |
| Neo-Rapid CARB from PBC | 90–120 min | subjective | Classes A, B, D | 99 | 91.4 | blood culture fluid | [56] | |
| Rapidec carba NP test (bioMérieux) | 5–120 min | subjective | Classes A, B, D | 99 | 100 | blood culture fluid | [59] | |
| β-CARBA test (Bio-Rad) | 30 min | subjective | Classes A, B, D | 64.9–84.9 | 90–95.6 | bacterial colonies | [60,61] | |
| β-CARBA test from PBC | 30 min | subjective | Classes A, B, D | 100 | 95.1 | blood culture fluid | [56] | |
| CARBA PAcE (MAST GROUP) | 10 min | subjective | Classes A, B, D | 72 | 91 | bacterial colonies | [62] | |
| Blue Carba test | 30–120 min | subjective | Classes A, B, D | 100 | 100 | bacterial cultures | [63] | |
| Rapid Carb Blue kit (Rosco Diagnostica) | 15-60 min | subjective | Classes A, B, D | 93.3 | 100 | blood culture fluid, urine | [64] | |
| CNPt-direct test | 2 h | subjective | Classes A, B, D | 98 | 100 | bacterial colonies | [65] | |
| Carba-H-assay | 2 h | subjective | Classes A, B, D | n.a. | n.a. | bacterial colonies, spiked urine samples | [66] | |
| Immunochromatographic assays | RESIST-3 O.K.N. assays (Coris BioConcept) | 20–45 min | easy | KPC, NDM, OXA-48-like | 100 | 100 | blood culture fluid | [67] |
| RESIST-4 O.K.N.V. assays (Coris BioConcept) | 15 min | easy | KPC, NDM, VIM, OXA-48-like | 84.2–99.2 | 100 | bacterial colonies | [55,68] | |
| RESIST-5 O.O.K.N.V. assays (Coris BioConcept) | 15 min | easy | KPC, NDM, VIM, OXA-48-like, OXA-163 | 99.4 | 100 | bacterial colonies | [69] | |
| NG-Test Carba 5 (NG Biotech) from PBC | 30 min | easy | KPC, NDM, VIM, OXA-48-like, IMP | 97.7 | 96.1 | blood culture fluid | [70] | |
| NG-Test Carba 5 (NG Biotech) | 15 min | easy | KPC, NDM, VIM, OXA-48-like, IMP | 88.2–100 | 95.3–100 | bacterial colonies | [55,69,71,72] | |
| Mass spectrometry | MALDI-TOF MS | 4.5 h | complex | Classes A, B, D | 100 | 90 | blood culture fluid | [73] |
| MBT STAR-Carba IVD Kit (Bruker DALTONICS) | 30–60 min | moderate | mass shift of hydrolyzed carbapenem | 98–100 | 97–100 | bacterial colonies | [74,75,76,77] | |
| MBT STAR-Carba IVD Kit (Bruker DALTONICS) from PBC | 1 h | moderate | mass shift of hydrolyzed carbapenem | 100 | 100 | blood culture fluid | [76] | |
| Direct-on-target microdroplet growth assay (DOT MGA), RUO | 4–6 h | moderate | bacterial growth in presence of defined carbapenem concentration | 100 | 100 | bacterial colonies | [78] | |
| Direct-on-target microdroplet growth assay (DOT MGA), RUO, from PBC | 4–6 h | moderate | bacterial growth in presence of defined carbapenem concentration | 91.7 | 100 | blood culture fluid | [79] | |
| Electrochemical assay | BYG Carba test (not yet commercially available) | 5–30 min | easy | Classes A, B, D | 95 | 100 | bacterial colonies | [80] |
| BL-RED test (Coris BioConcept) | 20 min | easy | 3GC resistance by hydrolysis | 46.7 (83.8 for detection of class A β-lactamases) | 100 | blood culture fluid | [81] | |
| Other molecular methods | Eazyplex Superbug CRE (Amplex Diagnostics) | 15 min | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like | 100 | 100 | blood culture fluid, rectal swab, urine | [82] |
| Multiplex PCR-based assays | BD MAX TM Check-Points CPO assay (Check Points) | <180 min | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | 97.1 | 98.8 | rectal swab | [83] |
| Check-MDR CT103 (Check Points) | 6 h | relatively easy | among others blaKPC2,3, blaNDM1,2,3, blaOXA-48, blaOXA-181, blaVIM1,2,3,4,19, blaIMP1,4,8,13 | 95–100 | 100 | DNA extracted from bacterial colonies | [84] | |
| CRE ELITe MGB ® kit (ELITechGroup) | <180 | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | 100 | 100 | PBC, rectal swab, respiratory sample | [85] | |
| Amplidiag ® CarbaR + VRE (Mobidiag Ltd.) | <180 min | relatively easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP, ISAba1-OXA-51, blaOXA-23, blaOXA-24/40, blaOXA-58 | 100 | 99 | DNA extracted from stool samples, rectal swab, pure culture | [86] | |
| Amplidiag ® CarbaR + MCR (Mobidiag Ltd.) | <180 min | relatively easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP, ISAba1-OXA-51, blaOXA-23, blaOXA-24/40, blaOXA-58, blaGES-2, blaGES4 throughblaGES-6, blaGES-13 throughblaGES-16, blaGES-18, blaGES-20/21, blaGES-24 | 92–100 | 86–100 | DNA extracted from stool samples, rectal swab, pure culture | [87] | |
| Xpert-Carba-R assay ® (Cepheid) | 50 min | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | 97.8 | 95.3 | rectal swab | [88] | |
| GenePOC/Revogene Carba C assay ® (Meridian) | 70 min | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | 100 | 100 | bacterial colonies | [46] | |
| Verigene BC-GN (Nanosphere) | 2–2.5 h | relatively easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP, blaOXA-23, blaOXA-24/40, blaOXA-58 | 100 | 100 | blood culture fluid | [89] | |
| Biofire ® Filmarray ® Blood Culture Identification (BCID) Panel (bioMérieux) | 1 h | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | n.a. | n.a. | blood culture fluid | [90] | |
| Biofire ® Filmarray ® Pneumonia plus Panel (bioMérieux) | 1 h | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | n.a. | n.a. | Respiratory specimen | [91] | |
| ePlex ® Blood Culture Identification Gram Negative Panel (GenMark) | 90 min | easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaOXA-23, blaIMP | n.a. | n.a. | blood culture fluid | [92] | |
| Unyvero (Curetis) | 4–5 h | relatively easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaOXA-23, blaOXA-24/40, blaOXA-58, blaIMP | n.a. | n.a. | sputum, tissue, bone fragment, pus, blood culture fluid | [93] | |
| Hyplex SuperBug ID test system (Amplex Diagnostics) | relatively easy | blaKPC, blaVIM, blaNDM, blaOXA-48-like, blaIMP | 96.7 | ≥99 | sputum, urine, blood culture fluid, swab specimens | [94] | ||
| Luminex xTAG ® assay (Luminex Corporation) | 5 h | relatively easy | blaKPC, blaVIM, blaNDM, blaIMI, blaGES, blaOXA-23, blaOXA-51, blaSEM, blaVEM, blaOXA-48-like, blaIMP | 100 | 99.4 | DNA extracted from bacterial colonies | [95] | |
| VAPChip ® (EppendorfArray Technologies) | 4 h | relatively easy | blaKPC, blaVIM, blaOXA-23, blaOXA-48-like, blaIMP, blaOXA-24/40, blaOXA-58 | 100 | 100 | Respiratory sample | [96] | |
| Next generation sequencing | MinION (Oxford Nanopore Technologies) | >8 h | complex | Classes A, B, D | 100 | 100 | extracted DNA | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noster, J.; Thelen, P.; Hamprecht, A. Detection of Multidrug-Resistant Enterobacterales—From ESBLs to Carbapenemases. Antibiotics 2021, 10, 1140. https://doi.org/10.3390/antibiotics10091140
Noster J, Thelen P, Hamprecht A. Detection of Multidrug-Resistant Enterobacterales—From ESBLs to Carbapenemases. Antibiotics. 2021; 10(9):1140. https://doi.org/10.3390/antibiotics10091140
Chicago/Turabian StyleNoster, Janina, Philipp Thelen, and Axel Hamprecht. 2021. "Detection of Multidrug-Resistant Enterobacterales—From ESBLs to Carbapenemases" Antibiotics 10, no. 9: 1140. https://doi.org/10.3390/antibiotics10091140
APA StyleNoster, J., Thelen, P., & Hamprecht, A. (2021). Detection of Multidrug-Resistant Enterobacterales—From ESBLs to Carbapenemases. Antibiotics, 10(9), 1140. https://doi.org/10.3390/antibiotics10091140






