What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance?
Abstract
1. Introduction
2. Efflux Pump-Mediated Multi-Drug Resistance in Bacteria
2.1. Underlying Biochemical Basis of Bacterial Resistance: An Overview
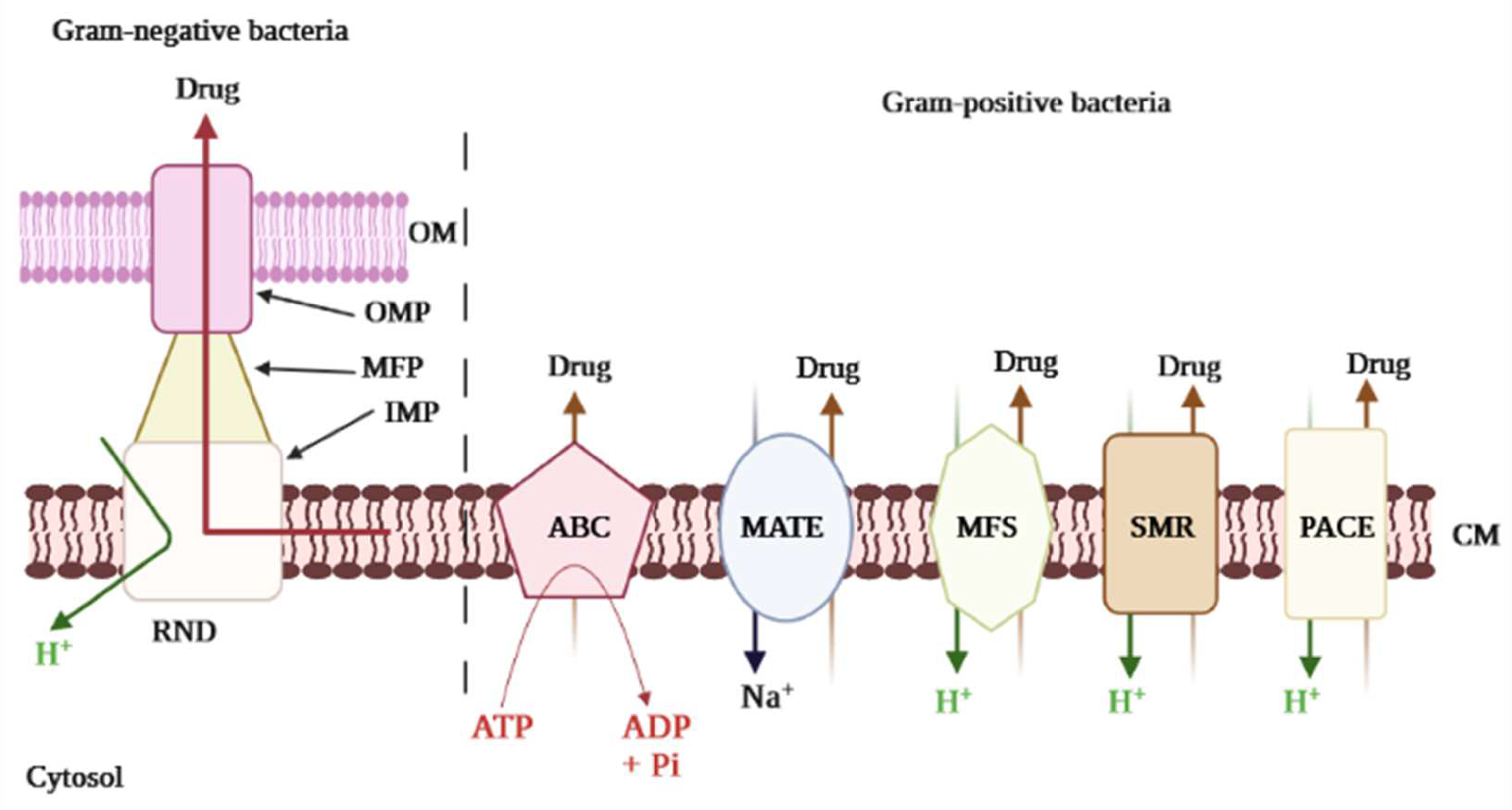
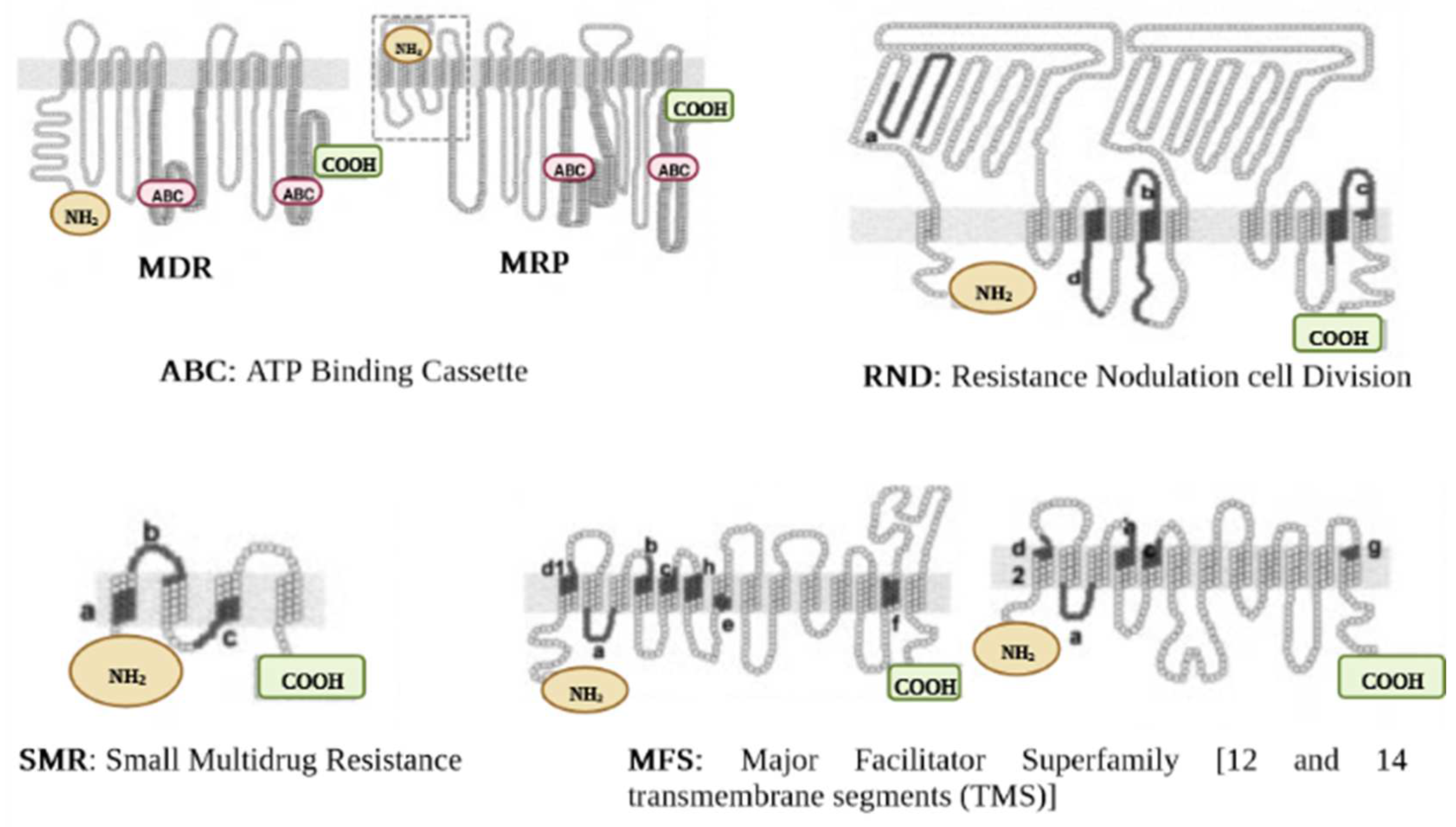
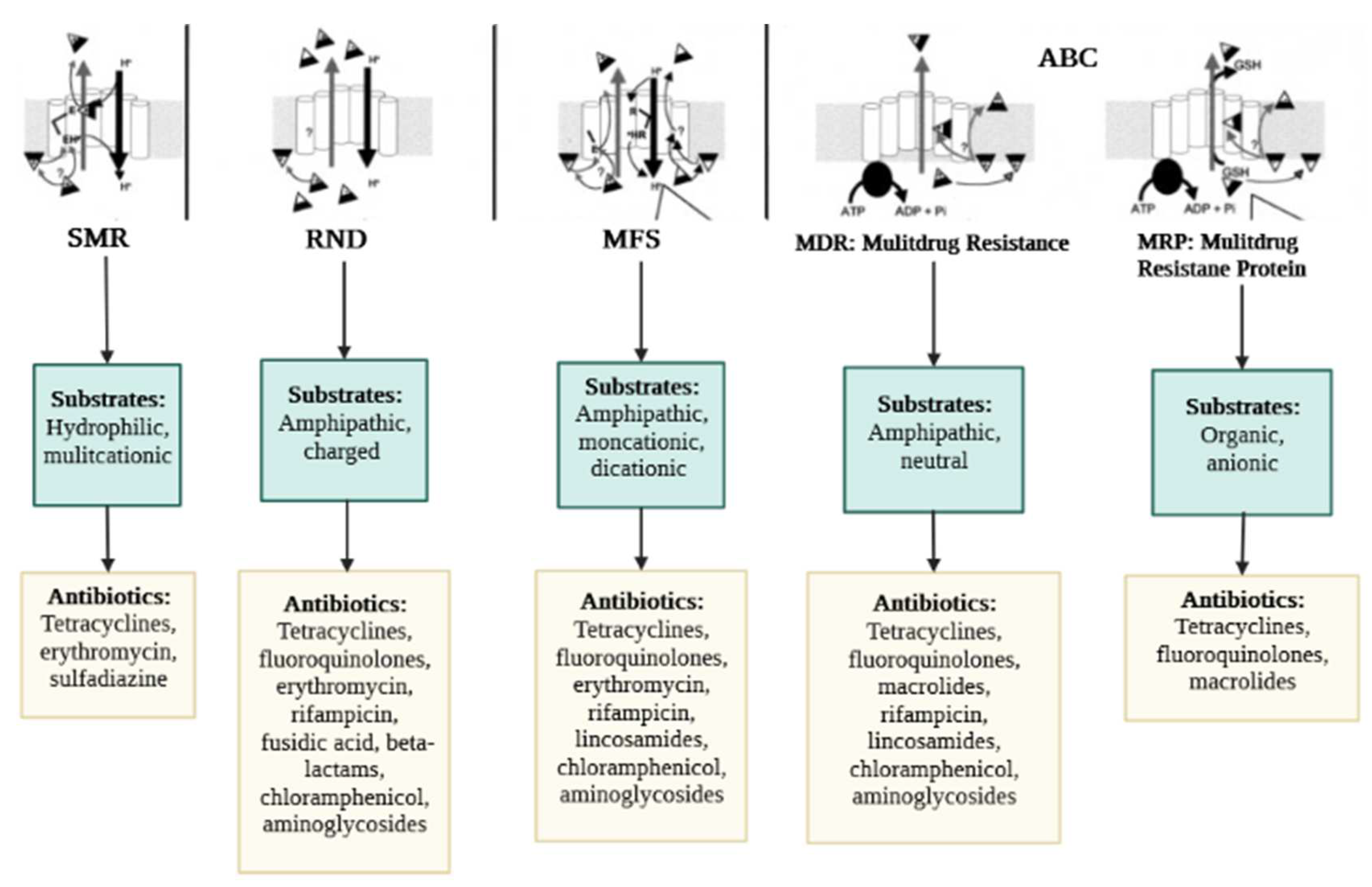
2.2. Efflux Pumps Superfamilies and Their Role in Imparting Multidrug Resistance in Bacteria
2.2.1. ABC Superfamily
2.2.2. MFS Superfamily
2.2.3. MATE Superfamily
2.2.4. SMR Superfamily
2.2.5. RND Superfamily
2.2.6. PACE Superfamily
2.3. Expression of Efflux Pump Gene, Detection of Antimicrobial Resistance, and Clinical Therapy
3. Strategies to Thwart Efflux Pump-Mediated Bacterial Resistance
3.1. Antibiotic Resistance Breakers to Stop Active Efflux in Bacteria: The Efflux Pump Inhibitors and Membrane Permeabilizers
3.1.1. Efflux Pump Inhibitors
3.1.2. Membrane Permeabilizers
3.2. Nanoparticle’s Carriers
3.3. Biologics
3.4. Bacteriophage Therapy
3.5. Bacteriocins
3.6. De Novo Strategies
4. Obstacles in the Development of AMR Blockers: The Case of Efflux Pump Inhibitors
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AA | Amino Acid |
| ABC | ATP Binding Cassette |
| AMP | Antimicrobial Peptide |
| AMR | Antimicrobial Resistance |
| ARB | Antibiotic Resistance Breaker |
| CCCP | Carbonyl Cyanide m-Chlorophenylhydrazine |
| DS | Dynamic Simulation |
| EP | Efflux Pump |
| EPI | Efflux Pump Inhibitor |
| GNB | Gram-Negative Bacteria |
| GPB | Gram-Positive Bacteria |
| HTS | High-Throughput Screening |
| MATE | Multidrug and Toxin Extrusion |
| MD | Molecular Dynamics |
| MDR | Multidrug Resistance |
| MFP | Membrane Fusion Protein |
| MFS | Major Facilitator Superfamily |
| MIC | Minimal Inhibitory Concentration |
| OM | Outer Membrane |
| OMP | Outer Membrane channel Protein |
| PACE | Proteobacterial Antimicrobial Compound Efflux |
| PCR | Polymerase Chain Reaction |
| PPI | Proton Pump Inhibitor |
| PT | Phage Therapy |
| QMD | Quantum Molecular Dynamics |
| RiPP | Ribosomally generated and Post-translationally modified Peptides |
| RND | Resistance Nodulation Cell Division |
| ROS | Reactive Oxygen Species |
| SAR | Structure-Activity Relationship |
| SMR | Small Multidrug Resistance |
| WHO | World Health Organization |
References
- O’Neill, J. Review on Antimicrobial Resistance: Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- World Health Organization. Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Genève, Switzerland, 2022. [Google Scholar]
- Sobierajski, T.; Mazińska, B.; Chajęcka-Wierzchowska, W.; Śmiałek, M.; Hryniewicz, W. Antimicrobial and Antibiotic Resistance from the Perspective of Polish Veterinary Students: An Inter-University Study. Antibiotics 2022, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, W.E.; Yang, Q. Clinical perspective of antimicrobial resistance in bacteria. Infect. Drug Resist. 2022, 15, 735–746. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 30 July 2022).
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Henderson, P.J. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 2015, 6, e01982-14. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, Y.; Nikaido, H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 2009, 6, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- Schindler, B.D.; Kaatz, G.W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updates 2016, 27, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Piddock, L.J. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, R.; Bhattacharyya, T.; Bhando, T.; Pathania, R. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother. 2017, 72, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.; Sutton, J.M.; Rahman, K.M. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in Gram-Negative (ESKAPEE) bacteria. Antibiotics 2019, 8, 229. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [PubMed]
- Seukep, A.J.; Dongmo, F.-M.C.; Mbuntcha, G.H.; Chen, G.; Assob, N.J.C.; Tenniswood, M.; Sarker, S.D.; Kuete, V.; Guo, M.-Q. Bacterial drug efflux pump inhibitors from plants. In Antimicrobial Resistance—Underlying Mechanisms and Therapeutic Approaches, 1st ed.; Kumar, V., Shriram, V., Paul, A., Thakur, M., Eds.; Springer: Singapore, 2022; pp. 487–532. [Google Scholar]
- Spengler, G.; Kincses, A.; Gajdács, M.; Amaral, L. New roads leading to old destinations: Efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 2017, 22, 468. [Google Scholar] [CrossRef]
- Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Mech. Antibiot. Resist. 2016, 4, 1–37. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: An overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Blanco, P.; Alcalde-Rico, M.; Corona, F.; Reales-Calderon, J.A.; Sanchez, M.B.; Martinez, J.L. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Updates 2016, 28, 13–27. [Google Scholar] [CrossRef]
- Blair, J.M.; Piddock, L.J. How to measure export via bacterial multidrug resistance efflux pumps. mBio 2016, 7, e00840-16. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martinez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef]
- Krishnamoorthy, G.; Weeks, J.W.; Zhang, Z.; Chandler, C.E.; Xue, H.; Schweizer, H.P.; Ernst, R.K.; Zgurskaya, H.I. Efflux pumps of Burkholderia thailandensis control the permeability barrier of the outer membrane. Antimicrob. Agents Chemother. 2019, 63, e00956-19. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A.; et al. Pacing across the membrane: The novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 2018, 69, 450–454. [Google Scholar] [CrossRef]
- Verchere, A.; Broutin, I.; Picard, M. Photo-induced proton gradients for the in vitro investigation of bacterial efflux pumps. Sci. Rep. 2012, 2, 306. [Google Scholar] [CrossRef]
- Neuberger, A.; Du, D.; Luisi, B.F. Structure and mechanism of bacterial tripartite efflux pumps. Res. Microbiol. 2018, 169, 401–413. [Google Scholar] [CrossRef]
- Hellmich, U.A.; Monkemeyer, L.; Velamakanni, S.; van Veen, H.W.; Glaubitz, C. Effects of nucleotide binding to LmrA: A combined MAS-NMR and solution NMR study. Biochim. Biophys. Acta 2015, 1848, 3158–3165. [Google Scholar] [CrossRef]
- Lerma, L.L.; Benomar, N.; Valenzuela, A.S.; Mdel, C.C.M.; Galvez, A.; Abriouel, H. Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 2014, 44, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Hellmich, U.A.; Lyubenova, S.; Kaltenborn, E.; Doshi, R.; van Veen, H.W.; Prisner, T.F.; Glaubitz, C. Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 2012, 134, 5857–5862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tatsuno, I.; Okada, R.; Hata, N.; Matsumoto, M.; Isaka, M.; Isobe, K.; Hasegawa, T. Predominant role of msr(D) over mef(A) in macrolide resistance in Streptococcus pyogenes. Microbiology 2016, 162, 46–52. [Google Scholar] [CrossRef]
- Tatsuno, I.; Isaka, M.; Masuno, K.; Hata, N.; Matsumoto, M.; Hasegawa, T. Functional Predominance of msr(D), Which Is More Effective as mef(A)-Associated Than mef(E)-Associated, Over mef(A)/mef(E) in Macrolide Resistance in Streptococcus pyogenes. Microb. Drug Resist. 2018, 24, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Baylay, A.J.; Piddock, L.J. Clinically relevant fluoroquinolone resistance due to constitutive overexpression of the PatAB ABC transporter in Streptococcus pneumoniae is conferred by disruption of a transcriptional attenuator. J. Antimicrob. Chemother. 2015, 70, 670–679. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Llabrés, S.; Neuberger, A.; Blaza, J.N.; Bai, X.C.; Okada, U.; Murakami, S.; van Veen, H.W.; Zachariae, U.; Scheres, S.H.W.; et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017, 2, 17070. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zgurskaya, H.I. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J. Bacteriol. 2013, 195, 4865–4872. [Google Scholar] [CrossRef]
- Shirshikova, T.V.; Sierra-Bakhshi, C.G.; Kamaletdinova, L.K.; Matrosova, L.E.; Khabipova, N.N.; Evtugyn, V.G.; Khilyas, I.V.; Danilova, I.V.; Mardanova, A.M.; Sharipova, M.R.; et al. The ABC-Type Efflux Pump MacAB Is Involved in Protection of Serratia marcescens against Aminoglycoside Antibiotics, Polymyxins, and Oxidative Stress. mSphere 2021, 6, e00033-21. [Google Scholar] [CrossRef]
- Shi, K.; Cao, M.; Li, C.; Huang, J.; Zheng, S.; Wang, G. Efflux proteins MacAB confer resistance to arsenite and penicillin/macrolidetype antibiotics in Agrobacterium tumefaciens 5A. World J. Microbiol. Biotechnol. 2019, 35, 115. [Google Scholar] [CrossRef]
- Du, D.; van Veen, H.W.; Murakami, S.; Pos, K.M.; Luisi, B.F. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr. Opin. Struct. Biol. 2015, 33, 76–91. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, L.; Gao, D.; Wang, Y.; Wang, D.; Zhang, Z.; Chen, M.; Su, Y.; Li, L.; Yan, H.; et al. Expression of efflux pump gene lde in ciprofloxacin-resistant foodborne isolates of Listeria monocytogenes. Microbiol. Immunol. 2012, 56, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Viveiros, M.; Amaral, L.; Couto, I. Multidrug efflux pumps in Staphylococcus aureus: An update. Open Microbiol. J. 2013, 7, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Bley, C.; van der Linden, M.; Reinert, R.R. mef(A) is the predominant macrolide resistance determinant in Streptococcus pneumoniae and Streptococcus pyogenes in Germany. Int. J. Antimicrob. Agents 2011, 37, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Chancey, S.T.; Bai, X.H.; Kumar, N.; Drabek, E.F.; Daugherty, S.C.; Colon, T.; Ott, S.; Sengamalay, N.; Sadzewicz, L.; Tallon, L.J.; et al. Transcriptional attenuation controls macrolide inducible efflux and resistance in Streptococcus pneumoniae and in other Gram-positive bacteria containing mef/mel (msr(D)) elements. PLoS ONE 2015, 10, e0116254. [Google Scholar] [CrossRef]
- Nunez-Samudio, V.; Chesneau, O. Functional interplay between the ATP binding cassette Msr(D) protein and the membrane facilitator superfamily Mef(E) transporter for macrolide resistance in Escherichia coli. Res. Microbiol. 2013, 164, 226–235. [Google Scholar] [CrossRef]
- Pasqua, M.; di Patti, M.C.B.; Fanelli, G.; Utsumi, R.; Eguchi, Y.; Trirocco, R.; Prosseda, G.; Grossi, M.; Colonna, B. Host-Bacterial pathogen communication: The wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 2021, 8, 723274. [Google Scholar] [CrossRef]
- Pérez-Varela, M.; Corral, J.; Aranda, J.; Barbé, J. Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 2019, 63, e02190-18. [Google Scholar] [CrossRef] [PubMed]
- Lu, M. Structures of multidrug and toxic compound extrusion transporters and their mechanistic implications. Channels 2016, 10, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Kusakizako, T.; Miyauchi, H.; Ishitani, R.; Nureki, O. Structural biology of the multidrug and toxic compound extrusion superfamily transporters. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183154. [Google Scholar] [CrossRef] [PubMed]
- Rouquette-Loughlin, C.E.; Dhulipala, V.; Reimche, J.L.; Raterman, E.; Begum, A.A.; Jerse, A.E.; Shafer, W.M. cis- and trans-acting factors influence expression of the norM-encoded efflux pump of Neisseria gonorrhoeae and levels of Gonococcal susceptibility to substrate antimicrobials. Antimicrob. Agents Chemother. 2018, 62, e00821-18. [Google Scholar] [CrossRef]
- Guelfo, J.R.; Rodríguez-Rojas, A.; Matic, I.; Blázquez, J. A MATE-family efflux pump rescues the Escherichia coli 8-oxoguaninerepair-deficient mutator phenotype and protects against H2O2 killing. PLoS Genet. 2010, 6, e1000931. [Google Scholar] [CrossRef] [PubMed]
- Tocci, N.; Iannelli, F.; Bidossi, A.; Ciusa, M.L.; Decorosi, F.; Viti, C.; Pozzi, G.; Ricci, S.; Oggioni, M.R. Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter with quinolone susceptibility. Antimicrob. Agents Chemother. 2013, 57, 248–253. [Google Scholar] [CrossRef]
- Bay, D.C.; Rommens, K.L.; Turner, R.J. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim. Biophys. Acta 2008, 1778, 1814–1838. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Tu, C.C.; Lan, C.Y. Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infect. 2017, 50, 224–231. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef]
- Padariya, M.; Kalathiya, U.; Baginski, M. Structural and dynamic insights on the EmrE protein with TPP+ and related substrates through molecular dynamics simulations. Chem. Phys. Lipids 2017, 212, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jaglic, Z.; Cervinkova, D. Genetic basis of resistance to quaternary ammonium compounds—The qac genes and their role: A review. Vet. Med. 2012, 57, 275–281. [Google Scholar] [CrossRef]
- Bay, D.C.; Turner, R.J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 2009, 9, 14. [Google Scholar] [CrossRef]
- Buffet-Bataillon, S.; Tattevin, P.; Maillard, J.Y.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol. 2016, 11, 81–92. [Google Scholar] [CrossRef]
- Leus, I.V.; Weeks, J.W.; Bonifay, V.; Smith, L.; Richardson, S.; Zgurskaya, H.I. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 2018, 200, e00049-18. [Google Scholar] [CrossRef] [PubMed]
- Perez-Boto, D.; Acebo, P.; Garcia-Pena, F.J.; Abad, J.C.; Echeita, M.A.; Amblar, M. Isolation of a point mutation associated with altered expression of the CmeABC efflux pump in a multidrug-resistant Campylobacter jejuni population of poultry origin. J. Glob. Antimicrob. Resist. 2015, 3, 115–122. [Google Scholar] [CrossRef]
- Dreier, J.; Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 660. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Jones, R.N.; Farrell, D.J. Evaluation of quinolone resistance-determining region mutations and efflux pump expression in Neisseria meningitidis resistant to fluoroquinolones. Diagn. Microbiol. Infect. Dis. 2012, 72, 263–266. [Google Scholar] [CrossRef]
- Yuan, J.; Xu, X.; Guo, Q.; Zhao, X.; Ye, X.; Guo, Y.; Wang, M. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 2012, 67, 1655–1659. [Google Scholar] [CrossRef]
- Wong, M.H.; Chan, E.W.; Xie, L.; Li, R.; Chen, S. IncHI2 Plasmids Are the Key Vectors Responsible for oqxAB Transmission among Salmonella Species. Antimicrob. Agents Chemother. 2016, 60, 6911–6915. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.H.; Sung, J.Y.; Kwon, K.C.; Koo, S.H. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann. Lab. Med. 2012, 32, 38–43. [Google Scholar] [CrossRef]
- Basler, G.; Thompson, M.; Tullman-Ercek, D.; Keasling, J. A Pseudomonas putida efflux pump acts on short-chain alcohols. Biotechnol. Biofuels 2018, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Maurya, P.; Tiwari, M.; Tiwari, V. In-silico interaction studies suggest RND efflux pump mediates polymyxin resistance in Acinetobacter baumannii. J. Biomol. Struct. Dyn. 2017, 37, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Bavro, V.N.; Ricci, V.; Modi, N.; Cacciotto, P.; Kleinekathfer, U.; Ruggerone, P.; Vargiu, A.V.; Baylay, A.J.; Smith, H.E.; et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. USA 2015, 112, 3511–3516. [Google Scholar] [CrossRef] [PubMed]
- Manjasetty, B.A.; Halavaty, A.S.; Luan, C.H.; Osipiuk, J.; Mulligan, R.; Kwon, K.; Anderson, W.F.; Joachimiak, A. Loop-to-helix transition in the structure of multidrug regulator AcrR at the entrance of the drug-binding cavity. J. Struct. Biol. 2016, 194, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Routh, M.D.; Su, C.C.; Zhang, Q.; Yu, E.W. Structures of AcrR and CmeR: Insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. Biochim. Biophys. Acta 2009, 1794, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, D.; Zhou, W.; Sang, H.; Liu, X.; Ge, Z.; Zhang, J.; Lan, L.; Yang, C.G.; Chen, H. Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa. Mol. Microbiol. 2016, 100, 749–758. [Google Scholar] [CrossRef]
- Fernandez-Escamilla, A.M.; Fernandez-Ballester, G.; Morel, B.; Casares-Atienza, S.; Ramos, J.L. Molecular Binding Mechanism of TtgR Repressor to Antibiotics and Antimicrobials. PLoS ONE 2015, 10, e0138469. [Google Scholar]
- Hernandez, A.; Mate, M.J.; Sanchez-Diaz, P.C.; Romero, A.; Rojo, F.; Martinez, J.L. Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J. Biol. Chem. 2009, 284, 14428–14438. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.J.; Shafer, W.M. The Transcriptional Repressor, MtrR, of the mtrCDE Efflux Pump Operon of Neisseria gonorrhoeae Can Also Serve as an Activator of “off Target” Gene (glnE) Expression. Antibiotics 2015, 4, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Anandapadamanaban, M.; Pilstal, R.; Andresen, C.; Trewhella, J.; Moche, M.; Wallner, B.; Sunnerhagen, M. Mutation-Induced Population Shift in the MexR Conformational Ensemble Disengages DNA Binding: A Novel Mechanism for MarR Family Derepression. Structure 2016, 24, 1311–1321. [Google Scholar] [CrossRef]
- Xu, S.; Chen, G.; Liu, Z.; Xu, D.; Wu, Z.; Li, Z.; Hong, M. Site-Directed Mutagenesis Reveals Crucial Residues in Escherichia coli Resistance-Nodulation-Division Efflux Pump OqxB. Microb. Drug Resist. 2020, 26, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Zhao, M.; Wang, W.; Wang, Q.; Huang, M.; Li, C.; Lian, Q.; Xia, J.; Qi, J.; Xiang, C.; et al. Changing Gly311 to an acidic amino acid in the MATE family protein DTX6 enhances Arabidopsis resistance to the dihydropyridine herbicides. Mol. Plant 2021, 14, 2115–2125. [Google Scholar] [CrossRef]
- Delmar, J.A.; Su, C.C.; Yu, E.W. Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 2014, 43, 93–117. [Google Scholar] [CrossRef]
- Long, F.; Su, C.C.; Lei, H.T.; Bolla, J.R.; Do, S.V.; Yu, E.W. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos. Trans. R Soc. Lond. B Biol. Sci. 2012, 367, 1047–1058. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef]
- Zwama, M.; Nishino, K. Ever-Adapting RND Efflux Pumps in Gram-Negative Multidrug-Resistant Pathogens: A Race against Time. Antibiotics 2021, 10, 774. [Google Scholar] [CrossRef]
- Bolla, J.R.; Howes, A.C.; Fiorentino, F.; Robinson, C.V. Assembly and regulation of the chlorhexidine-specific efflux pump AceI. Proc. Natl. Acad. Sci. USA 2020, 117, 17011–17018. [Google Scholar] [CrossRef]
- Hassan, K.A.; Elbourne, L.D.; Li, L.; Gamage, H.K.; Liu, Q.; Jackson, S.M.; Sharples, D.; Kolsto, A.B.; Henderson, P.J.; Paulsen, I.T. An ace up their sleeve: A transcriptomic approach exposes the AceI efflux protein of Acinetobacter baumannii and reveals the drug efflux potential hidden in many microbial pathogens. Front. Microbiol. 2015, 6, 333. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plesiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Murugan, N.; Malathi, J.; Therese, K.L.; Madhavan, H.N. Application of six multiplex PCR’s among 200 clinical isolates of Pseudomonas aeruginosa for the detection of 20 drug resistance encoding genes. Kaohsiung J. Med. Sci. 2018, 34, 79–88. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; He, X.; Ding, F.; Wu, W.; Luo, Y.; Fan, B.; Cao, H. Overproduction of efflux pumps caused reduced susceptibility to carbapenem under consecutive imipenem-selected stress in Acinetobacter baumannii. Infect. Drug Resist. 2017, 11, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Gerson, S.; Nowak, J.; Zander, E.; Ertel, J.; Wen, Y.; Krut, O.; Seifert, H.; Higgins, P.G. Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2018, 73, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K.; Nikaido, H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. USA 2009, 106, 5854–5858. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, Z.W.; Zhi, C.P.; Yang, T.; Zhao, J.J.; Chen, X.J.; Zeng, L.; Lv, L.C.; Zeng, Z.L.; Liu, J.H. Impact of plasmid-borne oqxAB on the development of fluoroquinolone resistance and bacterial fitness in Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.J.; Hu, H.Y.; Wu, Y.H.; Wei, B.; Lu, Y. Effect of chlorination and ultraviolet disinfection on tetA-mediated tetracycline resistance of Escherichia coli. Chemosphere 2013, 90, 2247–2253. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Ruggerone, P.; Opperman, T.J.; Nguyen, S.T.; Nikaido, H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014, 58, 6224–6234. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Li, X.Z.; Zhang, L.; Poole, K. Interplay between the MexAMexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000, 45, 433–436. [Google Scholar] [CrossRef]
- Iino, R.; Nishino, K.; Noji, H.; Yamaguchi, A.; Matsumoto, Y. A microfluidic device for simple and rapid evaluation of multidrug efflux pump inhibitors. Front. Microbiol. 2012, 3, 40. [Google Scholar] [CrossRef]
- Dupont, M.; De, E.; Chollet, R.; Chevalier, J.; Pages, J.M. Enterobacter aerogenes OmpX, a cation-selective channel mar- and osmo-regulated. FEBS Lett. 2004, 569, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J.-M.; Masi, M.; Barbe, J. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 2005, 11, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Molnár, J.; Engi, H.; Hohmann, J.; Molnár, P.; Deli, J.; Wesolowska, O.; Michalak, K.; Wang, Q. Reversal of multidrug resistance by natural substances from plants. Curr. Top. Med. Chem. 2010, 10, 1757–1768. [Google Scholar] [PubMed]
- Banoee, M.; Seif, S.; Nazari, Z.E.; Jafari-Fesharaki, P.; Shahverdi, H.R.; Moballegh, A.; Moghaddam, K.M.; Shahverdi, A.R. ZnO nanoparticles enhanced antibacterial activity of ciprofloxacin against Staphylococcus aureus and Escherichia coli. J. Biomed. Mater. Res. B Appl. Biomater. 2010, 93, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Telhig, S.; Said, L.B.; Zirah, S.; Fliss, I.; Rebuffat, S. Bacteriocins to thwart bacterial resistance in Gram-negative bacteria. Front. Microbiol. 2020, 11, 586433. [Google Scholar] [CrossRef]
- Hind, C.K.; Dowson, C.G.; Sutton, J.M.; Jackson, T.; Clifford, M.; Garner, R.C.; Czaplewski, L. Evaluation of a library of FDA-approved drugs for their ability to potentiate antibiotics against multidrug-resistant gram-negative pathogens. Antimicrob. Agents Chemother. 2019, 63, e00769-19. [Google Scholar] [CrossRef]
- Alessio, V. Can We Reverse Antibiotic Resistance? Available online: https://ec.europa.eu/research-and-innovation/en/horizon-magazine/can-we-reverse-antibiotic-resistance (accessed on 13 July 2022).
- Kalan, L.; Wright, G.D. Antibiotic adjuvants: Multicomponent anti-infective strategies. Expert Rev. Mol. Med. 2011, 13, e5. [Google Scholar] [CrossRef] [PubMed]
- Drawz, S.M.; Bonomo, R.A. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gill, E.E.; Franco, O.L.; Hancock, R.E. Antibiotic adjuvants: Diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 2015, 85, 56–78. [Google Scholar] [CrossRef]
- Jamshidi, S.; Sutton, J.M.; Rahman, K.M. An overview of bacterial efflux pumps and computational approaches to study efflux pump inhibitors. Future Med. Chem. 2016, 8, 195–210. [Google Scholar] [CrossRef]
- Laws, M.; Shaaban, A.; Rahman, K.M. Antibiotic resistance breakers: Current approaches and future directions. FEMS Microbiol. Rev. 2019, 43, 490–516. [Google Scholar] [CrossRef]
- Laws, M.; Jamshidi, S.; Nahar, K.; Sutton, M.; Rahman, K.M. Antibiotic Resistance Breakers. UK Patent Application Number: GB1708606.6; PCT Patent Application No. PCT/GB2018/051468, 2017. [Google Scholar]
- Stavri, M.; Piddock, L.J.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef]
- Mahmood, H.Y.; Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr. Med. Chem. 2016, 23, 1062–1081. [Google Scholar] [CrossRef] [PubMed]
- Kaatz, G.W.; Seo, S.M.; Ruble, C.A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1993, 37, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Neyfakh, A.A.; Borsch, C.M.; Kaatz, G.W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993, 37, 128–129. [Google Scholar] [CrossRef]
- Khan, I.A.; Mirza, Z.M.; Kumar, A.; Verma, V.; Qazi, G.N. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 810–812. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, J.Y.; Guo, N.; Sheng, H.; Li, L.; Liang, J.C.; Wang, X.L.; Li, Y.; Liu, M.Y.; Wu, X.P.; et al. Farnesol, a potential efflux pump inhibitor in Mycobacterium smegmatis. Molecules 2010, 15, 7750–7762. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.D.; Shah, S.; Anderson, J.C.; Hara, Y.; Hamilton-Miller, J.M.; Taylor, P.W. Modulation of betalactam resistance in Staphylococcus aureus by catechins and gallates. Int. J. Antimicrob. Agents 2004, 23, 462–467. [Google Scholar] [CrossRef]
- Gibbons, S.; Moser, E.; Kaatz, G.W. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med. 2004, 70, 1240–1242. [Google Scholar] [CrossRef] [PubMed]
- Oluwatuyi, M.; Kaatz, G.W.; Gibbons, S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004, 65, 3249–3254. [Google Scholar] [CrossRef]
- Su, F.; Wang, J. Berberine inhibits the MexXY-OprM efflux pump to reverse imipenem resistance in a clinical carbapenem resistant Pseudomonas aeruginosa isolate in a planktonic state. Exp. Ther. Med. 2018, 15, 467–472. [Google Scholar] [CrossRef]
- Zechini, B.; Versace, I. Inhibitors of multidrug resistant efflux systems in bacteria. Recent Pat. Antiinfect. Drug Discov. 2009, 4, 37–50. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Lorenz, P.; Tawara, J.N.; Zenewicz, L.A.; Lewis, K. Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5′- methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 2000, 97, 1433–1437. [Google Scholar] [CrossRef]
- Roy, S.K.; Kumari, N.; Gupta, S.; Pahwa, S.; Nandanwar, H.; Jachak, S.M. 7-Hydroxy-(E)-3- phenylmethylene-chroman-4-one analogues as efflux pump inhibitors against Mycobacterium smegmatis mc2 155. Eur. J. Med. Chem. 2013, 66, 499–507. [Google Scholar] [CrossRef]
- Lee, M.D.; Galazzo, J.L.; Staley, A.L.; Lee, J.C.; Warren, M.S.; Fuernkranz, H.; Chamberland, S.; Lomovskaya, O.; Miller, G.H. Microbial fermentation derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 2001, 56, 81–85. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]
- Lomovskaya, O. Antibiotics and Gram-Negative Membranes: The Ins and Outs; American Society for Microbiology: Atlanta, GA, USA, 2018. [Google Scholar]
- Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Mapping the dynamic functions and structural features of acrb efflux pump transporter using accelerated molecular dynamics simulations. Sci. Rep. 2018, 8, 10470. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, J.A.; Kern, W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005, 49, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Pannek, S.; Higgins, P.G.; Steinke, P.; Jonas, D.; Akova, M.; Bohnert, J.A.; Seifert, H.; Kern, W.V. Multidrug efflux inhibition in Acinetobacter baumannii: Comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-betanaphthylamide. J. Antimicrob. Chemother. 2006, 57, 970–974. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Steinke, P.; Bohnert, J.A.; Akova, M.; Jonas, D.; Kern, W.V. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 344–348. [Google Scholar] [CrossRef]
- Bina, X.R.; Philippart, J.A.; Bina, J.E. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-betanaphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob. Chemother. 2009, 63, 103–108. [Google Scholar] [CrossRef]
- Aron, Z.; Opperman, T.J. Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol. 2016, 33, 1–6. [Google Scholar] [CrossRef]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 722–733. [Google Scholar] [CrossRef]
- Opperman, T.J. MBX-4191. In A Novel Pyranopyridine RND Efflux Pump Inhibitor; American Society for Microbiology: Atlanta, GA, USA, 2018. [Google Scholar]
- Grossman, T.H.; Shoen, C.M.; Jones, S.M.; Jones, P.L.; Cynamon, M.H.; Locher, C.P. The efflux pump inhibitor timcodar improves the potency of antimycobacterial agents. Antimicrob. Agents Chemother. 2015, 59, 1534–1541. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Brown, D. Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 2015, 14, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Adamson, D.H.; Krikstopaityte, V.; Coote, P.J. Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J. Antimicrob. Chemother. 2015, 70, 2271–2278. [Google Scholar] [CrossRef] [PubMed]
- Kaatz, G.W.; Moudgal, V.V.; Seo, S.M.; Hansen, J.B.; Kristiansen, J.E. Phenylpiperidine selective serotonin reuptake inhibitors interfere with multidrug efflux pump activity in Staphylococcus aureus. Int. J. Antimicrob. Agents 2003, 22, 254–261. [Google Scholar] [CrossRef]
- Nzakizwanayo, J.; Scavone, P.; Jamshidi, S.; Hawthorne, J.A.; Pelling, H.; Dedi, C.; Salvage, J.P.; Hind, C.K.; Guppy, F.K.; Barnes, L.M.; et al. Fluoxetine and thioridazine inhibit efflux and attenuate crystalline biofilm formation by Proteus mirabilis. Sci. Rep. 2017, 7, 12222. [Google Scholar] [CrossRef]
- Aeschlimann, J.R.; Dresser, L.D.; Kaatz, G.W.; Rybak, M.J. Effects of NorA inhibitors on in vitro antibacterial activities and postantibiotic effects of levofloxacin, ciprofloxacin, and norfloxacin in genetically related strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 335–3340. [Google Scholar] [CrossRef] [PubMed]
- Pule, C.M.; Sampson, S.L.; Warren, R.M.; Black, P.A.; van Helden, P.D.; Victor, T.C.; Louw, G.E. Efflux pump inhibitors: Targeting mycobacterial efflux systems to enhance TB therapy. J. Antimicrob. Chemother. 2016, 71, 17–26. [Google Scholar] [CrossRef]
- Chien, J.Y.; Yu, C.J.; Hsueh, P.R. High incidence of fluoroquinolone resistance and effect of efflux pump inhibitors on moxifloxacin resistance among Mycobacterium tuberculosis isolates causing urinary tract infection in Taiwan. Int. J. Antimicrob. Agents 2017, 50, 491–495. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Moudgal, V.V.; Seo, S.M.; Kristiansen, J.E. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Torres, I.M.; Bento, E.B.; Almeida, L.C.; de Sá, L.Z.; Lima, E.M. Preparation, characterization and in vitro antimicrobial activity of liposomal ceftazidime and cefepime against Pseudomonas aeruginosa strains. Braz. J. Microbiol. 2012, 43, 984–992. [Google Scholar] [CrossRef]
- Zabawa, T.P.; Pucci, M.J.; Parr, T.R., Jr.; Lister, T. Treatment of gram-negative bacterial infections by potentiation of antibiotics. Curr. Opin. Microbiol. 2016, 33, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.L.; Powers, J.P.; Pflegerl, K.; Vasil, M.L.; Hancock, R.E.; Hodges, R.S. Effects of single D-amino acid substitutions on disruption of beta-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J. Pept. Res. 2004, 63, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Thamphiwatana, S.; Angsantikul, P.; Zhang, L. Nanoparticle approaches against bacterial infections. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Morrow, T.; Felcone, L.H. Defining the difference: What Makes Biologics Unique. Biotechnol. Healthc. 2004, 1, 24–29. [Google Scholar] [PubMed]
- Mariathasan, S.; Tan, M.W. Antibody-antibiotic conjugates: A novel therapeutic platform against bacterial infections. Trends Mol. Med. 2017, 23, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Morisaki, J.H.; et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Smith, H.W.; Huggins, M.B. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. J. Gen. Microbiol. 1982, 128, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Soothill, J.S. Treatment of experimental infections of mice with bacteriophages. J. Med. Microbiol. 1992, 37, 258–261. [Google Scholar] [CrossRef]
- Bogovazova, G.G.; Voroshilova, N.N.; Bondarenko, V.M. The efficacy of Klebsiella pneumoniae bacteriophage in the therapy of experimental Klebsiella infection. Zh. Mikrobiol. Epidemiol. Immunobiol. 1991, 4, 5–8. [Google Scholar]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Chaliis, G.L.; Clardy, J. Ribosomally synthesized and post-translationally modified peptide natural products, overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.I.; Rahman, R.; van Heel, A.J.; Viel, J.H.; Bandaraian, V.; Dittmann, E.; Genilloud, O.; Goto, Y. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.B.; Worobo, R.W. Chemical and genetic characterization of bacteriocins, antimicrobial peptides for food safety. J. Sci. Food Agric. 2014, 94, 28–44. [Google Scholar] [CrossRef]
- Egan, K.; Ross, R.P.; Hill, C. Bacteriocins, antibiotics in the age of the microbiome. Emerg. Top. Life Sci. 2017, 1, 55–63. [Google Scholar] [PubMed]
- Rebuffat, S. Microcins in action, amazing defence strategies of Enterobacteria. Biochem. Soc. Trans. 2012, 40, 1456–1462. [Google Scholar] [CrossRef]
- Baquero, F.; Lanza, V.F.; Baquero, M.R.; del Campo, R.; Bravo-Vazquez, D.A. Microcins in Enterobacteriaceae, Peptide antimicrobials in the eco-active intestinal chemosphere. Front. Microbiol. 2019, 10, 2261. [Google Scholar] [CrossRef]
- Li, Y.; Rebuffat, S. The manifold roles of microbial ribosomal peptidebased natural products in physiology and ecology. J. Biol. Chem. 2020, 295, 34–54. [Google Scholar]
- Adelman, K.; Yuzenkova, J.; la Porta, A.; Zenkin, N.; Lee, J.; Lis, J.T.; Borukhov, S.; Wang, M.D.; Severinov, K. Molecular mechanism of transcription inhibition by peptide antibiotic Microcin J25. Mol. Cell. 2004, 14, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Metlitskaya, A.; Kazakov, T.; Kommer, A.; Pavlova, O.; Praetorius-Ibba, M.; Ibba, M.; Krasheninnikov, I.; Kolb, V.; Khmel, I.; Severinov, K. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J. Biol. Chem. 2006, 281, 18033–18042. [Google Scholar] [CrossRef] [PubMed]
- Vizán, J.L.; Hernández-Chico, C.; del Castillo, I.; Moreno, F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli. DNA gyrase. EMBO J. 1991, 10, 467–476. [Google Scholar] [PubMed]
- Bieler, S.; Estrada, L.; Lagos, R.; Baeza, M.; Castilla, J.; Soto, C. Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 2005, 280, 26880–26885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Eberhart, L.J.; Orfe, L.H.; Lu, S.Y.; Besser, T.E.; Call, D.R. Genome-wide screening identifies six genes that are associated with susceptibility to Escherichia coli microcin PDI. Appl. Environ. Microbiol. 2015, 81, 6953–6963. [Google Scholar] [CrossRef]
- Palmer, J.D.; Mortzfeld, B.M.; Piattelli, E.; Silby, M.W.; McCormick, B.A.; Bucci, V. Microcin H47, A Class IIb microcin with potent activity against multidrug resistant Enterobacteriaceae. ACS Infect. Dis. 2020, 6, 672–679. [Google Scholar] [CrossRef]
- Lu, S.Y.; Graça, T.; Avillan, J.J.; Zhao, Z.; Call, D.R. Microcin PDI inhibits antibiotic-resistant strains of Escherichia coli and Shigella through a mechanism of membrane disruption and protection by homotrimer selfimmunity. Appl. Environ. Microbiol. 2019, 85, e0371-19. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.K.; Cacciotto, P.; Malloci, G.; Vargiu, A.V.; Ruggerone, P. Computational modelling of efflux pumps and their inhibitors. Essays Biochem. 2017, 61, 141–156. [Google Scholar] [PubMed]
- Chaskar, P.; Zoete, V.; Rohrig, U.F. On-the-Fly QM/MM Docking with Attracting Cavities. J. Chem. Inf. Model. 2017, 57, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Preskill, J. Quantum Computing in the NISQ era and beyond. Quantum 2018, 2, 79. [Google Scholar] [CrossRef]
- Marquez, B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie 2005, 87, 1137–1147. [Google Scholar] [CrossRef]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 12, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Opperman, T.J.; Nguyen, S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015, 6, 421. [Google Scholar] [CrossRef]
- Pfeifer, H.J.; Greenblatt, D.K.; Koch-Wester, J. Clinical toxicity of reserpine in hospitalized patients: A report from the Boston Collaborative Drug Surveillance Program. Am. J. Med. Sci. 1976, 271, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Corbett, D.; Wise, A.; Langley, T.; Skinner, K.; Trimby, E.; Birchall, S.; Dorali, A.; Sandiford, S.; Williams, J.; Warn, P.; et al. Potentiation of antibiotic activity by a novel cationic peptide: Potency and spectrum of activity of SPR741. Antimicrob. Agents Chemother. 2017, 61, e00200-17. [Google Scholar] [CrossRef] [PubMed]
- Zurawski, D.V.; Reinhart, A.A.; Alamneh, Y.A.; Pucci, M.J.; Si, Y.; Abu-Taleb, R.; Shearer, J.P.; Demons, S.T.; Tyner, S.D.; Lister, T. SPR741, an antibiotic adjuvant, potentiates the in vitro and in vivo activity of rifampin against clinically relevant extensively drug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017, 61, e01239-17. [Google Scholar] [CrossRef]

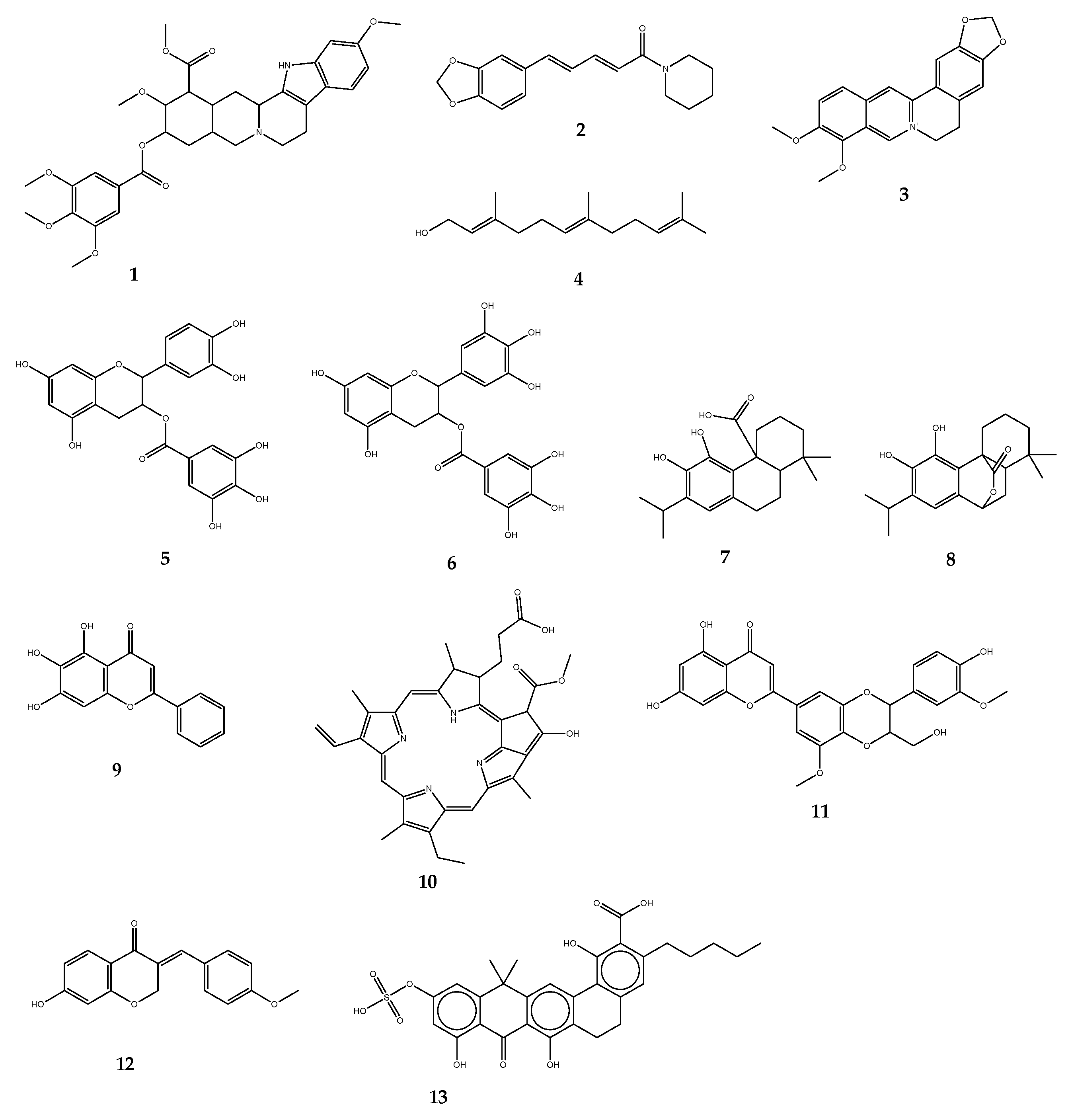
| Efflux Pump Inhibitors | Origin | Target Pumps (Bacteria) | Antibiotic Substrates | References |
|---|---|---|---|---|
| Reserpine | Rauvolfia sp. | NorA, Bmr, TetK, LmrA, PmrA, MepA (B. subtilis, S. aureus, S. pneumoniae, Lactococcus lactis) | Norfloxacin, ciprofloxacin, tetracycline | [115,116] |
| Piperine | Piper sp. | NorA, MdeA, Rv1258c (E. coli, S. aureus, Mycobacterium spp.) | Ciprofloxacin, norfloxacin | [117] |
| Berberine | Berberis sp. | MexAB-OprM, NorA (P. aeruginosa, S. aureus) | Imipenem | [122] |
| Epicatechin gallate | Camellia sinensis | NorA, TetK (S. aureus, S. epidermidis) | Oxacillin, norfloxacin | [119,120] |
| Epigallocatechin gallate | Camellia sinensis | NorA, TetK (S. aureus, S. epidermidis) | Oxacillin, norfloxacin | [119,120] |
| Carnosic acid | Rosmarinus officinalis | NorA, MsrA (S. aureus) | Erythromycin, tetracycline | [121] |
| Carnosol | Rosmarinus officinalis | NorA, MsrA (S. aureus) | Erythromycin, tetracycline | [121] |
| Baicalein | Thymus vulgaris | NorA, TetK (S. aureus, Salmonella enteridis, E. coli) | Tetracycline, ampicillin, oxacillin, ciprofloxacin | [18] |
| Porphyrin pheophorbide A | Berberis sp. | NorA (S. aureus) | Berberine | [123] |
| 5′-methoxyhydnocarpin | Berberis sp. | NorA (S. aureus) | Norfloxacin | [124] |
| EA-371α | Streptomyces | MexAB-OprM (P. aeruginosa) | Levofloxacin | [126] |
| EA-371δ | Streptomyces | MexAB-OprM (P. aeruginosa) | Levofloxacin | [126] |
| PAβN | Synthetic | MexAB-OprM (P. aeruginosa) | Levofloxacin | [127] |
| 1-(1-Naphthylmethyl)-piperazine | Synthetic | AcrAB, AcrEF (E. coli) | Levofloxacin | [130,131,132,133] |
| D13-9001 | Synthetic | AcrAB-TolC (E. coli), MexAB-OprM (P. aeruginosa) | Wide variety | [114] |
| MBX-2319 | Synthetic | AcrAB (E. coli) | ciprofloxacin, levofloxacin, piperacillin | [93,134,135] |
| Trimethoprim, sertraline | Drug repurposing | Efflux systems of P. aeruginosa | piperacillin, levofloxacin, meropenem | [140] |
| Paroxetine, fluoxetine | Drug repurposing | NorA, Bcr/CflA (S. aureus, Proteus mirabilis) | Norfloxacin, ethidium bromide | [141,142] |
| Omeprazole, lansoprazole | Drug repurposing | NorA (S. aureus) | Ciprofloxacin, norfloxacin | [142] |
| Verapamil | Drug repurposing | ABC (M. tuberculosis) | Bedaquiline | [144,145] |
| Chlorpromazine, prochlorperazine | Drug repurposing | NorA (S. aureus) | - | [146] |
| Biricodar, timcodar | Synthetic/Drug repurposing | EtBr efflux (S. aureus, E. faecalis, S. pneumoniae) | Ciprofloxacin, tetracycline, gentamicin, rifampicin, moxifloxacin, bedaquiline | [137] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seukep, A.J.; Mbuntcha, H.G.; Kuete, V.; Chu, Y.; Fan, E.; Guo, M.-Q. What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance? Antibiotics 2022, 11, 1287. https://doi.org/10.3390/antibiotics11101287
Seukep AJ, Mbuntcha HG, Kuete V, Chu Y, Fan E, Guo M-Q. What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance? Antibiotics. 2022; 11(10):1287. https://doi.org/10.3390/antibiotics11101287
Chicago/Turabian StyleSeukep, Armel Jackson, Helene Gueaba Mbuntcha, Victor Kuete, Yindi Chu, Enguo Fan, and Ming-Quan Guo. 2022. "What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance?" Antibiotics 11, no. 10: 1287. https://doi.org/10.3390/antibiotics11101287
APA StyleSeukep, A. J., Mbuntcha, H. G., Kuete, V., Chu, Y., Fan, E., & Guo, M.-Q. (2022). What Approaches to Thwart Bacterial Efflux Pumps-Mediated Resistance? Antibiotics, 11(10), 1287. https://doi.org/10.3390/antibiotics11101287





