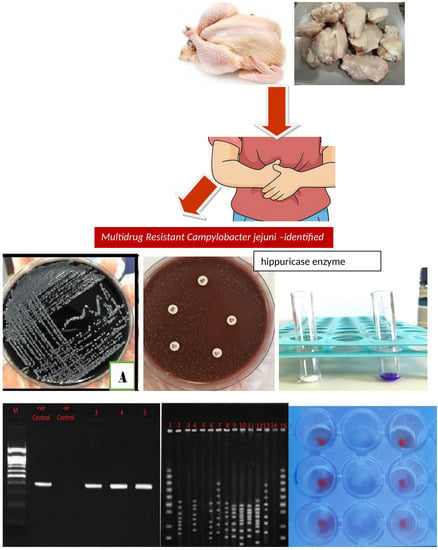
Genetic Signature and Serocompatibility Evidence for Drug Resistant Campylobacter jejuni
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Raw Milk Samples
2.3. Chicken Samples
2.4. Clinical Human Stool Samples
2.5. Vegetable Samples
2.6. Traditional Identification of Campylobacter
2.7. Molecular Identification of Campylobacter
2.8. Antimicrobial Susceptibility Test
2.9. Serotyping using Heat-Stable (HS) Lipopolysaccharide Antiserum
2.10. Pulsed-Field Gel Electrophoresis (PFGE)
2.11. Statistical Analysis
3. Results
3.1. Traditional Techniques for Identification of Campylobacter Species
3.2. Molecular Identification of Campylobacter
3.3. Antibiotic Susceptibility Using the Disc Diffusion Method
3.4. Serotyping Using HS-LPS Antiserum
3.5. Pulsed-Field-Gel-Electrophoresis (PFGE)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahrizaila, N.; Yuki, N. Guillain-barré syndrome animal model: The first proof of molecular mimicry in human autoimmune disorder. J. Biomed. Biotechnol. 2010, 2011, 829129. [Google Scholar] [CrossRef] [Green Version]
- Esan, O.B.; Pearce, M.; Hecke, O.V.; Roberts, N.; Collins, D.R.J.; Violato, M.; McCarthy, N.; Perera, R.; Fanshawe, T.R. Factors Associated with Sequelae of Campylobacter and Non-typhoidal Salmonella Infections: A Systematic Review. BioMedicine 2016, 15, 100–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igwarana, A.; Okoha, A.I. Human campylobacteriosis. A public health concern of global importance. Heliyon 2019, 5, e02814. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Vos, T.; Lozano, R.; Naghavi, M.; Flaxman, A.D.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e0592. [Google Scholar] [CrossRef] [Green Version]
- USDA Economic Research Center, Total Cost of Foodborne Illness Estimates for 15 Leading Foodborne Pathogens. Available online: https://www.ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses.aspx (accessed on 10 August 2022).
- Center for Disease Control and Prevention (CDC). 2021. Available online: https://www.cdc.gov/campylobacter/index.html (accessed on 1 September 2022).
- US Food and Drug Administration (USFDA), 2019. NARMS Update: Integrated Report Summary. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2019-narms-update-integrated-report-summary (accessed on 15 August 2022).
- Wieczorek, K.; Wołkowicz, I.T.; Osek, J. Antimicrobial Resistance and Virulence-Associated Traits of Campylobacter jejuni Isolated from Poultry Food Chain and Humans with Diarrhea. Front. Microbiol. 2018, 9, 1508. [Google Scholar] [CrossRef] [PubMed]
- Paintsil, E.K.; Ofori, L.A.; Akenten, C.W.; Fosu, D.; Ofori, S.; Lamshöft, M. Antimicrobial Usage in Commercial and Domestic Poultry Farming in Two communities in the Ashanti Region of Ghana. Antibiotics 2021, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Samie, S.; Moropeng, R.C.; Tanih, N.F.; Dillingham, R.; Guerrant, R.; Bessong, P.O. Epidemiology of Campylobacter infections among children of 0–24 months of age in South Africa. Arch. Public Health 2022, 80, 107. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Global View of Campylobacteriosis: Report of an Expert Consultation; WHO: Utrecht, The Netherlands, 2020. [Google Scholar]
- Hussain, I.; Mahmood, M.; Akhtar, M.; Khan, A. Prevalence of Campylobacter species in meat, milk and other food commodities in Pakistan. Food Microbiol. 2007, 24, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, J.A.; Kosek, M. Update on the burden of Campylobacter in developing countries. Curr. Opin. Infect. Dis. 2014, 27, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.; Castaño-Rodríguez, N.; Mitchell, H.; Man, S. Global epidemiology of Campylobacter infection. J. Clin. Microbiol. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habib, I.; Mohamed, M.Y.I.; Khan, M. Current State of Salmonella, Campylobacter and Listeria in the Food Chain across the Arab Countries: A Descriptive Review. Foods 2021, 10, 2369. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Chui, L.; Bae, J.; Li, V.; Ma, A.; Mutschall, S.K. Frequent implication of multistress-tolerant Campylobacter jejuni in human infections. Emerg. Infect. Dis. 2018, 24, 1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plishka, M.; Sargeant, J.M.; Greer, A.L.; Hookey, S.; Winder, C. The Prevalence of Campylobacter in Live Cattle, Turkey, Chicken, and Swine in the United States and Canada: A Systematic Review and Meta-Analysis. Foodborne Pathog. Dis. 2021, 18, 23–242. [Google Scholar] [CrossRef] [PubMed]
- Rasschaert, G.; De Zutter, L.; Herman, L.; Heyndrickx, M. Campylobacter contamination of Broilers: The Role of Transport and Slaughterhouse. Int. J. Food. Microbiol. 2020, 322, 108564. [Google Scholar] [CrossRef] [PubMed]
- Elmi, A.; Nasher, F.; Dorrell, N.; Wren, B.; Gundogd, O. Revisiting Campylobacter jejuni Virulence and Fitness Factors: Role in Sensing, Adapting, and Competing. Front. Cell. Infect. Microbiol. 2021, 10, 607704. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, S.K.; Nishimura, S.; Okitsu, S.; Hayakawa, S.; Mizuguchi, M.; Ushijima, H. Comparison of immune-chromatography, PCR and culture methods for the detection of Campylobacter bacteria. J. Microbiol. Methods 2012, 91, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Bakhshi, B.; Kalantar, M.; Rastegar-Lari, A. PFGE genotyping and molecular characterization of Campylobacter spp. isolated from chicken meat. Iran. J. Vet. Res. 2016, 17, 177–183. [Google Scholar] [PubMed]
- McKay, D.; Fletcher, J.; Cooper, P.; Thomson-Carter, F. Comparison of Two Methods for Serotyping Campylobacter spp. J. Clin. Microbiol. 2001, 39, 1917–1921. [Google Scholar] [CrossRef] [PubMed]
- USFDA Bacteriological Analytical Manual (BAM). 2022. Available online: https://www.fda.gov/food/laboratory-methods-food/bacteriological-analytical-manual-bam (accessed on 1 May 2022).
- Mandal, J.; Salim, S.; Parija, S. Isolation of Campylobacter from human stool samples. Indian J. Med. Microbiol. 2014, 32, 35. [Google Scholar] [CrossRef]
- Tripathi, N.; Sapra, A. Gram Staining; The National Center for Biotechnology Information: Bethesda, MD, USA, 2021. [Google Scholar]
- Tarrand, J.J.; Gröschel, D.H. Rapid, modified oxidase test for oxidase-variable bacterial isolates. J. Clin. Microbiol. 1982, 16, 772–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, W.; Achanzar, D. Catalase Test as an Aid to the Identification of Enterobacteriaceae. Appl. Microbiol. 1972, 24, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Adzitey, F.; Corry, J. A Comparison between Hippurate Hydrolysis and Multiplex PCR for Differentiating Campylobacter coli and Campylobacter jejuni. Trop. Life Sci. Res. 2011, 22, 91–98. [Google Scholar] [PubMed]
- Wideman, P.A.; Citronbaum, D.M.; Sutter, V.L. Simple disk technique for detection of nitrate reduction by anaerobic bacteria. J. Clin. Microbiol. 1977, 5, 315–319. [Google Scholar] [CrossRef]
- Reddy, S.; Zishiri, O.T. Genetic characterization of virulence genes associated with adherence, invasion and cytotoxicity in Campylobacter spp. isolated from commercial chickens and human clinical cases. Onderstepoort J. Vet. Res. 2018, 85, a1507. [Google Scholar] [CrossRef] [Green Version]
- Bissong, M.E.A. Detection of virulent thermophilic Campylobacter species in communal chickens. S. Afr. J. Sci. 2019, 115, 10. [Google Scholar] [CrossRef] [Green Version]
- Bayot, M.L.; Bragg, B.N. Antimicrobial Susceptibility Testing; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK539714/ (accessed on 1 September 2021).
- The European Committee on Antimicrobial Susceptibility Testing. Calibration of Zone Diameter Breakpoints against MIC Values. 2021. Available online: https://www.eucast.org/ast_of_bacteria/calibration_and_validation/ (accessed on 1 September 2021).
- Lee, S.; Park, H.; Lee, J.; Lim, J.; Seo, K.; Heo, E. Distribution and molecular characterization of Campylobacter species at different processing stages in two poultry processing plants. Foodborne Pathog. Dis. 2017, 14, 141–147. [Google Scholar] [CrossRef]
- da Silva, D.; Tejada, T.; Blum-Menezes, D.; Dias, P.; Timm, C. Campylobacter species isolated from poultry and humans, and their analysis using PFGE in southern Brazil. Int. J. Food Microbiol. 2016, 217, 189–194. [Google Scholar] [CrossRef]
- Zhu, J.; Yao, B.; Song, X.; Wang, Y.; Cui, S.; Xu, H. Prevalence and quantification of Campylobacter contamination on raw chicken carcasses for retail sale in China. Food Control. 2017, 75, 196–202. [Google Scholar] [CrossRef]
- Tang, Y.; Jiang, Q.; Tang, H.; Wang, Z.; Yin, Y.; Ren, F. Characterization and prevalence of Campylobacter spp. from broiler chicken rearing period to the slaughtering process in Eastern China. Front. Vet. Sci. 2020, 7, 227. [Google Scholar] [CrossRef] [PubMed]
- Alaboudi, A.; Malkawi, I.; Osaili, T.; Abu-Basha, E.; Guitian, V. Prevalence, antibiotic resistance and genotypes of Campylobacter jejuni and Campylobacter coli isolated from chickens in Irbid governorate, Jordan. Int. J. Food Microbiol. 2020, 327, 108656. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Soncini, G.; Nuvoloni, R.D.; Giannatale, E.; Colavita, G.; Alberghini, L.; Giaccone, V. Prevalence and quantification of thermophilic Campylobacter spp. in Italian retail poultry meat: Analysis of influencing factors. Food Microbiol. 2017, 62, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Nisar, M.; Ahmad, M.; Mushtaq, M.; Shehzad, W.; Hussain, A.; Muhammad, J.; Nagaraja, K.V.; Goyal, S.M. Prevalence and antimicrobial resistance patterns of Campylobacter spp. isolated from retail meat in Lahore, Pakistan. Food Control. 2017, 80, 327. [Google Scholar] [CrossRef]
- Skarp, C.P.A.; Hänninen, M.L.; Rautelin, H.I.K. Campylobacteriosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 103–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damjanova, M.J.; Farkas, T.; Mészáros, J.; Galántai, Z.; Turcsányi, I.; Bistyák, A.; Juhász, A.; Pászti, J.; Kiss, I.; Kardos, G. From farm to fork follow-up of thermotolerant Campylobacters throughout the broiler production chain and in human cases in a Hungarian county during a ten-months period. Int. J. Food Microbiol. 2011, 150, 95–102. [Google Scholar] [CrossRef]
- Verwoerd, D.J. Ostrich diseases. Rev. Sci. Tech. 2000, 19, 638–661. [Google Scholar] [CrossRef]
- Huang, H.; Brooks, B.; Lowman, R.; Carrillo, C. Campylobacter species in animal, food, and environmental sources, and relevant testing programs in Canada. Can. J. Microbiol. 2015, 61, 150728144452000. [Google Scholar] [CrossRef] [Green Version]
- Verhoeff-Bakkenes, L.; Jansen, H.; In’t Veld, P.; Beumer, R. Consumption of raw vegetables and fruits: A risk factor for Campylobacter infections. Int. J. Food Microbiol. 2011, 144, 406–412. [Google Scholar] [CrossRef]
- Obaidat, M.M. Seroprevalence and risk factors for Campylobacter jejuni seropositivity in Jordan. Infect. Dis. 2019, 51, 140–146. [Google Scholar] [CrossRef]
- Mohammad, P.H.; Berizi, E.; Hosseinzadeh, S.; Majlesi, M.; Zare, M. The prevalence of Campylobacter spp. in vegetables, fruits, and fresh produce: A systematic review and meta-analysis. Gut Pathog. 2018, 10, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abadias, M.; Usall, J.; Anguera, M.; Solsona, C.; Viñas, I. Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int. J. Food Microbiol. 2008, 31, 121–129. [Google Scholar] [CrossRef]
- Sibanda, N.; McKenna, A.; Richmond, A.; Ricke, S.C.; Callaway, T.; Stratakos, A.C.H. A Review of the Effect of Management Practices on Campylobacter Prevalence in Poultry Farms. Front. Microbiol. 2018, 9, 2002. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.C.; Targino, B.N.; Mendonça, R.S.; Sant’Ana, A.S.; Hungaro, H.M. Campylobacter: An overview of cases, occurrence in food, contamination sources, and antimicrobial resistance in Brazil. Food Rev. Int. 2018, 34, 364–389. [Google Scholar] [CrossRef]
- Saeed, A.; Abd, H.; Sandstrom, G. Microbial etiology of acute diarrhea in children under five years of age in Khartoum, Sudan. J. Med. Microbiol. 2015, 64, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Hashi, A.; Kumie, A.; Gasana, J. Prevalence of diarrhea and associated factors among under-five children in Jigjiga District, Somali Region, Eastern Ethiopia. Open J. Prev. Med. 2016, 6, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, F.M.; Akram, M.; Noureen, N.; Noreen, Z.; Bokhari, H. Antibiotic susceptibility profiling and virulence potential of Campylobacter jejuni isolates from different sources in Pakistan. Asian Pac. J. Trop. Med. 2015, 8, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Paintsil, E.K.; Ofori, L.A.; Adobea, S.; Akenten, C.W.; Phillips, R.O.; Maiga-Ascofare, O. Prevalence and Antibiotic Resistance in Campylobacter spp. Isolated from Humans and Food-Producing Animals in West Africa: A Systematic Review and Meta-Analysis. Pathogens 2022, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Megraud, F.; Boudraa, G.; Bessaoud, K.; Bensid, S.; Dabis, F.; Soltana, R.; Touhami, M. Incidence of Campylobacter infection in infants in western Algeria and the possible protective role of breast feeding. Epidemiol. Infect. 1990, 105, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Ziprin, R.L.; Young, C.R.; Byrd, J.A.; Stanker, L.H.; Hume, M.E.; Gray, S. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 2001, 45, 549–557. [Google Scholar] [CrossRef]
- Talukder, K.A.; Aslam, M.; Islam, Z.; Azmi, I.J.; Dutta, D.K.; Hossain, S.; Nur-E-Kamal, A.; Nair, G.B.; Cravioto, A.; Sack, D.A.; et al. Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 2008, 46, 1485–1488. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, D.; da SilvaSergio, M.; AzevedoRosa, M.; Elide, P.; Scarcelli, G. Campylobacter spp. strains isolated from broiler carcasses and vegetables in São Paulo, Brazil. Braz. J. Microbiol. 2013, 44, 693–699. [Google Scholar] [CrossRef] [Green Version]
- Biswas, D.; Hannon, S.; Townsend, H.; Potter, A.; Allan, B.J. Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry. Int. Microbiol. 2011, 14, 25–32. [Google Scholar] [CrossRef]
- Wieczorek, K.; Szewczyk, R.; Osek, J. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter jejuni and C. coli isolated from retail raw meat in Poland. Vet. Med. 2012, 57, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Meloa, R.; Grazziotina, A.; Júniora, E.; Prado, R.; Eliane, M.; Peres, P.; Rossia, D. Evolution of Campylobacter jejuni of poultry origin in Brazil. Food Microbiol. 2019, 82, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Niwa, H.; Itoh, K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003, 52, 345–348. [Google Scholar] [CrossRef] [Green Version]
- Hawari, A.D.; Obeidat, M.; Awaisheh, S.; Al-Daghistani, H.I.; AlAbbadi, A.A.; Omar, A.S.; Qrunfleh, I.M.; Al-Dmoor, H.M.; El-Qudah, J. Prevalence of mastitis pathogens and their resistance against antimicrobial agents in awassi sheep in Al-Balqa province of Jordan. Am. J. Anim. Vet. Sci. 2014, 9, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Taremi, M.; Mehdisoltandallal, M.; Gachkar, L.; Moezardalan, S.; Zolfagharian, K.; Rezazali, M. Prevalence and antimicrobial resistance of Campylobacter isolated from retail raw chicken and beef meat, Tehran, Iran. Int. J. Food Microbiol. 2006, 108, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jang, S.; Choo, E.; Heu, S.; Ryu, S. Prevalence, genetic diversity, and antibiotic resistance patterns of Campylobacter jejuni from retail raw chickens in Korea. Int. J. Food Microbiol. 2007, 114, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Fraqueza, M.; Martins, A.; Borges, A.; Fernandes, M.; Fernandes, M.; Vaz, Y.; Bessa, R.J.B.; Barreto, A.S. Antimicrobial resistance among Campylobacter spp. strains isolated from different poultry production systems at slaughterhouse level. Poult. Sci. 2014, 93, 1578–1586. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial Resistance Mechanisms among Campylobacter. BioMed Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef] [Green Version]
- Hasan, A.A.; Hassawi, D.S.; Al-Daghistani, H.I.; Hawari, A.D. Molecular and Biochemical Identification of Coagulase Positive Staphylococcus Species Isolated from Human and Animal Sources in Jordan. Int. J. Med. Med. Sci. 2014, 47, 1491–1507. [Google Scholar]
- Mohamed, M.; AbdulAziz, S.; Abu, J.; Bejo, S. Occurrence of antibiotic resistant Campylobacter in wild birds and poultry. Malays. J. Microbiol. 2019, 15, 143–151. [Google Scholar] [CrossRef]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiol. Open 2021, 10, e1197. [Google Scholar] [CrossRef] [PubMed]
- Acheson, D.; Allos, B. Campylobacter jejuni infections: Update on emerging issues and trends. Clin. Infect. Dis. 2001, 8, 1201–1206. [Google Scholar] [CrossRef]
- Sarkar, D.J.; Mukherjee, I.; Shakil, N.A.; Rana, V.S.; Kaushik, P.; Debnath, S. Antibiotics in Agriculture: Use and Impact. Ind. J. Ethnophytopharm. 2018, 4, 4–19. [Google Scholar]
- Ge, B.; Wang, F.; Sjolund-Karlsson, M.; McDermott, P. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. J. Microbiol. 2013, 95, 57–67. [Google Scholar] [CrossRef]
- Narvaez-Bravo, C.; Taboada, E.; Mutschall, S.; Aslam, M. Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int. J. Food Microbiol. 2017, 253, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.L.; Selinger, L.B.; Taboada, E.N.; Inglis, G.D. Subtype-specific selection for resistance to fluoroquinolones but not to tetracycline is evident in Campylobacter jejuni isolates from beef cattle in confined feeding operations in Southern Al-berta, Canada. Appl. Environ. Microbiol. 2018, 84, e02713-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafez, H.; Attia, Y. Challenges to the Poultry Industry: Current Perspectives and Strategic Future After the COVID-19 Outbreak. Front. Vet. Sci. 2020, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Hotzel, H.; El-Adawy, H.; Tran, H.; Le, M.; Tomaso, H.; Neubauer, H.; Hafez, H.M. Genotyping and antibiotic resistance of thermophilic Campylobacter isolated from chicken and pig meat in Vietnam. Gut Pathog. 2016, 8, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczepanowski, R.; Krahn, I.; Linke, B.; Goesmann, A.; Pühler, A.; Schlüter, A. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Mirobiology 2004, 150, 3613–3630. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhadidy, M.; Ali, M.; El-Shibiny, A.; Miller, W.; Elkhatib, W.; Botteldoorn, N. Antimicrobial Resistance Patterns and Molecular Resistance Markers of Campylobacter jejuni Isolates from Human Diarrheal Cases. PLoS ONE 2020, 15, e0227833. [Google Scholar] [CrossRef] [Green Version]
- Munroe, D.; Prescott, J.; Penner, J. Campylobacter jejuni and Campylobacter coli serotypes isolated from chickens, cattle, and pigs. J. Clin. Microbiol. 1983, 18, 877–881. [Google Scholar] [CrossRef] [Green Version]
- Saito, S.; Yatsuyanagi, J.; Harata, S.; Ito, Y.; Shinagawa, K.; Suzuki, N. Campylobacter jejuni isolated from retail poultry meat, bovine feces and bile, and human diarrheal samples in Japan: Comparison of serotypes and genotypes. FEMS Immunol. Med. Microbiol. 2005, 45, 311–319. [Google Scholar] [CrossRef] [Green Version]
- Sahin, O.; Luo, N.; Huang, S.; Zhang, Q. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Appl. Environ. Microbiol. 2003, 69, 5372–5379. [Google Scholar] [CrossRef] [Green Version]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing. Ninth Informational Supplement NCCLS Document M100-S9; National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2008. [Google Scholar]
- Denis, M.; Tanguy, M.; Chidaine, B.; Laisney, M.J.; Mégraud, F.; Fravalo, F. Description and sources of contamination by Campylobacter spp. of river water destined for human consumption in Brittany, France. Pathol. Biol. 2009, 59, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Wafaa, A. Abd El-Ghany. One health approach of campylobacteriosis in Egypt: An emerging zoonotic disease. J. Infect. Dev. Ctries. 2019, 3, 956–960. [Google Scholar] [CrossRef]



| Biochemical Tests | Campylobacter Species n = 20 Isolates Total | |
|---|---|---|
| Campylobacter jejuni n (%) | Campylobacter coli n (%) | |
| Catalase test | 15 (100%) | 5 (100%) |
| Oxidase test | 15 (100%) | 5 (100%) |
| Motility test | 15 (100%) | 5 (100%) |
| Nitrate reduction test | 15 (100%) | 5 (100%) |
| TSI test | ||
| -Glucose utilization | 0 (0%) | 0 (0%) |
| -H2S production | 0 (0%) | 5 (100%) |
| Hippurate test | 15 (100%) | 0 (0%) |
| Sources | Type of Sample | Samples Colonized with C. jejuni n (%) | |
|---|---|---|---|
| Animals (n = 90) | Licensed slaughterhouse (n = 30) | Neck (n = 15) | 1 (6.66%) |
| Wing (n = 15) | 0 (0%) | ||
| Non-licensed slaughterhouses (n = 60) | Neck (n = 15) | 5 (33.33%) | |
| Wing (n = 15) | 1 (6.66%) | ||
| Cloaca (n = 30) | 6 (20%) | ||
| Humans (n = 70) | Clinical Samples | Human stool (n = 70) | 2 (2.85%) |
| Groups of Antibiotics | Type of Antibiotics | Human C. jejuni Isolates n = 2 | Chickens’ C. jejuni Isolates n = 13 | ||
|---|---|---|---|---|---|
| Susceptible n (%) | Resistant n (%) | Susceptible n (%) | Resistant n (%) | ||
| Fluoroquinolones | Ciprofloxacin (5 μg) | 0 (0%) | 2 (100%) | 0 (0%) | 13 (100%) |
| Beta lactam | Imipenem (10 μg) | 2 (100%) | 0 (0%) | 13 (100%) | 0 (0%) |
| Azteronam (30 μg) | 0 (0%) | 2 (100%) | 0 (0%) | 13 (100%) | |
| Ampicillin (10 μg) | 0 (0%) | 2 (100%) | 1 (7.6%) | 12 (92%) | |
| Aminoglycosides | Gentamicin (10 μg) | 0 (0%) | (100%) | 5 (38%) | 8 (61.5%) |
| Macrolide | Erythromycin (15 μg) | 0 (0%) | 2 (100%) | 2 (15%) | 11 (84.6%) |
| Tetracycline | Tetracycline (30 μg) | 0 (0%) | 2 (100%) | 0 (0%) | 13 (100%) |
| Antibiotic Resistance Profiles | Total MDR Isolates (n = 15) | Total Isolates n (%) | |
|---|---|---|---|
| Human Isolates n = 2 | Poultry Isolates n = 13 | ||
| cip + tet + atm + amp + ery + gen + | 1 | 7 | 8 (53.3%) |
| cip + tet + atm + amp + ery + | 1 | 3 | 4 (26.7%) |
| cip + tet + atm + amp + gen + | 0 | 2 | 2 (13.3%) |
| cip + tet + atm + ery + | 0 | 1 | 1 (6.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Khresieh, R.O.; Al-Daghistani, H.I.; Abu-Romman, S.M.; Abu-Niaaj, L.F. Genetic Signature and Serocompatibility Evidence for Drug Resistant Campylobacter jejuni. Antibiotics 2022, 11, 1421. https://doi.org/10.3390/antibiotics11101421
Al-Khresieh RO, Al-Daghistani HI, Abu-Romman SM, Abu-Niaaj LF. Genetic Signature and Serocompatibility Evidence for Drug Resistant Campylobacter jejuni. Antibiotics. 2022; 11(10):1421. https://doi.org/10.3390/antibiotics11101421
Chicago/Turabian StyleAl-Khresieh, Rozan O., Hala I. Al-Daghistani, Saeid M. Abu-Romman, and Lubna F. Abu-Niaaj. 2022. "Genetic Signature and Serocompatibility Evidence for Drug Resistant Campylobacter jejuni" Antibiotics 11, no. 10: 1421. https://doi.org/10.3390/antibiotics11101421
APA StyleAl-Khresieh, R. O., Al-Daghistani, H. I., Abu-Romman, S. M., & Abu-Niaaj, L. F. (2022). Genetic Signature and Serocompatibility Evidence for Drug Resistant Campylobacter jejuni. Antibiotics, 11(10), 1421. https://doi.org/10.3390/antibiotics11101421