Methicillin-Resistant Staphylococcus aureus from Peninsular Malaysian Animal Handlers: Molecular Profile, Antimicrobial Resistance, Immune Evasion Cluster and Genotypic Categorization
Abstract
:1. Introduction
2. Results
2.1. Carriage Rate of S. aureus and MRSA from Animal Handlers
2.2. Antibiotic Susceptibility Profile of S. aureus and MRSA from Animal Handlers
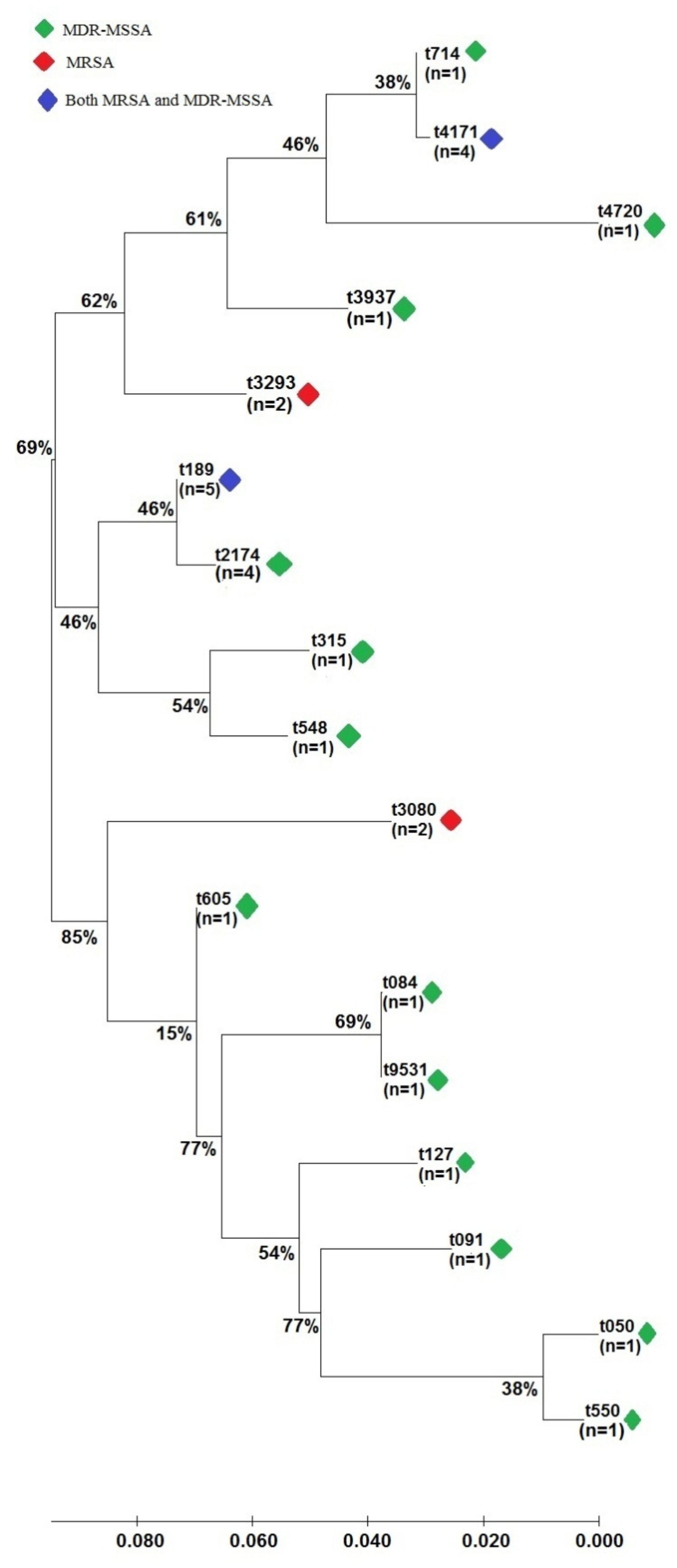
2.3. Genotypic Characterization of S. aureus
3. Discussion
4. Materials and Methods
4.1. Swab Sample Collection, Isolation and Genotypic Identification of Bacteria
4.2. Antibiotic Susceptibility Test (AST) of S. aureus
4.3. Genotypic Characterization of S. aureus
4.4. Spa Typing of MDRSA
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chai, M.H.; Sukiman, M.Z.; Najib, N.M.; Mohabbar, N.A.; Azizan, N.A.N.M.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Molecular detection and antibiogram of Staphylococcus aureus in rabbits, rabbit handlers, and rabbitry in Terengganu, Malaysia. J. Adv. Vet. Anim. Res. 2021, 8, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.H.; Faiq, T.A.M.; Ariffin, S.M.Z.; Suhaili, Z.; Sukiman, M.Z.; Ghazali, M.F. Prevalence of methicillin resistant Staphylococcus aureus in raw goat milks from selected farms in Terengganu, Malaysia. Trop. Anim. Sci. J. 2020, 43, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Infect. 2018, 31, 1–103. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Grúa, E.; Pérez-Fuentes, S.; Muñoz-Silvestre, A.; Viana, D.; Fernández-Ros, A.B.; Sanz-Tejero, C.; Corpa, J.M.; Selva, L. Characterization of livestock-associated methicillin-resistant Staphylococcus aureus isolates obtained from commercial rabbitries located in the Iberian Peninsula. Front. Microbiol. 2018, 9, 1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, M.H.; Majib, M.E.A.; Sukiman, M.Z.; Najib, N.M.; Mohabbar, N.A.; Azizan, N.A.N.M.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Molecular identification of multidrug-resistant Staphylococcus aureus (MDRSA) carriage and pattern of antibiotic resistance from rabbit, rabbit handler and rabbitry in the east coast region of Malaysia. Biosci. Res. 2021, 18, 380–387. [Google Scholar]
- Zomer, T.P.; Wielders, C.C.H.; Veenman, C.; Hengeveld, P.; Van der Hoek, W.; de Greeff, S.C.; Smit, L.A.M.; Heederik, D.J.; Yzermans, C.J.; Bosch, T.; et al. MRSA in persons not living or working on a farm in a livestock-dense area: Prevalence and risk factors. J. Antimicrob. Chemother. 2017, 72, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Oppliger, A.; Moreillon, P.; Charrière, N.; Giddey, M.; Morisset, D.; Sakwinska, O. Antimicrobial resistance of Staphylococcus aureus strains acquired by pig farmers from pigs. Appl. Environ. Microbiol. 2012, 78, 8010–8014. [Google Scholar] [CrossRef] [Green Version]
- Wardyn, S.E.; Forshey, B.M.; Farina, S.A.; Kates, A.E.; Nair, R.; Quick, M.K.; Wu, J.Y.; Hanson, B.M.; O’Malley, S.M.; Shows, H.W.; et al. Swine farming is a risk factor for infection with and high prevalence of carriage of multidrug-resistant Staphylococcus aureus. Clin. Infect. Dis. 2015, 61, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Haran, K.P.; Godden, S.M.; Boxrud, D.; Jawahir, S.; Bender, J.B.; Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012, 50, 688–695. [Google Scholar] [CrossRef] [Green Version]
- Fluit, A.C. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 2012, 18, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Suhaili, Z.; Rafee, P.A.; Mat Azis, N.; Yeo, C.C.; Nordin, S.A.; Abdul Rahim, A.R.; Al-Obaidi, M.M.J.; Mohd Desa, M.N. Characterization of resistance to selected antibiotics and Panton-Valentine leukocidin-positive Staphylococcus aureus in a healthy student population at a Malaysian University. Germs 2018, 8, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The impact on humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Kozajda, A.; Jeżak, K.; Kapsa, A. Airborne Staphylococcus aureus in different environments—A review. Environ. Sci. Pollut. Res. 2019, 26, 34741–34753. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Chai, M.H.; Sukiman, M.Z.; Chan, Y.F.; Liew, Y.W.; Lai, L.Z.H.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Detection, antibiogram and molecular characterization of MRSA and MSSA isolated from swine. IOP Conf. Ser. Earth Environ. Sci. 2021, 888, 012064. [Google Scholar] [CrossRef]
- Suhaili, Z.; Yeo, C.C.; Ghazali, M.F.; Chew, C.H.; Zainul, A.Z.; Mazen, M.J.A.-O.; Nordin, S.A.; Mohd Desa, M.N. Nasal colonisation, antimicrobial susceptibility and genotypic pattern of Staphylococcus aureus among agricultural biotechnology students in Besut, Terengganu, east coast of Malaysia. Trop. Med. Int. Health 2018, 23, 905–913. [Google Scholar] [CrossRef] [Green Version]
- Cuny, C.; Abdelbary, M.; Layer, F.; Werner, F.; Witte, W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 2015, 177, 219–223. [Google Scholar] [CrossRef]
- Van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; Van Kessel, K.P.M.; Van Strijp, J.A.G. The innate immune modulators Staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [Green Version]
- Hau, S.J.; Sun, J.; Davies, P.R.; Frana, T.S.; Nicholson, T.L. Comparative prevalence of immune evasion complex genes associated with β-hemolysin converting bacteriophages in MRSA ST5 isolates from swine, swine facilities, humans with swine contact, and humans with no swine contact. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef]
- Bien, J.; Sokolova, O.; Bozko, P. Characterization of virulence factors of Staphylococcus aureus: Novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J. Pathog. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohseni, M.; Rafiei, F.; Ghaemi, E.A. High frequency of exfoliative toxin genes among Staphylococcus aureus isolated from clinical specimens in the north of Iran: Alarm for the health of individuals under risk. Iran. J. Microbiol. 2018, 10, 158–165. [Google Scholar] [PubMed]
- Che Hamzah, A.M.; Yeo, C.C.; Puah, S.M.; Chua, K.H.; Chew, C.H. Staphylococcus aureus infections in Malaysia: A review of antimicrobial resistance and characteristics of the clinical isolates, 1990–2017. Antibiotics 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaweera, J.A.A.S.; Kumbukgolla, W.W. Antibiotic resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolated from livestock and associated farmers in Anuradhapura, Sri Lanka. Germs 2017, 7, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Fan, Y.; Wang, X.; Liu, W.; Yu, H.; Zhou, J.; Chen, S.; Yao, Z. Livestock-associated methicillin and multidrug resistant S. aureus in humans is associated with occupational pig contact, not pet contact. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Neela, V.; Ghaznavi-Rad, E.; Ghasemzadeh-Moghaddam, H.; Nor Shamsudin, M.; van Belkum, A.; Karunanidhi, A. Frequency of methicillin resistant Staphylococcus aureus in the noses of Malaysian chicken farmers and their chicken. Iran. J. Vet. Res. 2013, 14, 226–231. [Google Scholar]
- Aklilu, E.; Zakaria, Z.; Hassan, L.; Chen, H.C. Molecular relatedness of methicillin-resistant S. aureus isolates from staff, environment and pets at university veterinary hospital in Malaysia. PLoS ONE 2012, 7, e43329. [Google Scholar] [CrossRef]
- Che Hamzah, A.M.; Yeo, C.C.; Puah, S.M.; Chua, K.H.; Rahman, N.I.A.; Abdullah, F.H.; Othman, N.; Chew, C.H. Tigecycline and inducible clindamycin resistance in clinical isolates of methicillin-resistant Staphylococcus aureus from Terengganu, Malaysia. J. Med. Microbiol. 2019, 68, 1299–1305. [Google Scholar] [CrossRef]
- Jaja, I.F.; Jaja, C.I.; Chigor, N.V.; Anyanwu, M.U.; Maduabuchi, E.K.; Oguttu, J.W.; Green, E. Antimicrobial resistance phenotype of Staphylococcus aureus and Escherichia coli isolates obtained from meat in the formal and informal sectors in South Africa. Biomed. Res. Int. 2020, 2020, 3979482. [Google Scholar] [CrossRef]
- Ariffin, S.M.Z.; Hasmadi, N.; Syawari, N.M.; Sukiman, M.Z.; Ariffin, M.F.T.; Chai, M.H.; Ghazali, M.F. Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus, Streptococcus agalactiae and Escherichia coli in dairy goats with clinical and subclinical mastitis. J. Anim. Health Prod. 2019, 7, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Friese, A.; Schulz, J.; Zimmermann, K.; Tenhagen, B.A.; Fetsch, A.; Hartung, J.; Rösler, U. Occurrence of livestock-associated methicillin-resistant Staphylococcus aureus in turkey and broiler barns and contamination of air and soil surfaces in their vicinity. Appl. Environ. Microbiol. 2013, 79, 2759–2766. [Google Scholar] [CrossRef] [Green Version]
- Hetem, D.J.; Westh, H.; Boye, K.; Jarløv, J.O.; Bonten, M.J.M.; Bootsma, M.C.J. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish hospitals. J. Antimicrob. Chemother. 2012, 67, 1775–1780. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Ferreira, E.; Manageiro, V.; Reis, L.; Tejedor-Junco, M.T.; Sampaio, A.; Capelo, J.L.; Caniça, M.; Igrejas, G.; Poeta, P. Distribution and clonal diversity of Staphylococcus aureus and other staphylococci in surface waters: Detection of ST425-t742 and ST130-t843 mecC-positive MRSA strains. Antibiotics 2021, 10, 1416. [Google Scholar] [CrossRef]
- Mulders, M.N.; Haenen, A.P.J.; Geenen, P.L.; Vesseur, P.C.; Poldervaart, E.S.; Bosch, T.; Huijsdens, X.W.; Hengeveld, P.D.; Dam-Deisz, W.D.C.; Graat, E.A.M.; et al. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol. Infect. 2010, 138, 743. [Google Scholar] [CrossRef]
- Sineke, N.; Asante, J.; Amoako, D.G.; Luther, A.; Abia, K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Staphylococcus aureus in intensive pig production in South Africa: Antibiotic resistance, virulence determinants, and clonality. Pathogens 2021, 10, 317. [Google Scholar] [CrossRef]
- Ariffin, M.F.; Hasmadi, N.; Chai, M.H.; Ghazali, M.F.; Ariffin, S.M.Z. Prevalence and antimicrobial sensitivity pattern of Staphylococcus aureus isolated from clinical and subclinical mastitis in small ruminant in Besut and Setiu, Terengganu, Malaysia. Malays. J. Microbiol. 2020, 16, 104–110. [Google Scholar] [CrossRef]
- Corona, F.; Martinez, J.L. Phenotypic resistance to antibiotics. Antibiotics 2013, 2, 237–255. [Google Scholar] [CrossRef] [Green Version]
- Verkaik, N.J.; Benard, M.; Boelens, H.A.; De Vogel, C.P.; Nouwen, J.L.; Verbrugh, H.A.; Melles, D.C.; Van Belkum, A.; Van Wamel, W.J.B. Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin. Microbiol. Infect. 2011, 17, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Hirose, M.; Aung, M.S.; Fukuda, A.; Yahata, S.; Fujita, Y.; Saitoh, M. Antimicrobial resistance and molecular epidemiological characteristics of methicillin-resistant and susceptible Staphylococcal isolates from oral cavity of dental patients and staff in Northern Japan. Antibiotics 2021, 10, 1316. [Google Scholar] [CrossRef]
- Asadollahi, P.; Farahani, N.N.; Mirzaii, M.; Khoramrooz, S.S.; van Belkum, A.; Asadollahi, K.; Dadashi, M.; Darban-Sarokhalil, D. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: A review. Front. Microbiol. 2018, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.U.; Chua, K.H.; Chew, C.H.; Yeo, C.C.; Abdullah, F.H.; Othman, N.; Kee, B.P.; Puah, S.M. Spa diversity of methicillin-resistant and -susceptible Staphylococcus aureus in clinical strains from Malaysia: A high prevalence of invasive European spa-type. PeerJ 2021, 9, e11195. [Google Scholar] [CrossRef]
- Saiful, A.J.; Mastura, M.; Suhaili, Z.; Mazurah, M.I.; Shuhaimi, M.; Ali, A.M. Detection of methicillin-resistant Staphylococcus aureus using mecA/nuc genes and antibiotic susceptibility profile of Malaysian clinical isolates. World J. Microbiol. Biotechnol. 2006, 22, 1289–1294. [Google Scholar] [CrossRef]
- Stegger, M.; Liu, C.M.; Larsen, J.; Soldanova, K.; Aziz, M.; Contente-Cuomo, T.; Mammina, C.; van Belkum, A.; Salmenlinna, S.; Laurent, F.; et al. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS ONE 2013, 8, e79645. [Google Scholar] [CrossRef]
- Ng, L.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes. 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Tang, J.; Tang, C.; Chen, J.; Liu, J. Antimicrobial susceptibility and presence of resistance & enterotoxins/enterotoxin-likes genes in Staphylococcus aureus from food. CYTA J. Food 2018, 16, 76–84. [Google Scholar] [CrossRef]
- De Haas, C.J.C.; Veldkamp, K.E.; Peschel, A.; Weerkamp, F.; van Wamel, W.J.B.; Heezius, E.C.J.M.; Poppelier, M.J.J.G.; van Kessel, K.P.M.; van Strijp, J.A.G. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004, 199, 687–695. [Google Scholar] [CrossRef]
- Lina, G.; Quaglia, A.; Reverdy, M.E.; Leclercq, R.; Vandenesch, F.; Etienne, J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among Staphylococci. Antimicrob. Agents Chemother. 1999, 43, 1062–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, S.J.; Moore, C.E.; Justice, A.; Kantzanou, M.; Story, L.; Mackie, K.; O’Neill, G.; Day, N.P.J. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 2002, 70, 4987–4996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokajian, S.; Haddad, D.; Andraos, R.; Hashwa, F.; Araj, G. Toxins and antibiotic resistance in Staphylococcus aureus isolated from a major hospital in Lebanon. ISRN Microbiol. 2011, 2011, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 54–62. [Google Scholar]
- Harmsen, D.; Claus, H.; Witte, W.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphyloccocus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [Green Version]

| No. | Sample Groups | S. aureus Positive (%; 95% CI) | MRSA Positive (%; 95% CI) | MDRSA Positive (%; 95% CI) |
|---|---|---|---|---|
| 1. | Veterinarians (n = 10) | 3 (30.0; 1.0–58.4%) | 0 (0; 0%) | 2 (20.0; 0–44.8%) |
| 2. | Pet owners (n = 70) | 20 (28.6; 18.0–39.2%) | 1 (1.4; 0–4.2%) | 7 (10.0; 3.0–17.0%) |
| 3. | Animal farmers (n = 343) | 105 (30.6; 25.7–35.5%) | 4 (1.2; 0–2.4%) | 21 (6.1; 3.6–8.6%) |
| Total (n = 423) | 129 (30.5; 26.2–34.8%) | 5 (1.2; 0.2–2.2%) | 30 (7.1; 4.7–9.5%) |
| Antimicrobials | Number of Isolates (%) | ||
|---|---|---|---|
| Resistant | Intermediate | Susceptible | |
| Penicillin | 112 (72.3) | 0 (0) | 43 (27.7) |
| Amoxicillin | 83 (53.5) | 0 (0) | 72 (46.5) |
| Erythromycin | 22 (14.2) | 8 (5.2) | 125 (80.6) |
| Clindamycin | 19 (12.3) | 14 (9.0) | 122 (78.7) |
| Cefoxitin | 18 (11.6) | 0 (0) | 137 (88.4) |
| Tetracycline | 18 (11.6) | 4 (2.6) | 133 (85.8) |
| Quinupristin/Dalfopristin | 12 (7.7) | 1 (0.6) | 142 (91.6) |
| Chloramphenicol | 10 (6.5) | 4 (2.6) | 141 (90.9) |
| Cephalothin | 8 (5.2) | 0 (0) | 147 (94.8) |
| Cefotaxime | 7 (4.5) | 16 (10.3) | 132 (85.2) |
| Norfloxacin | 6 (3.9) | 2 (1.3) | 147 (94.8) |
| Doxycycline | 5 (3.2) | 4 (2.6) | 146 (94.2) |
| Ciprofloxacin | 5 (3.2) | 0 (0) | 150 (96.8) |
| Trimethoprim/sulfamethoxazole | 4 (2.6) | 2 (1.3) | 149 (96.1) |
| Amikacin | 2 (1.3) | 0 (0) | 153 (98.7) |
| Gentamicin | 0 (0) | 0 (0) | 155 (100) |
| Linezolid | 0 (0) | 0 (0) | 155 (100) |
| Number of Antibiotic | MARI Value | Number of Isolates | Total (%) |
|---|---|---|---|
| 0 | 0 | 31 | 20.0 |
| 1 | 0.06 | 33 | 21.3 |
| 2 | 0.11 | 51 | 32.9 |
| 3 | 0.16 | 14 | 9.0 |
| 4 | 0.22 | 9 | 5.8 |
| 5 | 0.27 | 6 | 3.8 |
| 6 and above | 0.33 | 11 | 7.1 |
| IEC Type | IEC Genes Composition | Number of Isolates (%) | Total Number of Isolates (%) | |
|---|---|---|---|---|
| MSSA (n = 149) | MRSA (n = 6) | |||
| A | scn, chp, sak, sea | 1 (6.7) | 0 (0) | 1 (0.6) |
| B | scn, chp, sak | 10 (6.7) | 0 (0) | 10 (6.5) |
| C | scn, chp | 0 (0) | 0 (0) | 0 (0) |
| D | scn, sak, sea | 0 (0) | 0 (0) | 0 (0) |
| E | scn, sak | 17 (11.4) | 2 (33.3) | 19 (12.3) |
| F | scn, chp, sak, sep | 0 (0) | 0 (0) | 0 (0) |
| G | scn, sak, sep | 1 (6.7) | 0 (0) | 1 (0.6) |
| H | Scn | 21 (14.1) | 1 ((16.7) | 21 (13.5) |
| Non-typable | Absent of scn gene | 17 (11.4) | 0 (0) | 17 (11.0) |
| No Type | Absent of all IEC genes | 68 (45.6) | 3 (50.0) | 71 (45.8) |
| No. | Primer | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) | References |
|---|---|---|---|---|---|
| 1. | nuc | F-GCGATTGATGGTGATACGGTT | 278 | 55 | [43] |
| R-AGCCAAGCCTTGACGAACTAAAGC | |||||
| 2. | mecA | F-AAAATCGATGGTAAAGGTTGGC | 533 | 55 | [43] |
| R-AGTTCTGCAGTACCGGATTTGC | |||||
| 3. | mecC | F-GAAAAAAAGGCTTAGAACGCCTC | 138 | 59 | [44] |
| R-GAAGATCTTTTCCGTTTTCAGC | |||||
| 4. | tetK | F-TCGATAGGAACAGCAGTA | 169 | 55 | [45] |
| R-CAGCAGATCCTACTCCTT | |||||
| 5. | tetO | F-AACTTAGGCATTCTGGCTCAC | 515 | 55 | [45] |
| R-TCCCACTGTTCCATATCGTCA | |||||
| 6. | tetM | F-GTGGACAAAGGTACAACGAG | 406 | 55 | [45] |
| R-CGGTAAAGTTCGTCACACAC | |||||
| 7. | tetL | F-TCGTTAGCGTGCTGTCATTC | 267 | 55 | [45] |
| R-GTATCCCACCAATGTAGCCG | |||||
| 8. | msrA | F-GGCACAATAAGAGTGTTTAAAGG | 940 | 50 | [46] |
| R-AAGTTATATCATGAATAGATTGTCCTGTT | |||||
| 9. | ermA | F-GTTCAAGAACAATCAATACAGAG | 421 | 52 | [46] |
| R-GGATCAGGAAAAGGACATTTTAC | |||||
| 10. | ermB | F-CCGTTTACGAAATTGGAACAGGTAAAGGGC | 359 | 55 | [46] |
| R-GAATCGAGACTTGAGTGTGC | |||||
| 11. | ermC | F-GCTAATATTGTTTAAATCGTCAATTCC | 572 | 52 | [46] |
| R-GGATCAGGAAAAGGACATTTTAC | |||||
| 12. | vanA | F-ATGAATAGAATAAAAGTTGC | 1032 | 62 | [46] |
| R-TCACCCCTTTAACGCTAATA | |||||
| 13. | scn | F-AGCACAAGCTTGCCAACATCG | 258 | 50 | [20] |
| R-TTAATATTTACTTTTTAGTGC | |||||
| 14. | sak | F-AAGGCGATGACGCGAGTTAT | 223 | 50 | [20] |
| R-GCGCTTGGATCTAATTCAAC | |||||
| 15. | sea | F-AGATCATTCGTGGTATAACG | 408 | 50 | [20] |
| R-TTAACCGAAGGTTCTGTAGA | |||||
| 16. | sep | F-AATCATAACCAACCGAATCA | 500 | 50 | [20] |
| R-TCATAATGGAAGTGCTATAA | |||||
| 17. | chp | F-GAAAAAGAAATTAGCAACAACAG | 410 | 50 | [47] |
| R-CATAAGATGATTTAGACTCTCC | |||||
| 18. | luk-PV | F-ATCATTAGGTAAAATGTCTGGACATGATCCA | 433 | 55 | [48] |
| R-GCATCAAGTGTATTGGATAGCAAAAGC | |||||
| 19. | tst | F-TTATCGTAAGCCCTTTGTTG | 398 | 60 | [49] |
| R-TAAAGGTAGTTCTATTGGAGTAGG | |||||
| 20. | eta | F-CTAGTGCATTTGTTATTCAA | 119 | 55 | [50] |
| R-TGCATTGACACCATAGTACT | |||||
| 21. | etb | F-ACGGCTATATACATTCAATT | 200 | 55 | [50] |
| R-TCCATCGATAATATACCTAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, M.; Sukiman, M.Z.; Kamarun Baharin, A.H.; Ramlan, I.; Lai, L.Z.; Liew, Y.; Malayandy, P.; Mohamad, N.M.; Choong, S.; Ariffin, S.M.Z.; et al. Methicillin-Resistant Staphylococcus aureus from Peninsular Malaysian Animal Handlers: Molecular Profile, Antimicrobial Resistance, Immune Evasion Cluster and Genotypic Categorization. Antibiotics 2022, 11, 103. https://doi.org/10.3390/antibiotics11010103
Chai M, Sukiman MZ, Kamarun Baharin AH, Ramlan I, Lai LZ, Liew Y, Malayandy P, Mohamad NM, Choong S, Ariffin SMZ, et al. Methicillin-Resistant Staphylococcus aureus from Peninsular Malaysian Animal Handlers: Molecular Profile, Antimicrobial Resistance, Immune Evasion Cluster and Genotypic Categorization. Antibiotics. 2022; 11(1):103. https://doi.org/10.3390/antibiotics11010103
Chicago/Turabian StyleChai, Minhian, Muhammad Zikree Sukiman, Amirah Huda Kamarun Baharin, Insyirah Ramlan, Lennard Zhunhoong Lai, Yeewen Liew, Pavitra Malayandy, Noor Muzamil Mohamad, Siewshean Choong, Siti Mariam Zainal Ariffin, and et al. 2022. "Methicillin-Resistant Staphylococcus aureus from Peninsular Malaysian Animal Handlers: Molecular Profile, Antimicrobial Resistance, Immune Evasion Cluster and Genotypic Categorization" Antibiotics 11, no. 1: 103. https://doi.org/10.3390/antibiotics11010103
APA StyleChai, M., Sukiman, M. Z., Kamarun Baharin, A. H., Ramlan, I., Lai, L. Z., Liew, Y., Malayandy, P., Mohamad, N. M., Choong, S., Ariffin, S. M. Z., & Ghazali, M. F. (2022). Methicillin-Resistant Staphylococcus aureus from Peninsular Malaysian Animal Handlers: Molecular Profile, Antimicrobial Resistance, Immune Evasion Cluster and Genotypic Categorization. Antibiotics, 11(1), 103. https://doi.org/10.3390/antibiotics11010103








