Antimicrobial Bacillus: Metabolites and Their Mode of Action
Abstract
:1. Introduction
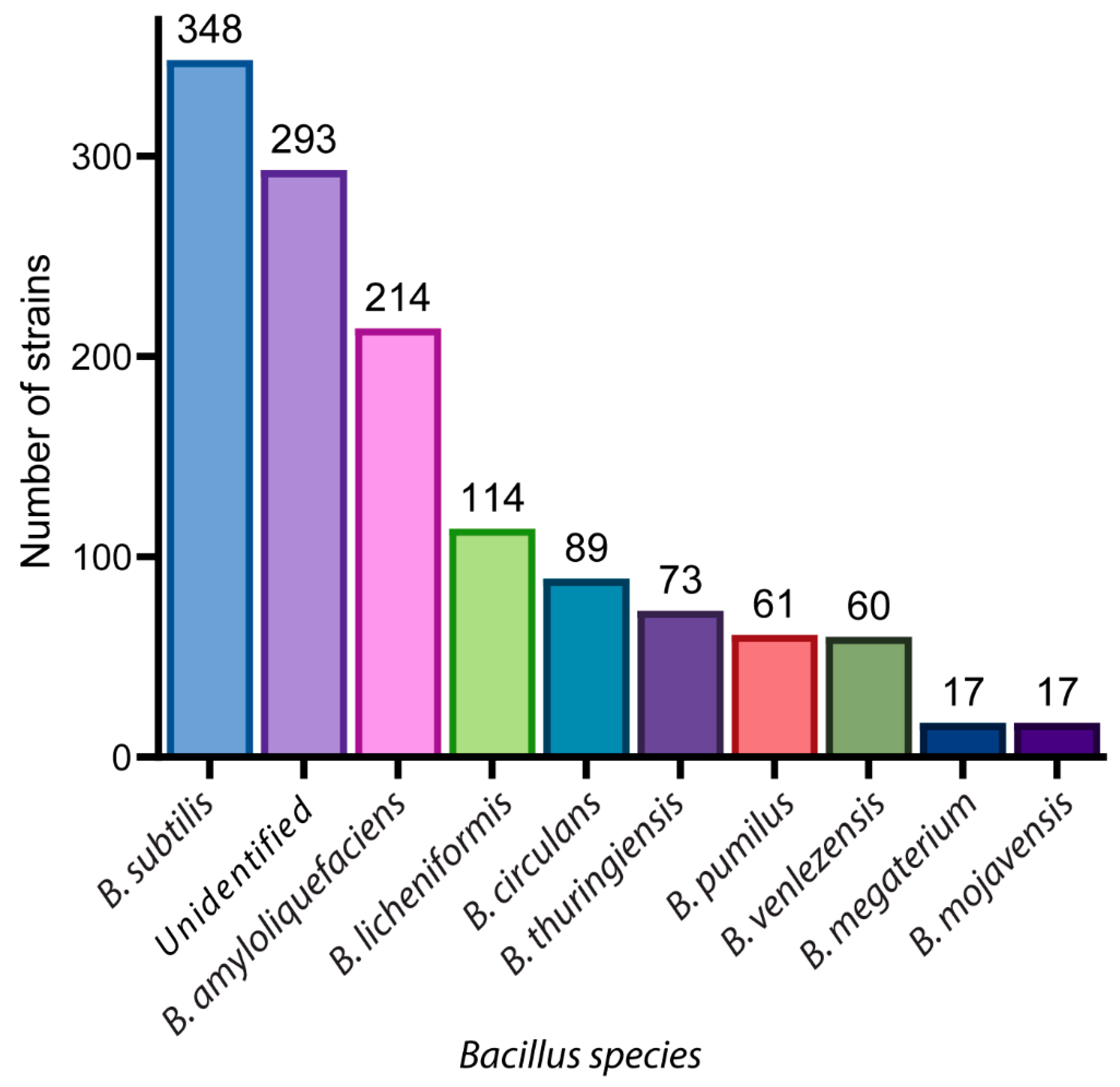
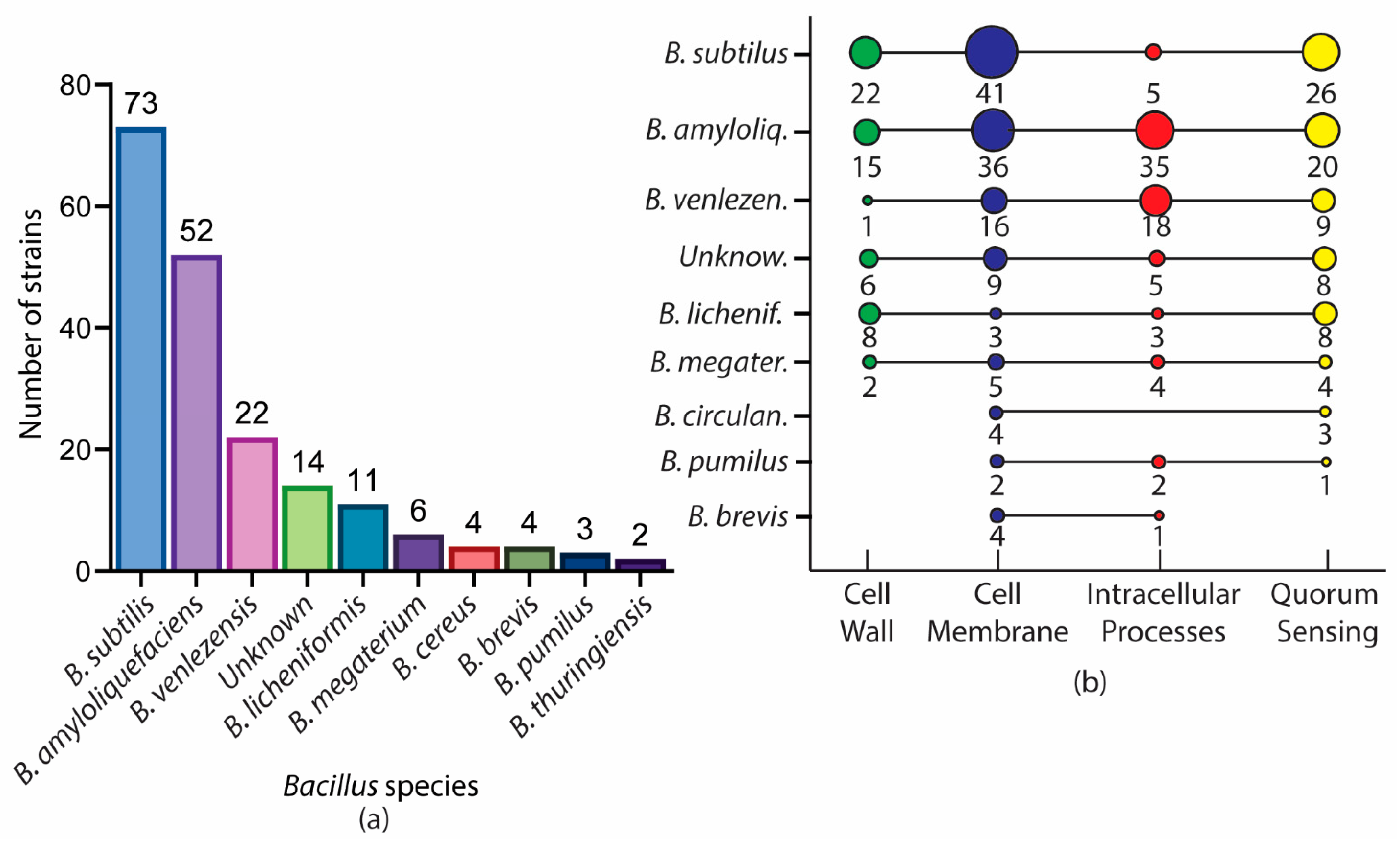
2. A Glance of Bioactive Bacillus and Their Antimicrobial Metabolites
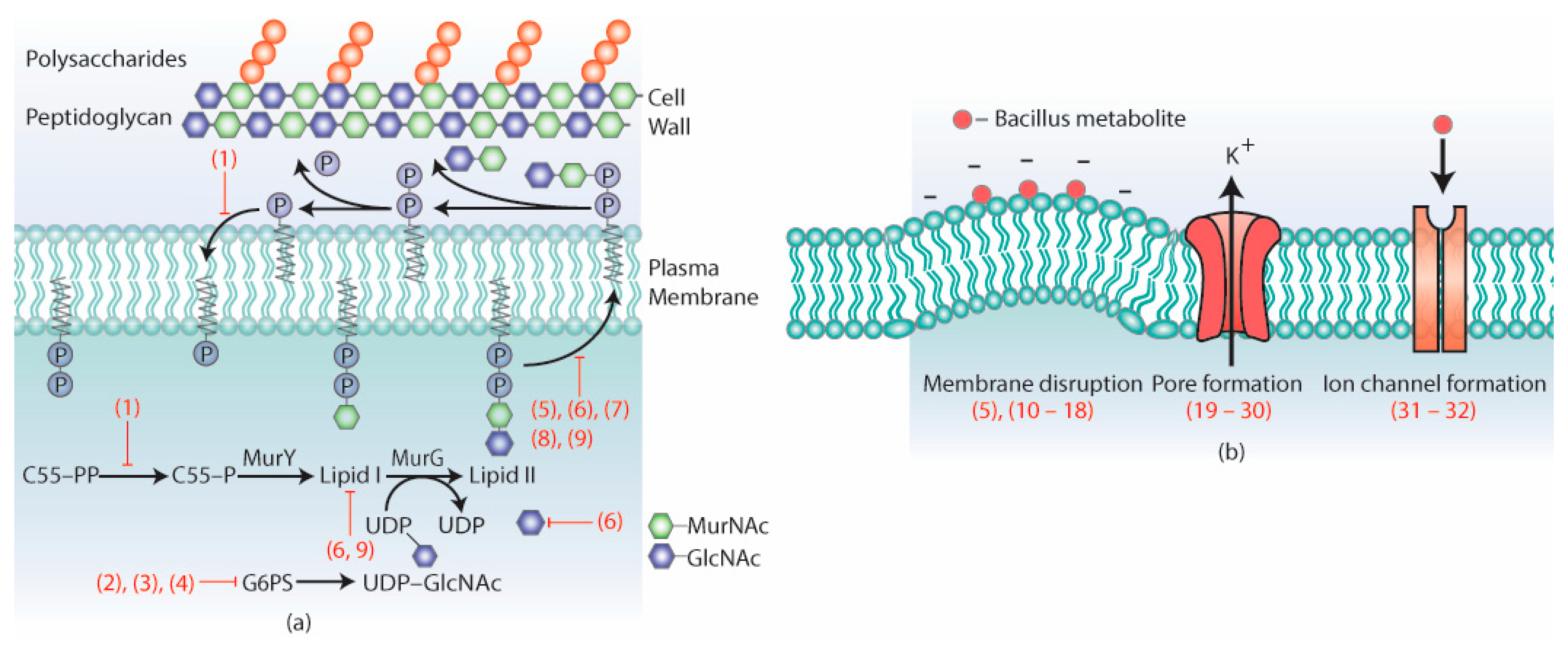
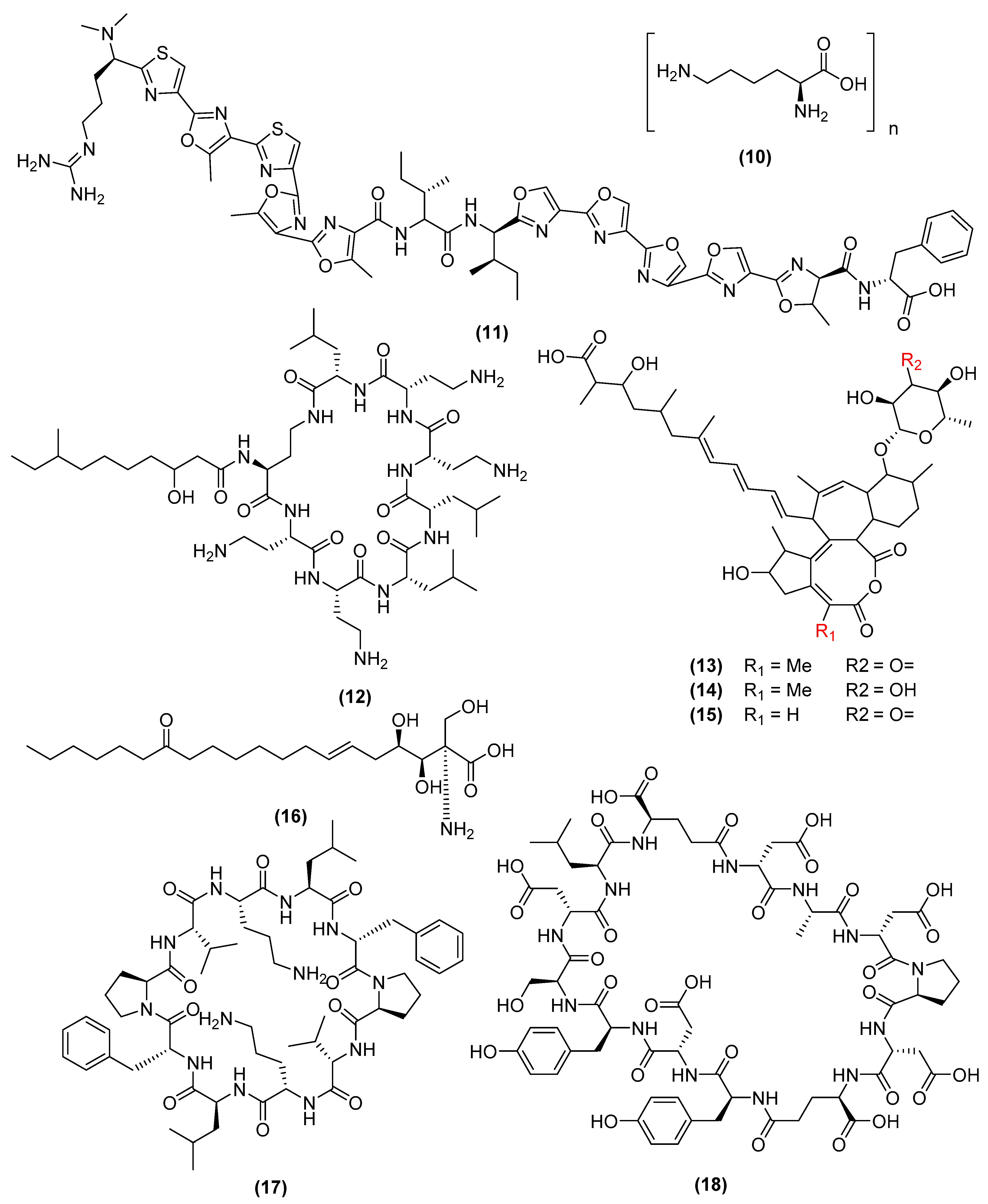
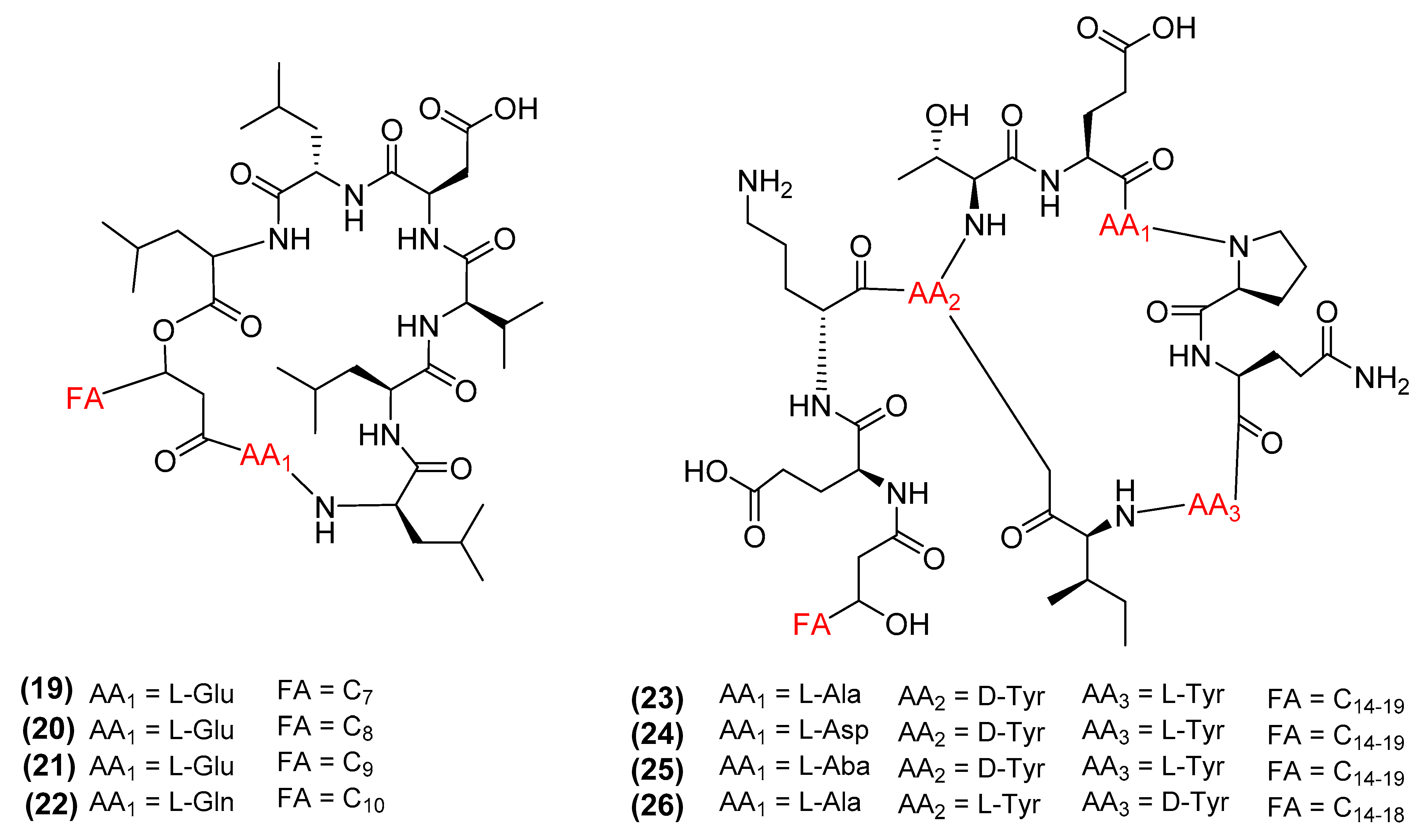
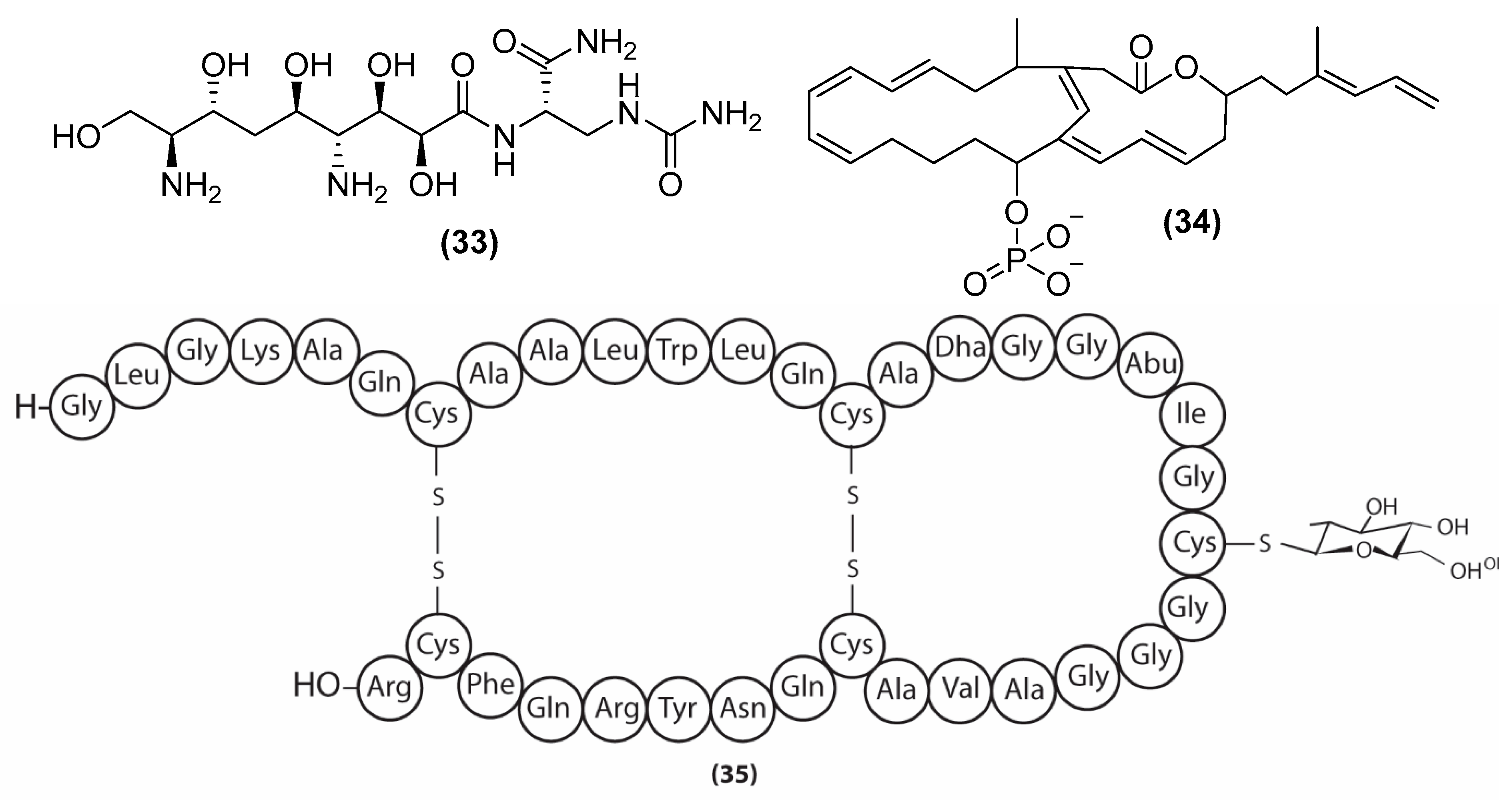
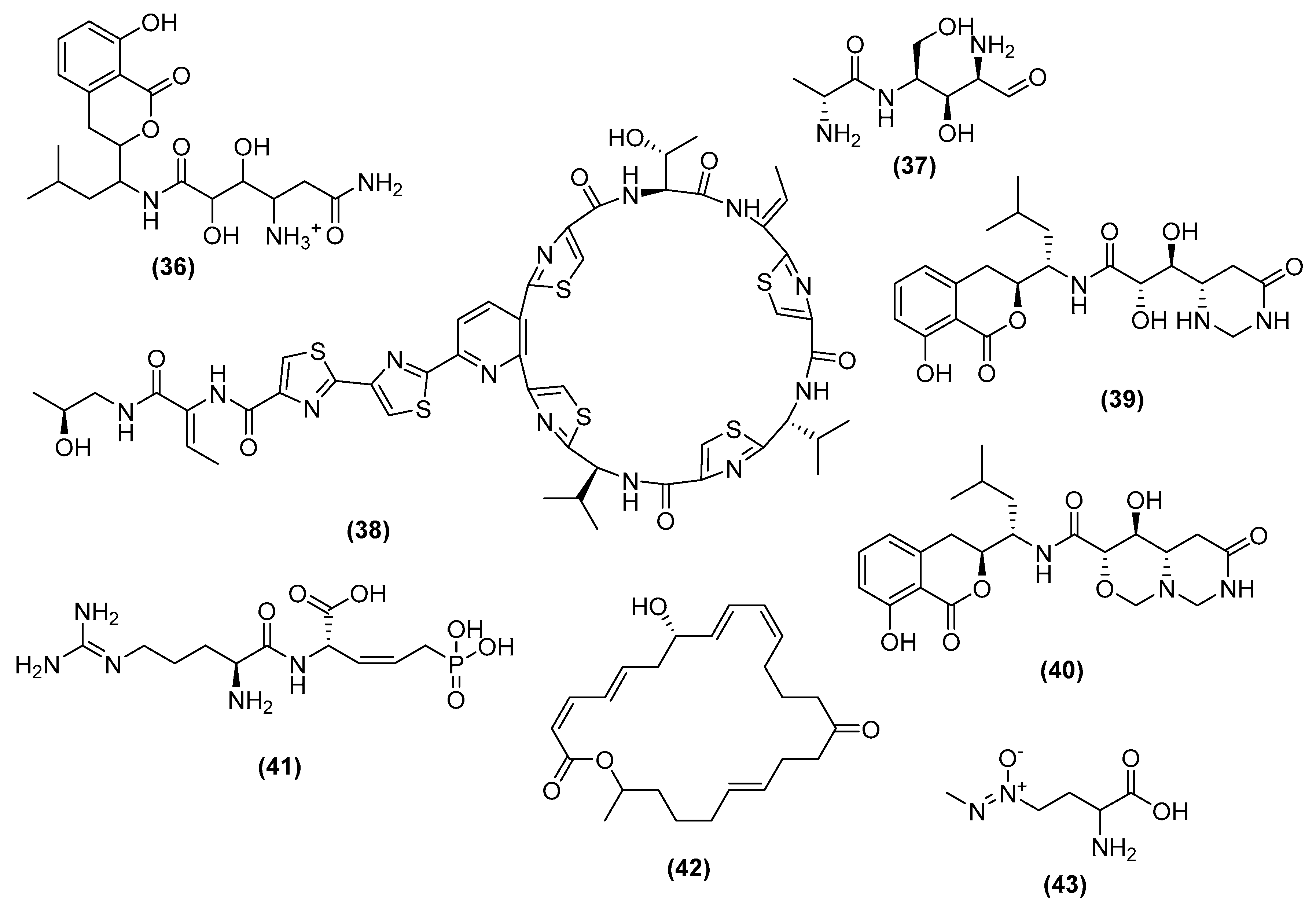
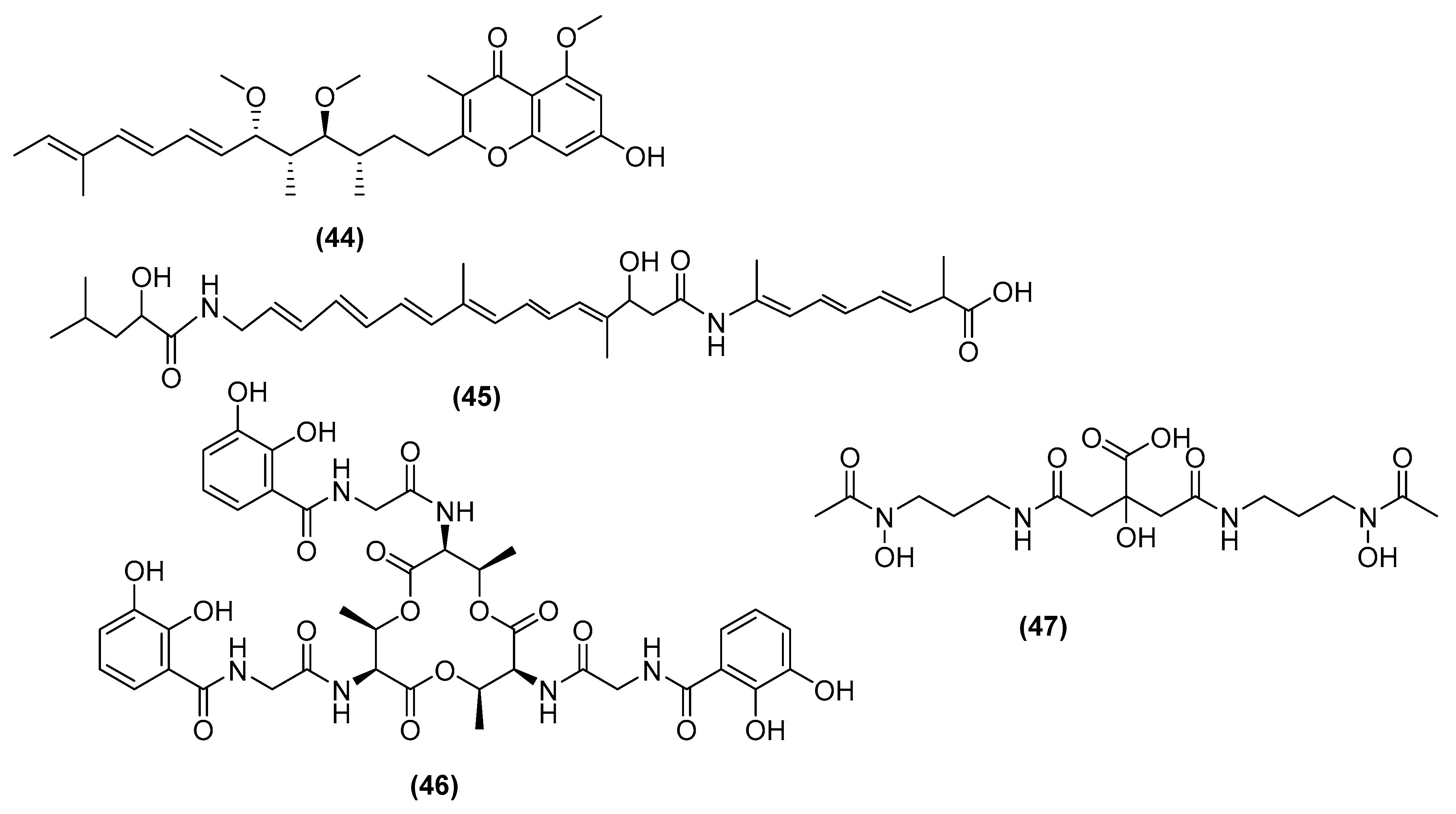
3. Antimicrobial Metabolites and Their Mechanism of Action
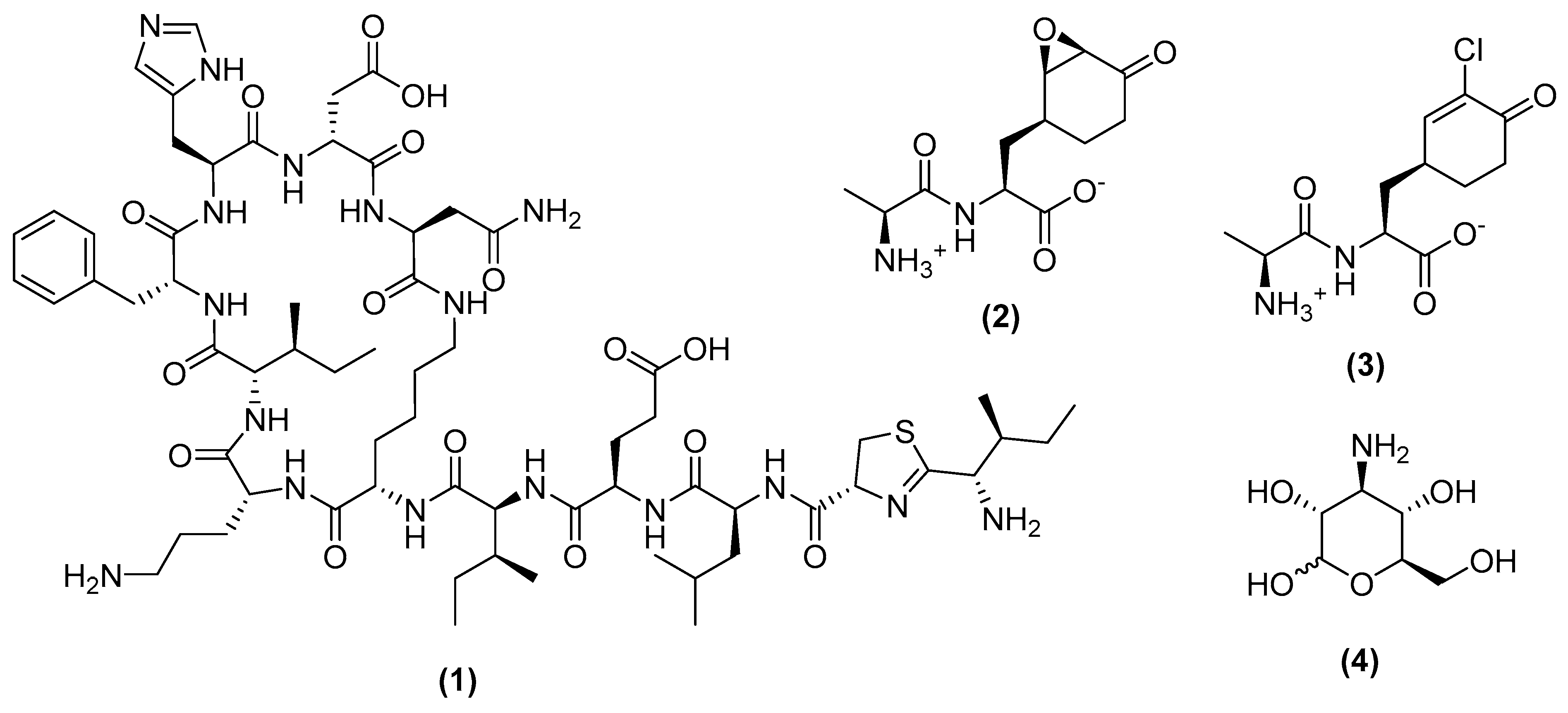
3.1. Metabolites Targeting the Cell Wall
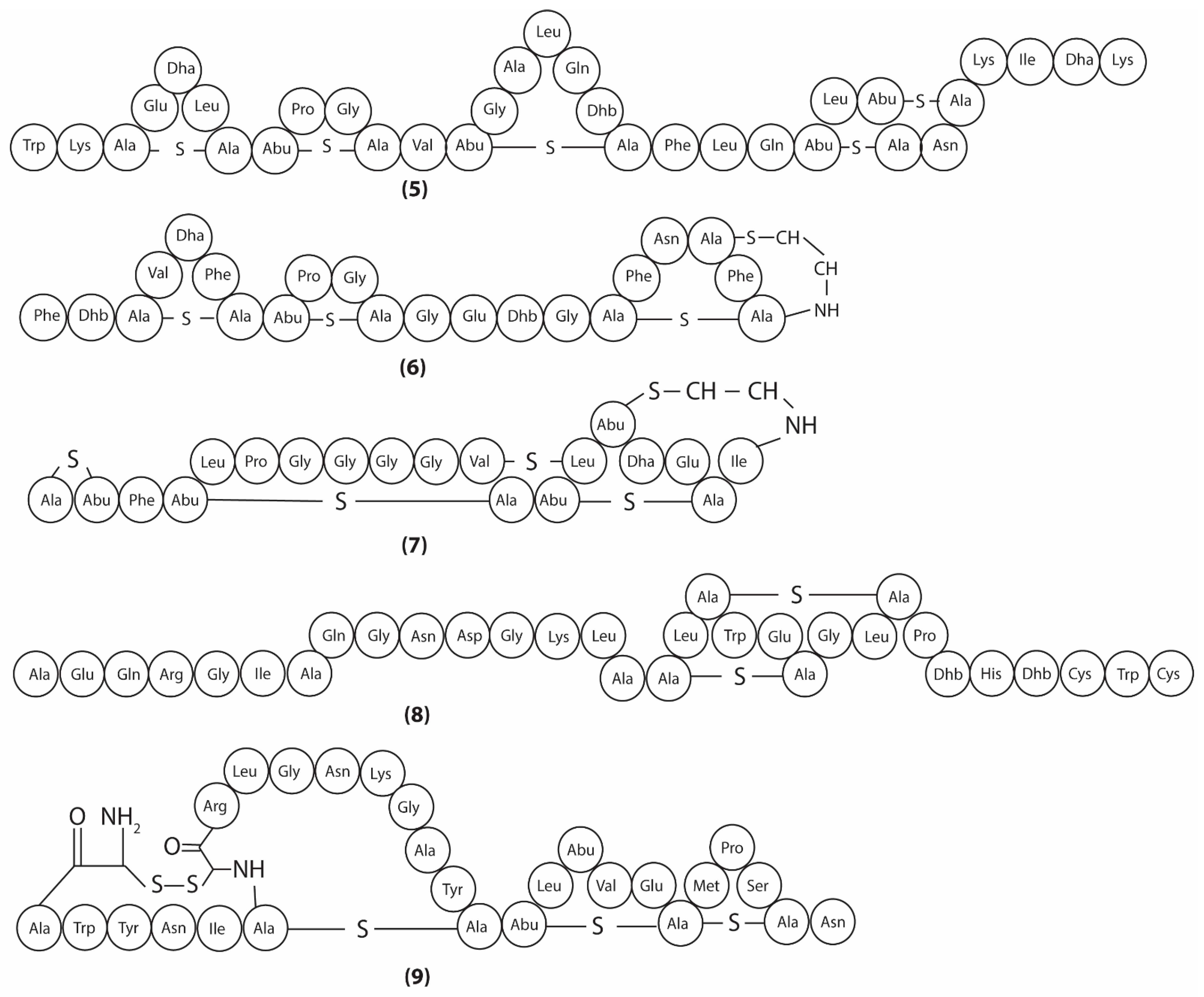
3.2. Metabolites Targeting Plasma Membrane
3.3. Metabolites Targeting Intracellular Processes
3.4. Metabolites Interacting with Other Emerging Targets
4. Conclusions Remarks and Future Directions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhao, J.; Guo, P.; Lu, W.; Geng, Z.; Levesque, C.L.; Johnston, L.J.; Wang, C.; Liu, L.; Zhang, J.; et al. Dietary Corn Bran Fermented by Bacillus subtilis MA139 Decreased Gut Cellulolytic Bacteria and Microbiota Diversity in Finishing Pigs. Front. Cell. Infect. Microbiol. 2017, 7, 526. [Google Scholar] [CrossRef]
- Kahn, L.H. Antimicrobial resistance: A One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Duong, M.C.; Hsu, L.Y.; Tam, C.C. Determinants influencing antibiotic use in Singapore’s small-scale aquaculture sectors: A qualitative study. PLoS ONE 2020, 15, e0228701. [Google Scholar] [CrossRef]
- Carlet, J. The gut is the epicentre of antibiotic resistance. Antimicrob. Resist. Infect. Control 2012, 1, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xin, H.; Jiang, N.; Lv, Z.; Shu, J.; Shi, H. Bacillus Amyloliquefaciens-9 as an Alternative Approach to Cure Diarrhea in Saanen Kids. Animals 2021, 11, 592. [Google Scholar] [CrossRef]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic Resistance of LACTOBACILLUS Strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Abel-Santos, E. Chapter 9—Endospores, Sporulation and Germination. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.-W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 163–178. [Google Scholar] [CrossRef]
- Adeniji, A.A.L.D.; Babalola, O.O. Bacillus velezensis: Phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019, 103, 3669–3682. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, A.; Sansinenea, E. Chemical Compounds Produced by Bacillus sp. Factories and Their Role in Nature. Mini Rev. Med. Chem. 2019, 19, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishino, K.; Hsu, F.-F.; Turk, J.; Cromie, M.J.; Wösten, M.M.S.M.; Groisman, E.A. Identification of the lipopolysaccharide modifications controlled by the Salmonella PmrA/PmrB system mediating resistance to Fe(III) and Al(III). Mol. Microbiol. 2006, 61, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, B.A.; Anker, H.; Meleney, F.L. Bacitracin: A new antibiotic produced by a member of the B. subtilis group. Science 1945, 102, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Singh, S.L.; Khan, A.U. Exploring antibiofilm potential of bacitracin against streptococcus mutans. Microb. Pathog. 2020, 149, 104279. [Google Scholar] [CrossRef]
- Siewert, G.; Strominger, J.L. Bacitracin: An inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell walls. Proc. Natl. Acad. Sci. USA 1967, 57, 767–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlstedt, S.A.; Walsh, C.T. Investigation of Anticapsin Biosynthesis Reveals a Four-Enzyme Pathway to Tetrahydrotyrosine in Bacillus subtilis. Biochemistry 2010, 49, 912–923. [Google Scholar] [CrossRef] [Green Version]
- Newton, G.G. Antibiotics from a strain of B. subtilis; bacilipin A and B and bacilysin. Br. J. Exp. Pathol. 1949, 30, 306–319. [Google Scholar]
- Wang, T.; Wu, M.B.; Chen, Z.J.; Lin, J.P.; Yang, L.R. Separation, determination and antifungal activity test of the products from a new Bacillus amyloliquefaciens. Nat. Prod. Res. 2016, 30, 1215–1218. [Google Scholar] [CrossRef]
- Kenig, M.; Abraham, E.P. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 1976, 94, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Kenig, M.; Vandamme, E.; Abraham, E.P. The mode of action of bacilysin and anticapsin and biochemical properties of bacilysin-resistant mutants. J. Gen. Microbiol. 1976, 94, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Milner, J.L.; Silo-Suh, L.; Lee, J.C.; He, H.; Clardy, J.; Handelsman, J. Production of kanosamine by Bacillus cereus UW85. Appl. Env. Microbiol. 1996, 62, 3061–3065. [Google Scholar] [CrossRef] [Green Version]
- Janiak, A.M.; Milewski, S. Mechanism of antifungal action of kanosamine. Med. Mycol. 2001, 39, 401–408. [Google Scholar] [CrossRef] [PubMed]
- van Heijenoort, J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 2007, 71, 620–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derouaux, A.; Turk, S.; Olrichs, N.K.; Gobec, S.; Breukink, E.; Amoroso, A.; Offant, J.; Bostock, J.; Mariner, K.; Chopra, I.; et al. Small molecule inhibitors of peptidoglycan synthesis targeting the lipid II precursor. Biochem. Pharm. 2011, 81, 1098–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chugunov, A.; Pyrkova, D.; Nolde, D.; Polyansky, A.; Pentkovsky, V.; Efremov, R. Lipid-II forms potential “landing terrain” for lantibiotics in simulated bacterial membrane. Sci. Rep. 2013, 3, 1678. [Google Scholar] [CrossRef] [Green Version]
- Ross, A.C.; Vederas, J.C. Fundamental functionality: Recent developments in understanding the structure–activity relationships of lantibiotic peptides. J. Antibiot. 2011, 64, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Chan, W.C.; Bycroft, B.W.; Leyland, M.L.; Lian, L.Y.; Roberts, G.C. A novel post-translational modification of the peptide antibiotic subtilin: Isolation and characterization of a natural variant from Bacillus subtilis A.T.C.C. 6633. Biochem. J. 1993, 291 Pt 1, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Parisot, J.; Carey, S.; Breukink, E.; Chan, W.C.; Narbad, A.; Bonev, B. Molecular mechanism of target recognition by subtilin, a class I lanthionine antibiotic. Antimicrob. Agents Chemother. 2008, 52, 612–618. [Google Scholar] [CrossRef] [Green Version]
- Bouhss, A.; Al-Dabbagh, B.; Vincent, M.; Odaert, B.; Aumont-Nicaise, M.; Bressolier, P.; Desmadril, M.; Mengin-Lecreulx, D.; Urdaci, M.C.; Gallay, J. Specific interactions of clausin, a new lantibiotic, with lipid precursors of the bacterial cell wall. Biophys. J. 2009, 97, 1390–1397. [Google Scholar] [CrossRef] [Green Version]
- Ahire, J.J.; Kashikar, M.S.; Lakshmi, S.G.; Madempudi, R. Identification and characterization of antimicrobial peptide produced by indigenously isolated Bacillus paralicheniformis UBBLi30 strain. 3 Biotech 2020, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Brötz, H.; Bierbaum, G.; Leopold, K.; Reynolds, P.E.; Sahl, H.G. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 1998, 42, 154–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sass, P.; Jansen, A.; Szekat, C.; Sass, V.; Sahl, H.G.; Bierbaum, G. The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 2008, 8, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brötz, H.; Bierbaum, G.; Reynolds, P.E.; Sahl, H.G. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur. J. Biochem. 1997, 246, 193–199. [Google Scholar] [CrossRef]
- Arguelles Arias, A.; Joris, B.; Fickers, P. Dual mode of action of amylolysin: A type-B lantibiotic produced by Bacillus amyloliquefaciens GA1. Protein Pept. Lett. 2014, 21, 336–340. [Google Scholar] [CrossRef]
- Arguelles Arias, A.; Ongena, M.; Devreese, B.; Terrak, M.; Joris, B.; Fickers, P. Characterization of amylolysin, a novel lantibiotic from Bacillus amyloliquefaciens GA1. PLoS ONE 2013, 8, e83037. [Google Scholar] [CrossRef] [Green Version]
- Oman, T.J.; van der Donk, W.A. Insights into the mode of action of the two-peptide lantibiotic haloduracin. ACS Chem. Biol. 2009, 4, 865–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oman, T.J.; Lupoli, T.J.; Wang, T.S.; Kahne, D.; Walker, S.; van der Donk, W.A. Haloduracin α binds the peptidoglycan precursor lipid II with 2:1 stoichiometry. J. Am. Chem. Soc. 2011, 133, 17544–17547. [Google Scholar] [CrossRef]
- Ferrante, T.; Viola, F.; Balliano, G.; Oliaro-Bosso, S. Difference in the late ergosterol biosynthesis between yeast spheroplasts and intact cells. Acta Biochim. Pol. 2016, 63, 371–375. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, B.; Morais, T.P.; Zaini, P.A.; Campos, C.S.; Almeida-Souza, H.O.; Dandekar, A.M.; Nascimento, R.; Goulart, L.R. Antimicrobial activity of Epsilon-Poly-l-lysine against phytopathogenic bacteria. Sci. Rep. 2020, 10, 11324. [Google Scholar] [CrossRef]
- El-Sersy, N.A.; Abdelwahab, A.E.; Abouelkhiir, S.S.; Abou-Zeid, D.M.; Sabry, S.A. Antibacterial and anticancer activity of ε-poly-L-lysine (ε-PL) produced by a marine Bacillus subtilis sp. J. Basic Microbiol. 2012, 52, 513–522. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Mygind, T.; Vad, B.S.; Stenvang, M.; Otzen, D.E.; Meyer, R.L. The antimicrobial mechanism of action of epsilon-poly-l-lysine. Appl. Environ. Microbiol. 2014, 80, 7758–7770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molohon, K.J.; Saint-Vincent, P.M.B.; Park, S.; Doroghazi, J.R.; Maxson, T.; Hershfield, J.R.; Flatt, K.M.; Schroeder, N.E.; Ha, T.; Mitchell, D.A. Plantazolicin is an ultra-narrow spectrum antibiotic that targets the Bacillus anthracis membrane. ACS Infect. Dis. 2016, 2, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Velkov, T.; Gallardo-Godoy, A.; Swarbrick, J.D.; Blaskovich, M.A.T.; Elliott, A.G.; Han, M.; Thompson, P.E.; Roberts, K.D.; Huang, J.X.; Becker, B.; et al. Structure, Function, and Biosynthetic Origin of Octapeptin Antibiotics Active against Extensively Drug-Resistant Gram-Negative Bacteria. Cell Chem. Biol. 2018, 25, 380–391.e385. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, K.S.; Ferguson, R.A.; Storm, D.R. Mechanism of action of EM 49, membrane-active peptide antibiotic. Antimicrob. Agents Chemother. 1977, 12, 665–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhu, X.; Cao, M.; Wang, C.; Zhang, C.; Lu, Z.; Lu, F. Genomics-Inspired Discovery of Three Antibacterial Active Metabolites, Aurantinins B, C, and D from Compost-Associated Bacillus subtilis fmb60. J. Agric. Food Chem. 2016, 64, 8811–8820. [Google Scholar] [CrossRef]
- Rollin-Pinheiro, R.; Bayona-Pacheco, B.; Domingos, L.T.S.; da Rocha Curvelo, J.A.; de Castro, G.M.M.; Barreto-Bergter, E.; Ferreira-Pereira, A. Sphingolipid Inhibitors as an Alternative to Treat Candidiasis Caused by Fluconazole-Resistant Strains. Pathogens 2021, 10, 856. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, J.M.; Clarke, D.J.; McMahon, S.A.; Lowther, J.P.; Beattie, A.E.; Langridge-Smith, P.R.; Broughton, H.B.; Dunn, T.M.; Naismith, J.H.; Campopiano, D.J. The chemical basis of serine palmitoyltransferase inhibition by myriocin. J. Am. Chem. Soc. 2013, 135, 14276–14285. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, Z.; Xu, W.; Wang, K. Comprehensive transcriptomic and proteomic analyses identify intracellular targets for myriocin to induce Fusarium oxysporum f. sp. niveum cell death. Microb. Cell Fact. 2021, 20, 69. [Google Scholar] [CrossRef]
- Takada, Y.; Itoh, H.; Paudel, A.; Panthee, S.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Discovery of gramicidin A analogues with altered activities by multidimensional screening of a one-bead-one-compound library. Nat. Commun. 2020, 11, 4935. [Google Scholar] [CrossRef]
- Hladky, S.B.; Haydon, D.A. Ion transfer across lipid membranes in the presence of gramicidin A. I. Studies of the unit conductance channel. Biochim. Biophys. Acta 1972, 274, 294–312. [Google Scholar] [CrossRef]
- Liou, J.W.; Hung, Y.J.; Yang, C.H.; Chen, Y.C. The antimicrobial activity of gramicidin A is associated with hydroxyl radical formation. PLoS ONE 2015, 10, e0117065. [Google Scholar] [CrossRef] [Green Version]
- Swierstra, J.; Kapoerchan, V.; Knijnenburg, A.; van Belkum, A.; Overhand, M. Structure, toxicity and antibiotic activity of gramicidin S and derivatives. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 763–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonin, S.; Dürr, U.H.; Wadhwani, P.; Salgado, J.; Ulrich, A.S. Solid State NMR Structure Analysis of the Antimicrobial Peptide Gramicidin S in Lipid Membranes: Concentration-Dependent Re-alignment and Self-Assembly as a β-Barrel. Top Curr. Chem. 2008, 273, 139–154. [Google Scholar] [CrossRef]
- Kaprel’iants, A.S.; Nikiforov, V.V.; Miroshnikov, A.I.; Snezhkova, L.G.; Eremin, V.A.; Ostrovskiĭ, D.N. Membranes of bacteria and mechanism of action of the antibiotic gramicidin S. Biokhimiia 1977, 42, 329–337. [Google Scholar] [PubMed]
- Frangou-Lazaridis, M.; Seddon, B. Effect of gramicidin S on the transcription system of the producer Bacillus brevis Nagano. J. Gen. Microbiol. 1985, 131, 437–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grau, A.; Ortiz, A.; de Godos, A.; Gómez-Fernández, J.C. A biophysical study of the interaction of the lipopeptide antibiotic iturin A with aqueous phospholipid bilayers. Arch. Biochem. Biophys. 2000, 377, 315–323. [Google Scholar] [CrossRef]
- Inès, M.; Dhouha, G. Lipopeptide surfactants: Production, recovery and pore forming capacity. Peptides 2015, 71, 100–112. [Google Scholar] [CrossRef]
- Qi, G.; Zhu, F.; Du, P.; Yang, X.; Qiu, D.; Yu, Z.; Chen, J.; Zhao, X. Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides 2010, 31, 1978–1986. [Google Scholar] [CrossRef]
- Ambroggio, E.E.; Separovic, F.; Bowie, J.H.; Fidelio, G.D.; Bagatolli, L.A. Direct visualization of membrane leakage induced by the antibiotic peptides: Maculatin, citropin, and aurein. Biophys. J. 2005, 89, 1874–1881. [Google Scholar] [CrossRef] [Green Version]
- Lei, S.; Zhao, H.; Pang, B.; Qu, R.; Lian, Z.; Jiang, C.; Shao, D.; Huang, Q.; Jin, M.; Shi, J. Capability of iturin from Bacillus subtilis to inhibit Candida albicans in vitro and in vivo. Appl. Microbiol. Biotechnol. 2019, 103, 4377–4392. [Google Scholar] [CrossRef] [PubMed]
- Peypoux, F.; Besson, F.; Michel, G.; Delcambe, L. Preparation and antibacterial activity upon Micrococcus luteus of derivatives of iturin A, mycosubtilin and bacillomycin L, antibiotics from Bacillus subtilis. J. Antibiot. 1979, 32, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, D.; de Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [Green Version]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Sheppard, J.D.; Jumarie, C.; Cooper, D.G.; Laprade, R. Ionic channels induced by surfactin in planar lipid bilayer membranes. Biochim. Biophys. Acta 1991, 1064, 13–23. [Google Scholar] [CrossRef]
- Liu, J.; Li, W.; Zhu, X.; Zhao, H.; Lu, Y.; Zhang, C.; Lu, Z. Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl. Microbiol. Biotechnol. 2019, 103, 4565–4574. [Google Scholar] [CrossRef]
- Farace, G.; Fernandez, O.; Jacquens, L.; Coutte, F.; Krier, F.; Jacques, P.; Clément, C.; Barka, E.A.; Jacquard, C.; Dorey, S. Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 2015, 16, 177–187. [Google Scholar] [CrossRef]
- Tao, Y.; Bie, X.M.; Lv, F.X.; Zhao, H.Z.; Lu, Z.X. Antifungal activity and mechanism of fengycin in the presence and absence of commercial surfactin against Rhizopus stolonifer. J. Microbiol. 2011, 49, 146–150. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Wang, S.; Guo, Y.; He, T.; Zhou, R. A Novel Adjuvant “Sublancin” Enhances Immune Response in Specific Pathogen-Free Broiler Chickens Inoculated with Newcastle Disease Vaccine. J. Immunol. Res. 2019, 2019, 1016567. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Sun, C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [Green Version]
- Johnson, V.L.; Ko, S.C.; Holmstrom, T.H.; Eriksson, J.E.; Chow, S.C. Effector caspases are dispensable for the early nuclear morphological changes during chemical-induced apoptosis. J. Cell Sci. 2000, 113 Pt 17, 2941–2953. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]
- Chung, L.K.; Raffatellu, M. Probiotic fengycins dis(Agr)ee with Staphylococcus aureus colonization. Cell Res. 2019, 29, 93–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Han, J.; Liu, H.; Qu, X.; Lu, Z.; Bie, X. Plipastatin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie Van Leeuwenhoek 2017, 110, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Cawoy, H.; Debois, D.; Franzil, L.; De Pauw, E.; Thonart, P.; Ongena, M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 2015, 8, 281–295. [Google Scholar] [CrossRef]
- Besson, F.; Peypoux, F.; Michel, G.; Delcambe, L. Antifungal activity upon Saccharomyces cerevisiae of iturin A, mycosubtilin, bacillomycin L and of their derivatives; inhibition of this antifungal activity by lipid antagonists. J. Antibiot. 1979, 32, 828–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, A.D.; Li, H.P.; Yuan, Q.S.; Song, X.S.; Yao, W.; He, W.J.; Zhang, J.B.; Liao, Y.C. Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, C.; Liang, J.; Wu, L.; Gao, W.; Jiang, J. Iturin A Extracted from Bacillus subtilis WL-2 Affects Phytophthora infestans via Cell Structure Disruption, Oxidative Stress, and Energy Supply Dysfunction. Front. Microbiol. 2020, 11, 2205. [Google Scholar] [CrossRef]
- Tunsagool, P.; Leelasuphakul, W.; Jaresitthikunchai, J.; Phaonakrop, N.; Roytrakul, S.; Jutidamrongphan, W. Targeted transcriptional and proteomic studies explicate specific roles of Bacillus subtilis iturin A, fengycin, and surfactin on elicitation of defensive systems in mandarin fruit during stress. PLoS ONE 2019, 14, e0217202. [Google Scholar] [CrossRef] [Green Version]
- Jin, P.; Wang, H.; Tan, Z.; Xuan, Z.; Dahar, G.Y.; Li, Q.X.; Miao, W.; Liu, W. Antifungal mechanism of bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides Penz. Pestic. Biochem. Physiol. 2020, 163, 102–107. [Google Scholar] [CrossRef]
- Peypoux, F.; Besson, F.; Michel, G.; Lenzen, C.; Dierickx, L.; Delcambe, L. Characterization of a new antibiotic of iturin group: Bacillomycin D. J. Antibiot. 1980, 33, 1146–1149. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Chen, M.; Zhou, L.; Lu, F.; Bie, X.; Lu, Z. Bacillomycin D effectively controls growth of Malassezia globosa by disrupting the cell membrane. Appl. Microbiol. Biotechnol. 2020, 104, 3529–3540. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D Produced by Bacillus amyloliquefaciens Is Involved in the Antagonistic Interaction with the Plant-Pathogenic Fungus Fusarium graminearum. Appl. Environ. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Mandic-Mulec, I.; Zhang, H.; Liu, Y.; Sun, X.; Feng, H.; Xun, W.; Zhang, N.; Shen, Q.; Zhang, R. Antibiotic Bacillomycin D Affects Iron Acquisition and Biofilm Formation in Bacillus velezensis through a Btr-Mediated FeuABC-Dependent Pathway. Cell Rep. 2019, 29, 1192–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Dong, C.; Shang, Q.; Cong, Y.; Kong, W.; Li, P. Purification and partial characterization of bacillomycin L produced by Bacillus amyloliquefaciens K103 from lemon. Appl. Biochem. Biotechnol. 2013, 171, 2262–2272. [Google Scholar] [CrossRef]
- Quentin, M.J.; Besson, F.; Peypoux, F.; Michel, G. Action of peptidolipidic antibiotics of the iturin group on erythrocytes. Effect of some lipids on hemolysis. Biochim. Biophys. Acta 1982, 684, 207–211. [Google Scholar] [CrossRef]
- Besson, F.; Michel, G. Action of the antibiotics of the iturin group on artificial membranes. J. Antibiot. 1984, 37, 646–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Qin, Y.; Han, Y.; Dong, C.; Li, P.; Shang, Q. Comparative proteomic analysis reveals intracellular targets for bacillomycin L to induce Rhizoctonia solani Kühn hyphal cell death. Biochim. Biophys. Acta 2016, 1864, 1152–1159. [Google Scholar] [CrossRef]
- Loison, C.; Nasir, M.N.; Benichou, E.; Besson, F.; Brevet, P.-F. Multi-scale modeling of mycosubtilin lipopeptides at the air/water interface: Structure and optical second harmonic generation. Phys. Chem. Chem. Phys. 2014, 16, 2136–2148. [Google Scholar] [CrossRef]
- Besson, F.; Michel, G. Action of mycosubtilin, an antifungal antibiotic of Bacillus subtilis, on the cell membrane of Saccharomyces cerevisiae. Microbios 1989, 59, 113–121. [Google Scholar]
- Bhattacharyya, P.; Bose, S.K. Effect of mycobacillin, an antifungal polypeptide antibiotic, on the producer Bacillus subtilis B3. Indian J. Med. Res. 1967, 55, 1025–1029. [Google Scholar] [PubMed]
- Chowdhury, B.; Das, S.K.; Bose, S.K. Use of resistant mutants to characterize the target of mycobacillin in Aspergillus niger membranes. Microbiology 1998, 144, 1123–1130. [Google Scholar] [CrossRef] [Green Version]
- Das, S.K.; Mukherjee, S.; Majumdar, S.; Basu, S.; Bose, S.K. Physico-chemical interaction of mycobacillin with Aspergillus niger protoplast membrane, the site of its action. J. Antibiot. 1987, 40, 1036–1043. [Google Scholar] [CrossRef] [Green Version]
- Babasaki, K.; Takao, T.; Shimonishi, Y.; Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985, 98, 585–603. [Google Scholar] [CrossRef]
- Turovskiy, Y.; Cheryian, T.; Algburi, A.; Wirawan, R.E.; Takhistov, P.; Sinko, P.J.; Chikindas, M.L. Susceptibility of Gardnerella vaginalis biofilms to natural antimicrobials subtilosin, ε-poly-L-lysine, and lauramide arginine ethyl ester. Infect. Dis. Obs. Gynecol. 2012, 2012, 284762. [Google Scholar] [CrossRef]
- Noll, K.S.; Sinko, P.J.; Chikindas, M.L. Elucidation of the Molecular Mechanisms of Action of the Natural Antimicrobial Peptide Subtilosin against the Bacterial Vaginosis-associated Pathogen Gardnerella vaginalis. Probiotics Antimicrob. Proteins 2011, 3, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Algburi, A.; Zehm, S.; Netrebov, V.; Bren, A.B.; Chistyakov, V.; Chikindas, M.L. Subtilosin Prevents Biofilm Formation by Inhibiting Bacterial Quorum Sensing. Probiotics Antimicrob. Proteins 2017, 9, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Yan, L.; Liu, J.; Song, F.; Zhang, J.; Li, X. Extraction of antibiotic zwittermicin A from Bacillus thuringiensis by macroporous resin and silica gel column chromatography. Biotechnol. Appl. Biochem. 2015, 62, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Stabb, E.V.; Handelsman, J. Genetic analysis of zwittermicin A resistance in Escherichia coli: Effects on membrane potential and RNA polymerase. Mol. Microbiol. 1998, 27, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Im, S.M.; Yu, N.H.; Joen, H.W.; Kim, S.O.; Park, H.W.; Park, A.R.; Kim, J.-C. Biological control of tomato bacterial wilt by oxydifficidin and difficidin-producing Bacillus methylotrophicus DR-08. Pestic. Biochem. Physiol. 2020, 163, 130–137. [Google Scholar] [CrossRef]
- Zweerink, M.M.; Edison, A. Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J. Antibiot. 1987, 40, 1692–1697. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Q.; Zeng, X.; Ye, Q.; Huang, S.; Yu, H.; Yang, T.; Qiao, S. Use of the Antimicrobial Peptide Sublancin with Combined Antibacterial and Immunomodulatory Activities To Protect against Methicillin-Resistant Staphylococcus aureus Infection in Mice. J. Agric. Food Chem. 2017, 65, 8595–8605. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Biswas, S.; Garcia De Gonzalo, C.V.; van der Donk, W.A. Investigations into the Mechanism of Action of Sublancin. ACS Infect. Dis 2019, 5, 454–459. [Google Scholar] [CrossRef]
- Maksimova, E.M.; Vinogradova, D.S.; Osterman, I.A.; Kasatsky, P.S.; Nikonov, O.S.; Milón, P.; Dontsova, O.A.; Sergiev, P.V.; Paleskava, A.; Konevega, A.L. Multifaceted Mechanism of Amicoumacin A Inhibition of Bacterial Translation. Front. Microbiol. 2021, 12, 172. [Google Scholar] [CrossRef] [PubMed]
- Lama, A.; Pané-Farré, J.; Chon, T.; Wiersma, A.M.; Sit, C.S.; Vederas, J.C.; Hecker, M.; Nakano, M.M. Response of methicillin-resistant Staphylococcus aureus to amicoumacin A. PLoS ONE 2012, 7, e34037. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Fukuda, M.; Amaki, Y.; Sakaguchi, T.; Inai, K.; Ishihara, A.; Nakajima, H. Importance of prumycin produced by Bacillus amyloliquefaciens SD-32 in biocontrol against cucumber powdery mildew disease. Pest. Manag. Sci. 2017, 73, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.L.; Katagiri, M.; Omura, S.; Tishler, M. The mechanism of prumycin action. J. Antibiot. 1974, 27, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Hata, T.; Omura, S.; Katagiri, M.; Atsumi, K.; Awaya, J. A new antifungal antibiotic, prumycin. J. Antibiot. 1971, 24, 900–901. [Google Scholar] [CrossRef] [Green Version]
- Chan, D.C.K.; Burrows, L.L. Thiocillin and micrococcin exploit the ferrioxamine receptor of Pseudomonas aeruginosa for uptake. J. Antimicrob. Chemother. 2021, 76, 2029–2039. [Google Scholar] [CrossRef]
- Bleich, R.; Watrous, J.D.; Dorrestein, P.C.; Bowers, A.A.; Shank, E.A. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 3086–3091. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Lu, Q.; Sun, C.; Lukianov, D.; Osterman, I.A.; Sergiev, P.V.; Dontsova, O.A.; Hu, X.; You, X.; Liu, S.; et al. Hetiamacin E and F, New Amicoumacin Antibiotics from Bacillus subtilis PJS Using MS/MS-Based Molecular Networking. Molecules 2020, 25, 4446. [Google Scholar] [CrossRef]
- Gahungu, M.; Arguelles-Arias, A.; Fickers, P.; Zervosen, A.; Joris, B.; Damblon, C.; Luxen, A. Synthesis and biological evaluation of potential threonine synthase inhibitors: Rhizocticin A and Plumbemycin A. Bioorg. Med. Chem. 2013, 21, 4958–4967. [Google Scholar] [CrossRef] [PubMed]
- Kugler, M.; Loeffler, W.; Rapp, C.; Kern, A.; Jung, G. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: Biological properties. Arch. Microbiol. 1990, 153, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.S.; Zheng, C.J.; Lee, S.; Kwak, J.H.; Kim, W.G. Macrolactin N, a new peptide deformylase inhibitor produced by Bacillus subtilis. Bioorg. Med. Chem. Lett. 2006, 16, 4889–4892. [Google Scholar] [CrossRef]
- Aoki, Y.; Yamamoto, M.; Hosseini-Mazinani, S.M.; Koshikawa, N.; Sugimoto, K.; Arisawa, M. Antifungal azoxybacilin exhibits activity by inhibiting gene expression of sulfite reductase. Antimicrob. Agents Chemother. 1996, 40, 127–132. [Google Scholar] [CrossRef] [Green Version]
- Fujiu, M.; Sawairi, S.; Shimada, H.; Takaya, H.; Aoki, Y.; Okuda, T.; Yokose, K. Azoxybacilin, a novel antifungal agent produced by Bacillus cereus NR2991. Production, isolation and structure elucidation. J. Antibiot. 1994, 47, 833–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeriouh, H.; de Vicente, A.; Pérez-García, A.; Romero, D. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 2014, 16, 2196–2211. [Google Scholar] [CrossRef]
- Caro-Astorga, J.; Frenzel, E.; Perkins, J.R.; Álvarez-Mena, A.; de Vicente, A.; Ranea, J.A.G.; Kuipers, O.P.; Romero, D. Biofilm formation displays intrinsic offensive and defensive features of Bacillus cereus. NPJ Biofilms Microbiomes 2020, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Karunakaran, E.; Biggs, C.A. Mechanisms of Bacillus cereus biofilm formation: An investigation of the physicochemical characteristics of cell surfaces and extracellular proteins. Appl. Microbiol. Biotechnol. 2011, 89, 1161–1175. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.A.; Singh, A.K.; Nerurkar, A. Disrupting the quorum sensing mediated virulence in soft rot causing Pectobacterium carotovorum by marine sponge associated Bacillus sp. OA10. World J. Microbiol. Biotechnol. 2021, 37, 5. [Google Scholar] [CrossRef]
- Boopathi, S.; Vashisth, R.; Manoharan, P.; Kandasamy, R.; Sivakumar, N. Stigmatellin Y—An anti-biofilm compound from Bacillus subtilis BR4 possibly interferes in PQS-PqsR mediated quorum sensing system in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2017, 27, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Han, X.; Dong, Y.; Xu, S.; Chen, C.; Feng, Y.; Cui, Q.; Li, W. Bacillaenes: Decomposition Trigger Point and Biofilm Enhancement in Bacillus. ACS Omega 2021, 6, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Tonziello, G.; Caraffa, E.; Pinchera, B.; Granata, G.; Petrosillo, N. Present and future of siderophore-based therapeutic and diagnostic approaches in infectious diseases. Infect. Dis. Rep. 2019, 11, 8208. [Google Scholar] [CrossRef] [Green Version]
- Dertz, E.A.; Xu, J.; Stintzi, A.; Raymond, K.N. Bacillibactin-mediated iron transport in Bacillus subtilis. J. Am. Chem. Soc. 2006, 128, 22–23. [Google Scholar] [CrossRef]
- Hu, X.; Boyer, G.L. Siderophore-Mediated Aluminum Uptake by Bacillus megaterium ATCC 19213. Appl. Environ. Microbiol. 1996, 62, 4044–4048. [Google Scholar] [CrossRef] [Green Version]
- Segond, D.; Abi Khalil, E.; Buisson, C.; Daou, N.; Kallassy, M.; Lereclus, D.; Arosio, P.; Bou-Abdallah, F.; Nielsen Le Roux, C. Iron acquisition in Bacillus cereus: The roles of IlsA and bacillibactin in exogenous ferritin iron mobilization. PLoS Pathog. 2014, 10, e1003935. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.; Bont, L.; Engelhard, D.; Moore, E.; Fernández, D.; Kreisberg-Greenblatt, R.; Oved, K.; Eden, E.; Hays, J.P. A multifaceted ‘omics’ approach for addressing the challenge of antimicrobial resistance. Future Microbiol. 2015, 10, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Zhang, W.; Cai, X.; Zhu, T.; Xue, Y.; Liu, C. Bacillus velezensis CC09: A Potential ‘Vaccine’ for Controlling Wheat Diseases. Mol. Plant-Microbe Interact. 2018, 31, 623–632. [Google Scholar] [CrossRef]
- Alkema, W.; Boekhorst, J.; Wels, M.; van Hijum, S.A. Microbial bioinformatics for food safety and production. Brief. Bioinform. 2016, 17, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, S.; Voigt, B.; Albrecht, D.; Bongaerts, J.; Evers, S.; Hecker, M.; Daniel, R.; Liesegang, H. Fermentation stage-dependent adaptations of Bacillus licheniformis during enzyme production. Microb. Cell Factories 2013, 12, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, A.; Khan, M.A.; Ehsanullah, M.; Haroon, U.; Azam, S.M.; Hameed, A. Production of peptide antibiotics by Bacillus sp. GU 057 indigenously isolated from saline soil. Braz. J. Microbiol. 2012, 43, 1340–1346. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Li, L.; Wu, F.; Wu, Y.; Wang, Z.; Chen, X.; Li, J.; Cai, D.; Chen, S. Metabolic Engineering of Aspartic Acid Supply Modules for Enhanced Production of Bacitracin in Bacillus licheniformis. ACS Synth. Biol. 2021, 10, 2243–2251. [Google Scholar] [CrossRef]
- Haavik, H.I.; Vessia, B. Bacitracin production by the high-yielding mutant bacillus licheniformis strain al: Stimulatory effect of l-leucine. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. 1978, 86B, 67–70. [Google Scholar] [CrossRef]
- Ganchev, K.; Kozhukharova, L. Bacitracin biosynthesis by Bacillus licheniformis 16. Acta Microbiol. Bulg. 1984, 15, 38–42. [Google Scholar] [PubMed]
- Egorov, N.S.; Loriia, Z.K.; Vybornykh, S.N.; Khamrun, R. Effect of bacitracin on the sporulation of Bacillus licheniformis 28 KA. Nauchnye Dokl. vysshei shkoly. Biol. Nauk. 1985, 6, 89–91. [Google Scholar]
- Vitković, L.; Sadoff, H.L. In vitro production of bacitracin by proteolysis of vegetative Bacillus licheniformis cell protein. J. Bacteriol. 1977, 131, 897–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamil, B.; Hasan, F.; Hameed, A.; Ahmed, S. Isolation of Bacillus subtilis MH-4 from soil and its potential of polypeptidic antibiotic production. Pak. J. Pharm. Sci. 2007, 20, 26–31. [Google Scholar] [PubMed]
- Amin, A.; Khan, M.A.; Ahmad, T. Optimized antimicrobial peptide (Bacitracin) production by immobilized and free cells and of Bacillus Spp. GU215 using Wood chips and silicon polymer beads. Pak. J. Pharm. Sci. 2013, 26, 1077–1082. [Google Scholar] [PubMed]
- Lu, J.Y.; Zhou, K.; Huang, W.T.; Zhou, P.; Yang, S.; Zhao, X.; Xie, J.; Xia, L.; Ding, X. A comprehensive genomic and growth proteomic analysis of antitumor lipopeptide bacillomycin Lb biosynthesis in Bacillus amyloliquefaciens X030. Appl. Microbiol. Biotechnol. 2019, 103, 7647–7662. [Google Scholar] [CrossRef]
- Rogers, H.J.; Newton, G.G.F.; Abraham, E.P. Production and purification of bacilysin. Biochem. J. 1965, 97, 573–578. [Google Scholar] [CrossRef] [Green Version]
- Ozcengiz, G.; Alaeddinoglu, N.G.; Demain, A.L. Regulation of biosynthesis of bacilysin by Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 1990, 6, 91–100. [Google Scholar] [CrossRef]
- Chung, S.; Kong, H.; Buyer, J.S.; Lakshman, D.K.; Lydon, J.; Kim, S.-D.; Roberts, D.P. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl. Microbiol. Biotechnol. 2008, 80, 115–123. [Google Scholar] [CrossRef]
- Phister, T.G.; O’Sullivan, D.J.; McKay, L.L. Identification of bacilysin, chlorotetaine, and iturin a produced by Bacillus sp. strain CS93 isolated from pozol, a Mexican fermented maize dough. Appl. Environ. Microbiol. 2004, 70, 631–634. [Google Scholar] [CrossRef] [Green Version]
- Vairagkar, U.; Mirza, Y. Antagonistic Activity of Antimicrobial Metabolites Produced from Seaweed-Associated Bacillus amyloliquefaciens MTCC 10456 Against Malassezia spp. Probiotics Antimicrob. Proteins 2021, 13, 1228–1237. [Google Scholar] [CrossRef]
- Panneerselvam, P.; Senapati, A.; Kumar, U.; Sharma, L.; Lepcha, P.; Prabhukarthikeyan, S.R.; Jahan, A.; Parameshwaran, C.; Govindharaj, G.P.P.; Lenka, S.; et al. Antagonistic and plant-growth promoting novel Bacillus species from long-term organic farming soils from Sikkim, India. 3 Biotech 2019, 9, 416. [Google Scholar] [CrossRef]
- AlGburi, A.; Alazzawi, S.A.; Al-Ezzy, A.I.A.; Weeks, R.; Chistyakov, V.; Chikindas, M.L. Potential Probiotics Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895 Co-Aggregate with Clinical Isolates of Proteus mirabilis and Prevent Biofilm Formation. Probiotics Antimicrob. Proteins 2020, 12, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Ahire, J.J.; Kashikar, M.S.; Madempudi, R.S. Survival and Germination of Bacillus clausii UBBC07 Spores in in vitro Human Gastrointestinal Tract Simulation Model and Evaluation of Clausin Production. Front. Microbiol. 2020, 11, 1010. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Markus, A.; Molitor, E.; Sahl, H.G. Mode of action of the lantibiotic mersacidin: Inhibition of peptidoglycan biosynthesis via a novel mechanism? Antimicrob. Agents Chemother. 1995, 39, 714–719. [Google Scholar] [CrossRef] [Green Version]
- Appleyard, A.N.; Choi, S.; Read, D.M.; Lightfoot, A.; Boakes, S.; Hoffmann, A.; Chopra, I.; Bierbaum, G.; Rudd, B.A.; Dawson, M.J.; et al. Dissecting Structural and Functional Diversity of the Lantibiotic Mersacidin. Chem. Biol. 2009, 16, 490–498. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Hao, K.; Blom, J.; Rückert, C.; Vater, J.; Mao, Z.; Wu, Y.; Hou, M.; He, P.; He, Y.; et al. Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601-Y2 and expression of mersacidin and other secondary metabolites. J. Biotechnol. 2013, 164, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Molinatto, G.; Puopolo, G.; Sonego, P.; Moretto, M.; Engelen, K.; Viti, C.; Ongena, M.; Pertot, I. Complete genome sequence of Bacillus amyloliquefaciens subsp. plantarum S499, a rhizobacterium that triggers plant defences and inhibits fungal phytopathogens. J. Biotechnol. 2016, 238, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Scholz, R.; Molohon, K.J.; Nachtigall, J.; Vater, J.; Markley, A.L.; Süssmuth, R.D.; Mitchell, D.A.; Borriss, R. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2011, 193, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyers, E.; Brown, W.E.; Principe, P.A.; Rathnum, M.L.; Parker, W.L. EM49, a new peptide antibiotic. I. Fermentation, isolation, and preliminary characterization. J. Antibiot. 1973, 26, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, Z.; Liu, Z.; Wang, K.; Xu, W. Membrane disruption of Fusarium oxysporum f. sp. niveum induced by myriocin from Bacillus amyloliquefaciens LZN01. Microb. Biotechnol. 2021, 14, 517–534. [Google Scholar] [CrossRef]
- Marahiel, M.A.; Lurz, R.; Kleinkauf, H. Characterization of chromosomal and membrane associated plasmid in Bacillus brevis ATCC 9999. J. Antibiot. 1981, 34, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Udalova, T.P.; Gus’Kova, T.M.; Silaev, A.B. Growth of Bacillus brevis var. G.B. and formation of gramicidin S in relation to the intensity of aeration. Mikrobiologiia 1972, 41, 280–286. [Google Scholar]
- Jiang, J.; Gao, L.; Bie, X.; Lu, Z.; Liu, H.; Zhang, C.; Lu, F.; Zhao, H. Identification of novel surfactin derivatives from NRPS modification of Bacillus subtilis and its antifungal activity against Fusarium moniliforme. BMC Microbiol. 2016, 16, 31. [Google Scholar] [CrossRef] [Green Version]
- Kim, P.I.; Ryu, J.; Kim, Y.H.; Chi, Y.-T. Production of Biosurfactant Lipopeptides Iturin A, Fengycin and Surfactin A from Bacillus subtilis CMB32 for Control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 2010, 20, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Wang, J.; Zhu, Y.; Liu, B.; Yang, W.; Ruan, C. Antibacterial activity against Ralstonia solanacearum of the lipopeptides secreted from the Bacillus amyloliquefaciens strain FJAT-2349. J. Appl. Microbiol. 2019, 126, 1519–1529. [Google Scholar] [CrossRef]
- Perez, K.J.; Viana, J.D.S.; Lopes, F.C.; Pereira, J.Q.; Dos Santos, D.M.; Oliveira, J.S.; Velho, R.V.; Crispim, S.M.; Nicoli, J.R.; Brandelli, A.; et al. Bacillus spp. Isolated from Puba as a Source of Biosurfactants and Antimicrobial Lipopeptides. Front. Microbiol. (Orig. Res.) 2017, 8, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Wu, Y.; Chong, X.; Xin, Q.; Wang, D.; Bian, K. Seed-borne endophytic Bacillus velezensis LHSB1 mediate the biocontrol of peanut stem rot caused by Sclerotium rolfsii. J. Appl. Microbiol. 2020, 128, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Pan, L.; Gong, L.; Yang, Y.; He, H.; Li, Y.; Peng, Y.; Li, N.; Yan, L.; Ding, X.; et al. Interaction of a novel Bacillus velezensis (BvL03) against Aeromonas hydrophila in vitro and in vivo in grass carp. Appl. Microbiol. Biotechnol. 2019, 103, 8987–8999. [Google Scholar] [CrossRef]
- Zidour, M.; Belguesmia, Y.; Cudennec, B.; Grard, T.; Flahaut, C.; Souissi, S.; Drider, D. Genome Sequencing and Analysis of Bacillus pumilus ICVB403 Isolated from Acartia tonsa Copepod Eggs Revealed Surfactin and Bacteriocin Production: Insights on Anti-Staphylococcus Activity. Probiotics Antimicrob. Proteins 2019, 11, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Zhang, Y.; Qiao, H.; He, J.; Yuan, Q.; Chen, X.; Fan, J. High-cell-density culture enhances the antimicrobial and freshness effects of Bacillus subtilis S1702 on table grapes (Vitis vinifera cv. Kyoho). Food Chem. 2019, 286, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wei, Z.; Tan, S.; Mei, X.; Shen, Q.; Xu, Y. Suppression of Bacterial Wilt of Tomato by Bioorganic Fertilizer Made from the Antibacterial Compound Producing Strain Bacillus amyloliquefaciens HR62. J. Agric. Food Chem. 2014, 62, 10708–10716. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Lehner, S.; Bacher, M.; Compant, S.; Nikolic, B.; Plesko, M.; Schuhmacher, R.; Sessitsch, A.; Brader, G. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ. Microbiol. 2016, 18, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Pueyo, M.T.; Bloch, C.; Carmona-Ribeiro, A.M.; di Mascio, P. Lipopeptides Produced by a Soil Bacillus Megaterium Strain. Microb. Ecol. 2009, 57, 367–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Quan, C.; Wang, Y.; Wang, J.; Fan, S. Bacillus amyloliquefaciens Q-426 as a potential biocontrol agent against Fusarium oxysporum f. sp. spinaciae. J. Basic Microbiol. 2014, 54, 448–456. [Google Scholar] [CrossRef]
- Malfanova, N.; Kamilova, F.; Validov, S.; Shcherbakov, A.; Chebotar, V.; Tikhonovich, I.; Lugtenberg, B. Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microb. Biotechnol. 2011, 4, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Vanittanakom, N.; Loeffler, W.; Koch, U.; Jung, G. Fengycin—A novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. 1986, 39, 888–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Abdallah, D.; Frikha-Gargouri, O.; Tounsi, S. Bacillus amyloliquefaciens strain 32a as a source of lipopeptides for biocontrol of Agrobacterium tumefaciens strains. J. Appl. Microbiol. 2015, 119, 196–207. [Google Scholar] [CrossRef]
- Pecci, Y.; Rivardo, F.; Martinotti, M.G.; Allegrone, G. LC/ESI-MS/MS characterisation of lipopeptide biosurfactants produced by the Bacillus licheniformis V9T14 strain. Biol. Mass Spectrom. 2010, 45, 772–778. [Google Scholar] [CrossRef]
- Li, X.-Y.; Mao, Z.-C.; Wang, Y.-H.; Wu, Y.-X.; He, Y.-Q.; Long, C.-L. ESI LC-MS and MS/MS Characterization of Antifungal Cyclic Lipopeptides Produced by Bacillus subtilis XF-1. J. Mol. Microbiol. Biotechnol. 2012, 22, 83–93. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, N.; Hu, J.; Wang, S. Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J. Antibiot. 2012, 65, 317–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Guo, W.; Chen, X. Exogenous addition of alkanoic acids enhanced production of antifungal lipopeptides in Bacillus amyloliquefaciens Pc3. Appl. Microbiol. Biotechnol. 2019, 103, 5367–5377. [Google Scholar] [CrossRef] [PubMed]
- Nastro, R.A.; Arguelles-Arias, A.; Ongena, M.; Di Costanzo, A.; Trifuoggi, M.; Guida, M.; Fickers, P. Antimicrobial Activity of Bacillus amyloliquefaciens ANT1 Toward Pathogenic Bacteria and Mold: Effects on Biofilm Formation. Probiotics Antimicrob. Proteins 2013, 5, 252–258. [Google Scholar] [CrossRef]
- Malfanova, N.; Franzil, L.; Lugtenberg, B.; Chebotar, V.; Ongena, M. Cyclic lipopeptide profile of the plant-beneficial endophytic bacterium Bacillus subtilis HC8. Arch. Microbiol. 2012, 194, 893–899. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Morales, A.; Martínez-Peniche, R.A.; Arvizu-Gómez, J.L.; Arvizu-Medrano, S.M.; Rodríguez-Ontiveros, A.; Ramos-López, M.A.; Pacheco-Aguilar, J.R. Production of a Mixture of Fengycins with Surfactant and Antifungal Activities by Bacillus sp. MA04, a Versatile PGPR. Indian J. Microbiol. 2018, 58, 208–213. [Google Scholar] [CrossRef]
- Escobar, V.V.; Ceballos, I.; Mira, J.J.; Argel, L.E.; Peralta, S.O.; Romero-Tabarez, M. Fengycin C Produced by Bacillus subtilis EA-CB0015. J. Nat. Prod. 2013, 76, 503–509. [Google Scholar] [CrossRef]
- Klich, M.A.; Lax, A.R.; Bland, J. Inhibition of some mycotoxigenic fungi by iturin A, a peptidolipid produced by Bacillus subtilis. Mycopathologia 1991, 116, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Zhao, F.; Liu, X.; Fan, X.; Huang, R.; Gao, W.; Wang, S.; Yang, C. Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb. Cell Factories 2019, 18, 68. [Google Scholar] [CrossRef]
- Cho, S.J.; Lee, S.K.; Cha, B.J.; Kim, Y.H.; Shin, K.S. Detection and characterization of the Gloeosporium gloeosporioides growth inhibitory compound iturin A from Bacillus subtilis strain KS03. FEMS Microbiol. Lett. 2003, 223, 47–51. [Google Scholar] [CrossRef] [Green Version]
- Mizumoto, S.; Hirai, M.; Shoda, M. Production of lipopeptide antibiotic iturin A using soybean curd residue cultivated with Bacillus subtilis in solid-state fermentation. Appl. Microbiol. Biotechnol. 2006, 72, 869–875. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, C.; Cai, K.; Zheng, Z.; Yu, Z. Purification and identification of iturin A from Bacillus subtilis JA by electrospray ionization mass spectrometry. Acta Microbiol. Sin. 2008, 48, 116–120. [Google Scholar]
- Thasana, N.; Prapagdee, B.; Rangkadilok, N.; Sallabhan, R.; Aye, S.L.; Ruchirawat, S.; Loprasert, S. Bacillus subtilis SSE4 produces subtulene A, a new lipopeptide antibiotic possessing an unusual C15 unsaturated β-amino acid. FEBS Lett. 2010, 584, 3209–3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, X.; Li, K.; Niu, Y.; Guo, M.; Hu, C.; Wan, X.; Gong, Y.; Huang, F. Direct Bio-Utilization of Untreated Rapeseed Meal for Effective Iturin A Production by Bacillus subtilis in Submerged Fermentation. PLoS ONE 2014, 9, e111171. [Google Scholar] [CrossRef]
- Yamamoto, S.; Shiraishi, S.; Suzuki, S. Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 2015, 60, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, X.; Li, C.; Tian, W.; Shen, Q.; Shen, B. Isolation of Bacillus amyloliquefaciens S20 and its application in control of eggplant bacterial wilt. J. Environ. Manag. 2014, 137, 120–127. [Google Scholar] [CrossRef]
- Saechow, S.; Thammasittirong, A.; Kittakoop, P.; Prachya, S.; Thammasittirong, S.N.-R. Antagonistic Activity against Dirty Panicle Rice Fungal Pathogens and Plant Growth-Promoting Activity of Bacillus amyloliquefaciens BAS23. J. Microbiol. Biotechnol. 2018, 28, 1527–1535. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, B.; Shen, Q.; You, C.; Yu, Y.; Li, P.; Shang, Q. Purification and Identification of Two Antifungal Cyclic Peptides Produced by Bacillus amyloliquefaciens L-H15. Appl. Biochem. Biotechnol. 2015, 176, 2202–2212. [Google Scholar] [CrossRef] [PubMed]
- Calvo, H.; Mendiara, I.; Arias, E.; Blanco, D.; Venturini, M. The role of iturin A from B. amyloliquefaciens BUZ-14 in the inhibition of the most common postharvest fruit rots. Food Microbiol. 2019, 82, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ding, Z.; Zhong, J.; Zhou, J.; Shu, D.; Luo, D.; Yang, J.; Tan, H. Improvement of iturin A production in Bacillus subtilis ZK0 by overexpression of the comA and sigA genes. Lett. Appl. Microbiol. 2017, 64, 452–458. [Google Scholar] [CrossRef]
- Athukorala, S.N.; Fernando, W.G.; Rashid, K.Y. Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can. J. Microbiol. 2009, 55, 1021–1032. [Google Scholar] [CrossRef]
- Shi, J.; Zhu, X.; Lu, Y.; Zhao, H.; Lu, F.; Lu, Z. Improving Iturin A Production of Bacillus amyloliquefaciens by Genome Shuffling and Its Inhibition Against Saccharomyces cerevisiae in Orange Juice. Front. Microbiol. 2018, 9, 2683. [Google Scholar] [CrossRef]
- Kaushal, M.; Kumar, A.; Kaushal, R. Bacillus pumilus strain YSPMK11 as plant growth promoter and bicontrol agent against Sclerotinia sclerotiorum. 3 Biotech 2017, 7, 90. [Google Scholar] [CrossRef] [Green Version]
- Moryl, M.; Spętana, M.; Dziubek, K.; Paraszkiewicz, K.; Różalska, S.; Płaza, G.A.; Rozalski, A. Antimicrobial, antiadhesive and antibiofilm potential of lipopeptides synthesised by Bacillus subtilis, on uropathogenic bacteria. Acta Biochim. Pol. 2015, 62, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Taira, T.; Imura, T. Screening of a Bacillus subtilis Strain Producing Multiple Types of Cyclic Lipopeptides and Evaluation of Their Surface-tension-lowering Activities. J. Oleo Sci. 2017, 66, 785–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.-H.; Ye, Z.-W.; Zheng, Q.-W.; Wei, T.; Lin, J.-F.; Guo, L.-Q. Isolation and characterization of cyclic lipopeptides with broad-spectrum antimicrobial activity from Bacillus siamensis JFL15. 3 Biotech 2018, 8, 444. [Google Scholar] [CrossRef]
- Ambrico, A.; Trupo, M.; Magarelli, R.A. Influence of Phenotypic Dissociation in Bacillus subtilis Strain ET-1 on Iturin A Production. Curr. Microbiol. 2019, 76, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Park, D.; Kim, K.; Lim, S.M.; Yu, N.H.; Kim, S.; Kim, H.-Y.; Jung, K.S.; Jang, J.Y.; Park, J.-C.; et al. Characterization of Bacillus amyloliquefaciens DA12 Showing Potent Antifungal Activity against Mycotoxigenic Fusarium Species. Plant Pathol. J. 2017, 33, 499–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Li, X.; Ma, X.; Chen, S.; Kang, Y.; Yang, M.; Huang, F.; Wan, X. Simultaneous hydrolysis with lipase and fermentation of rapeseed cake for iturin A production by Bacillus amyloliquefaciens CX-20. BMC Biotechnol. 2019, 19, 98. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Tsai, C.-H.; Chen, P.-Y.; Wu, C.-Y.; Chang, Y.-L.; Yang, Y.-L.; Chen, Y.-L. Biological control of potato common scab by Bacillus amyloliquefaciens Ba01. PLoS ONE 2018, 13, e0196520. [Google Scholar] [CrossRef] [Green Version]
- Waewthongrak, W.; Leelasuphakul, W.; Mccollum, G. Cyclic Lipopeptides from Bacillus subtilis ABS–S14 Elicit Defense-Related Gene Expression in Citrus Fruit. PLoS ONE 2014, 9, e109386. [Google Scholar] [CrossRef]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.-R.; Zhang, R. Exploring Elicitors of the Beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 to Induce Plant Systemic Resistance and Their Interactions with Plant Signaling Pathways. Mol. Plant-Microbe Interactions 2018, 31, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Wang, Q.; Wang, K.; Brian, K.; Liu, C.; Gu, Y. Study of the antifungal activity of Bacillus vallismortis ZZ185 in vitro and identification of its antifungal components. Bioresour. Technol. 2010, 101, 292–297. [Google Scholar] [CrossRef]
- Mácha, H.; Marešová, H.; Juříková, T.; Švecová, M.; Benada, O.; Škríba, A.; Baránek, M.; Novotný, Č.; Palyzová, A. Killing Effect of Bacillus velezensis FZB42 on a Xanthomonas campestris pv. Campestris (Xcc) Strain Newly Isolated from Cabbage Brassica oleracea Convar. Capitata (L.): A Metabolomic Study. Microorganisms 2021, 9, 1410. [Google Scholar] [CrossRef]
- Elkahoui, S.; Djébali, N.; Karkouch, I.; Ibrahim, A.H.; Kalai, L.; Bachkovel, S.; Tabbene, O.; Limam, F. Mass spectrometry identification of antifungal lipopeptides from Bacillus sp. BCLRB2 against Rhizoctonia solani and Sclerotinia sclerotiorum. Microb. Pathog. 2014, 50, 184–188. [Google Scholar] [CrossRef]
- Nam, J.; Alam, S.T.; Kang, K.; Choi, J.; Seo, M.-H. Anti-staphylococcal activity of a cyclic lipopeptide, C15-bacillomycin D, produced by Bacillus velezensis NST. J. Appl. Microbiol. 2021, 131, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Soussi, S.; Essid, R.; Hardouin, J.; Gharbi, D.; Elkahoui, S.; Tabbene, O.; Cosette, P.; Jouenne, T.; Limam, F. Utilization of Grape Seed Flour for Antimicrobial Lipopeptide Production by Bacillus amyloliquefaciens C5 Strain. Appl. Biochem. Biotechnol. 2019, 187, 1460–1474. [Google Scholar] [CrossRef] [PubMed]
- Radovanovic, N.; Milutinović, M.; Mihajlovski, K.; Jović, J.; Nastasijević, B.; Rajilic-Stojanovic, M.; Dimitrijević-Branković, S. Biocontrol and plant stimulating potential of novel strain Bacillus sp. PPM3 isolated from marine sediment. Microb. Pathog. 2018, 120, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.B.; Woodruff, H.B. A crystalline antifungal agent, mycosubtilin, isolated from subtilin broth. J. Clin. Investig. 1949, 28, 924–926. [Google Scholar] [CrossRef]
- Fickers, P.; Guez, J.-S.; Damblon, C.; Leclère, V.; Béchet, M.; Jacques, P.; Joris, B. High-Level Biosynthesis of the Anteiso-C 17 Isoform of the Antibiotic Mycosubtilin in Bacillus subtilis and Characterization of Its Candidacidal Activity. Appl. Environ. Microbiol. 2009, 75, 4636–4640. [Google Scholar] [CrossRef] [Green Version]
- Chevanet, C.; Besson, F.; Michel, G. Effect of various growth conditions on spore formation and Bacillomycin L production in Bacillus subtilis. Can. J. Microbiol. 1986, 32, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Wei, Z.; Guan, Z.; Cai, Y.; Liao, X. Antifungal Activity of Isolated Bacillus amyloliquefaciens SYBC H47 for the Biocontrol of Peach Gummosis. PLoS ONE 2016, 11, e0162125. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, S.; Bose, S. Properties and localisation of mycobacillin-synthesising enzyme system in Bacillus subtilis B3. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 1971, 237, 120–122. [Google Scholar] [CrossRef]
- Velho, R.V.; Caldas, D.G.G.; Medina, L.F.C.; Tsai, S.M.; Brandelli, A. Real-time PCR investigation on the expression of sboA and ituD genes in Bacillus spp. Lett. Appl. Microbiol. 2011, 52, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lee, J.Y.; Jeong, S.-J.; Cho, K.M.; Kim, G.M.; Shin, J.-H.; Kim, J.-S.; Kim, J.H. Properties of a Bacteriocin Produced by Bacillus subtilis EMD4 Isolated from Ganjang (Soy Sauce). J. Microbiol. Biotechnol. 2015, 25, 1493–1501. [Google Scholar] [CrossRef] [Green Version]
- Sutyak, K.; Wirawan, R.; Aroutcheva, A.; Chikindas, M. Isolation of the Bacillus subtilis antimicrobial peptide subtilosin from the dairy product-derived Bacillus amyloliquefaciens. J. Appl. Microbiol. 2008, 104, 1067–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Gao, G.; Xu, H.; Qiao, M. Identification of the bacteriocin subtilosin A and loss of purL results in its high-level production in Bacillus amyloliquefaciens. Res. Microbiol. 2012, 163, 470–478. [Google Scholar] [CrossRef]
- Parveen Rani, R.; Anandharaj, M.; Hema, S.; Deepika, R.; David Ravindran, A. Purification of Antilisterial Peptide (Subtilosin A) from Novel Bacillus tequilensis FR9 and Demonstrate Their Pathogen Invasion Protection Ability Using Human Carcinoma Cell Line. Front Microbiol. 2016, 7, 1910. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yin, S.; An, L.; Zhang, G.; Cheng, H.; Xi, Y.; Cui, G.; Zhang, F.; Zhang, L. Complete genome sequence of Bacillus subtilis BSD-2, a microbial germicide isolated from cultivated cotton. J. Biotechnol. 2016, 230, 26–27. [Google Scholar] [CrossRef]
- Li, Q.; Liao, S.; Wei, J.; Xing, D.; Xiao, Y.; Yang, Q. Isolation of Bacillus subtilis strain SEM-2 from silkworm excrement and characterisation of its antagonistic effect against Fusarium spp. Can. J. Microbiol. 2020, 66, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Shi, H.; Heng, J.; Wang, D.; Bian, K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 2019, 218, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-Y.; Liu, Y.; Miao, L.-L.; Li, E.-W.; Sun, G.-X.; Liu, Z.-P. Characterization and mechanism of anti-Aeromonas salmonicida activity of a marine probiotic strain, Bacillus velezensis V4. Appl. Microbiol. Biotechnol. 2017, 101, 3759–3768. [Google Scholar] [CrossRef]
- Sharma, D.; Singh, S.S.; Baindara, P.; Sharma, S.; Khatri, N.; Grover, V.; Patil, P.B.; Korpole, S. Surfactin Like Broad Spectrum Antimicrobial Lipopeptide Co-produced With Sublancin From Bacillus subtilis Strain A52: Dual Reservoir of Bioactives. Front. Microbiol. 2020, 11, 1167. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.-Y. Five New Amicoumacins Isolated from a Marine-Derived Bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Park, H.B.; Perez, C.E.; Perry, E.; Crawford, J.M. Activating and Attenuating the Amicoumacin Antibiotics. Molecules 2016, 21, 824. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J.; Shomura, T.; Omoto, S.; Miyado, S.; Yuda, Y.; Shibata, U.; Inouye, S. Isolation, Physicochemical Properties and Biological Activities of Amicoumacins Produced by Bacillus pumilus. Agric. Biol. Chem. 1982, 46, 1255–1259. [Google Scholar] [CrossRef]
- Wang, D.; Li, J.; Zhu, G.; Zhao, K.; Jiang, W.; Li, H.; Wang, W.; Kumar, V.; Dong, S.; Zhu, W.; et al. Mechanism of the Potential Therapeutic Candidate Bacillus subtilis BSXE-1601 Against Shrimp Pathogenic Vibrios and Multifunctional Metabolites Biosynthetic Capability of the Strain as Predicted by Genome Analysis. Front. Microbiol. 2020, 11, 581802. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.; Duan, L.; Lei, C.; Pan, H.; Ding, Y.; Zhang, Q.; Chen, D.; Shen, B.; Yu, Y.; Liu, W. Thiopeptide Biosynthesis Featuring Ribosomally Synthesized Precursor Peptides and Conserved Posttranslational Modifications. Chem. Biol. 2009, 16, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Akasapu, S.; Hinds, A.B.; Powell, W.C.; Walczak, M.A. Total synthesis of micrococcin P1 and thiocillin I enabled by Mo(vi) catalyst. Chem. Sci. 2019, 10, 1971–1975. [Google Scholar] [CrossRef] [Green Version]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; Santos-Villalobos, S.D.L. Integrated omics approaches for deciphering antifungal metabolites produced by a novel Bacillus species, B. cabrialesii TE3T, against the spot blotch disease of wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef]
- Kino, K.; Kotanaka, Y.; Arai, T.; Yagasaki, M. A novel L-amino acid ligase from Bacillus subtilis NBRC3134, a microorganism producing peptide-antibiotic rhizocticin. Biosci. Biotechnol. Biochem. 2009, 73, 901–907. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.S.; Huang, S.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P.; Mayerl, F. Bacillaene, a Novel Inhibitor of Procaryotic Protein Synthesis Produced by Bacillus subtilis: Production, Taxonomy, Isolation, Physico-chemical Characterization and Biological Activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, J.; Da, R.; Cheng, Y.; Tuo, X.; Wei, J.; Jiang, K.; Monisayo, A.O.; Han, B. Mechanism of Antibacterial Activity of Bacillus amyloliquefaciens C-1 Lipopeptide toward Anaerobic Clostridium difficile. BioMed Res. Int. 2020, 2020, 3104613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Tian, Z.; Luo, Y.; Cheng, Y.; Long, C.-A. Antagonistic Activity and the Mechanism of Bacillus amyloliquefaciens DH-4 Against Citrus Green Mold. Phytopathology 2018, 108, 1253–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonejuie, P.; Trial, R.M.; Newton, G.L.; Lamsa, A.; Perera, V.R.; Aguilar, J.; Liu, W.-T.; Dorrestein, P.C.; Pogliano, J.; Pogliano, K. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J. Antibiot. 2016, 69, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Daas, M.S.; Acedo, J.; Rosana, A.R.; Orata, F.; Reiz, B.; Zheng, J.; Nateche, F.; Case, R.J.; Kebbouche-Gana, S.; Vederas, J.C. Bacillus amyloliquefaciens ssp. plantarum F11 isolated from Algerian salty lake as a source of biosurfactants and bioactive lipopeptides. FEMS Microbiol. Lett. 2018, 365, fnx248. [Google Scholar] [CrossRef] [PubMed]
- Butcher, R.A.; Schroeder, F.C.; Fischbach, M.A.; Straight, P.D.; Kolter, R.; Walsh, C.T.; Clardy, J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 1506–1509. [Google Scholar] [CrossRef] [Green Version]
- Dimopoulou, A.; Theologidis, I.; Benaki, D.; Koukounia, M.; Zervakou, A.; Tzima, A.; Diallinas, G.; Hatzinikolaou, D.G.; Skandalis, N. Direct Antibiotic Activity of Bacillibactin Broadens the Biocontrol Range of Bacillus amyloliquefaciens MBI600. mSphere 2021, 6, e0037621. [Google Scholar] [CrossRef]
- Peters, W.J.; Warren, R.A.J. Itoic Acid Synthesis in Bacillus subtilis. J. Bacteriol. 1968, 95, 360–366. [Google Scholar] [CrossRef] [Green Version]
- Ollinger, J.; Song, K.-B.; Antelmann, H.; Hecker, M.; Helmann, J.D. Role of the Fur Regulon in Iron Transport in Bacillus subtilis. J. Bacteriol. 2006, 188, 3664–3673. [Google Scholar] [CrossRef] [Green Version]

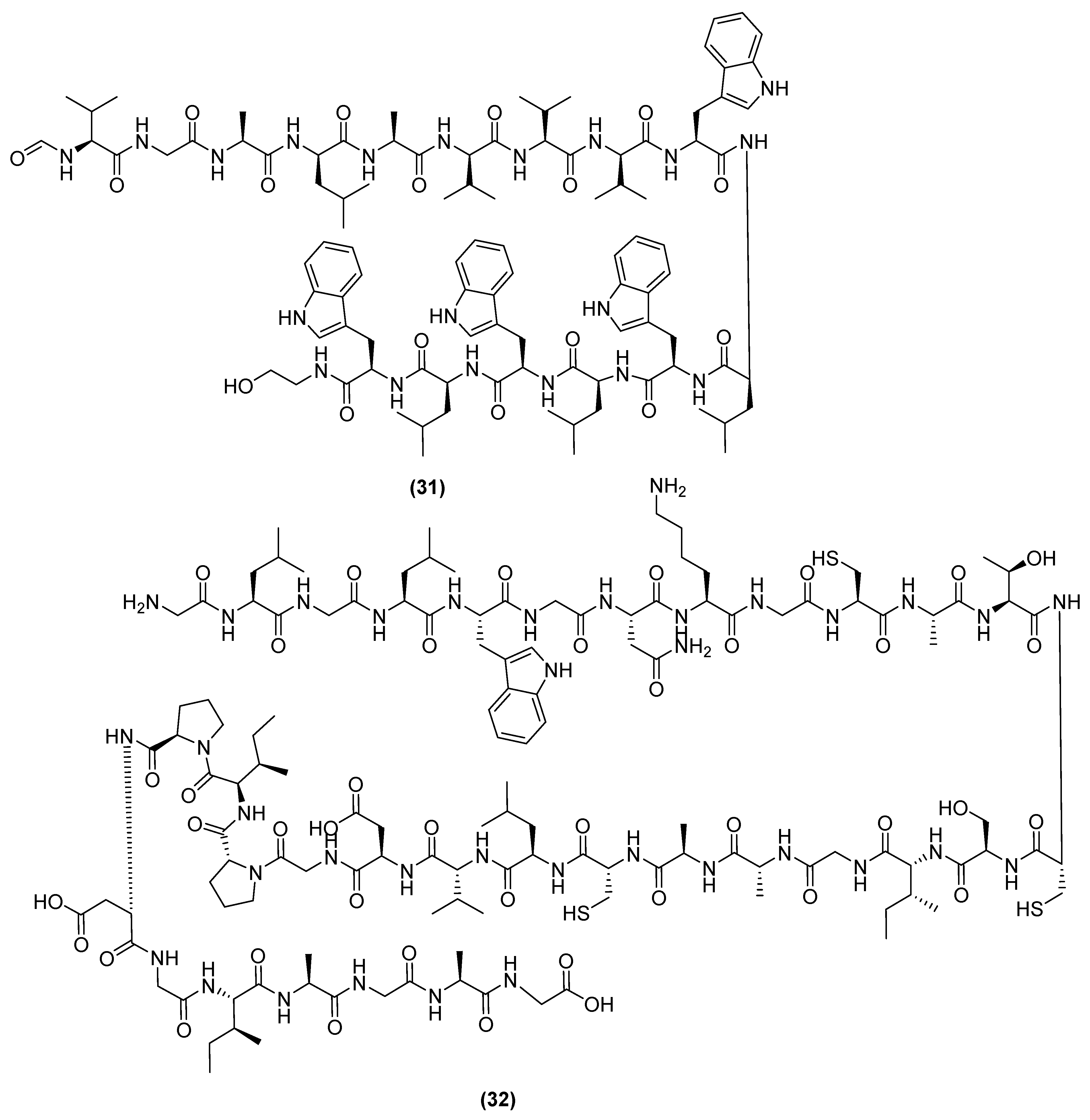
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics 2022, 11, 88. https://doi.org/10.3390/antibiotics11010088
Tran C, Cock IE, Chen X, Feng Y. Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics. 2022; 11(1):88. https://doi.org/10.3390/antibiotics11010088
Chicago/Turabian StyleTran, Charlie, Ian E. Cock, Xiaojing Chen, and Yunjiang Feng. 2022. "Antimicrobial Bacillus: Metabolites and Their Mode of Action" Antibiotics 11, no. 1: 88. https://doi.org/10.3390/antibiotics11010088
APA StyleTran, C., Cock, I. E., Chen, X., & Feng, Y. (2022). Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics, 11(1), 88. https://doi.org/10.3390/antibiotics11010088






