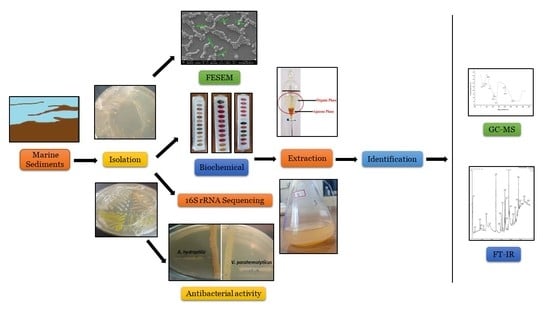
Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens
Abstract
1. Introduction
2. Results
2.1. Isolation of Actinomycetes
2.2. Characterization of Isolates
2.2.1. Screening of Potential Actinomycete Isolates
2.2.2. Screening of the Potential Isolate in Comparison to the Antibiotic
2.2.3. Morphological Characterization
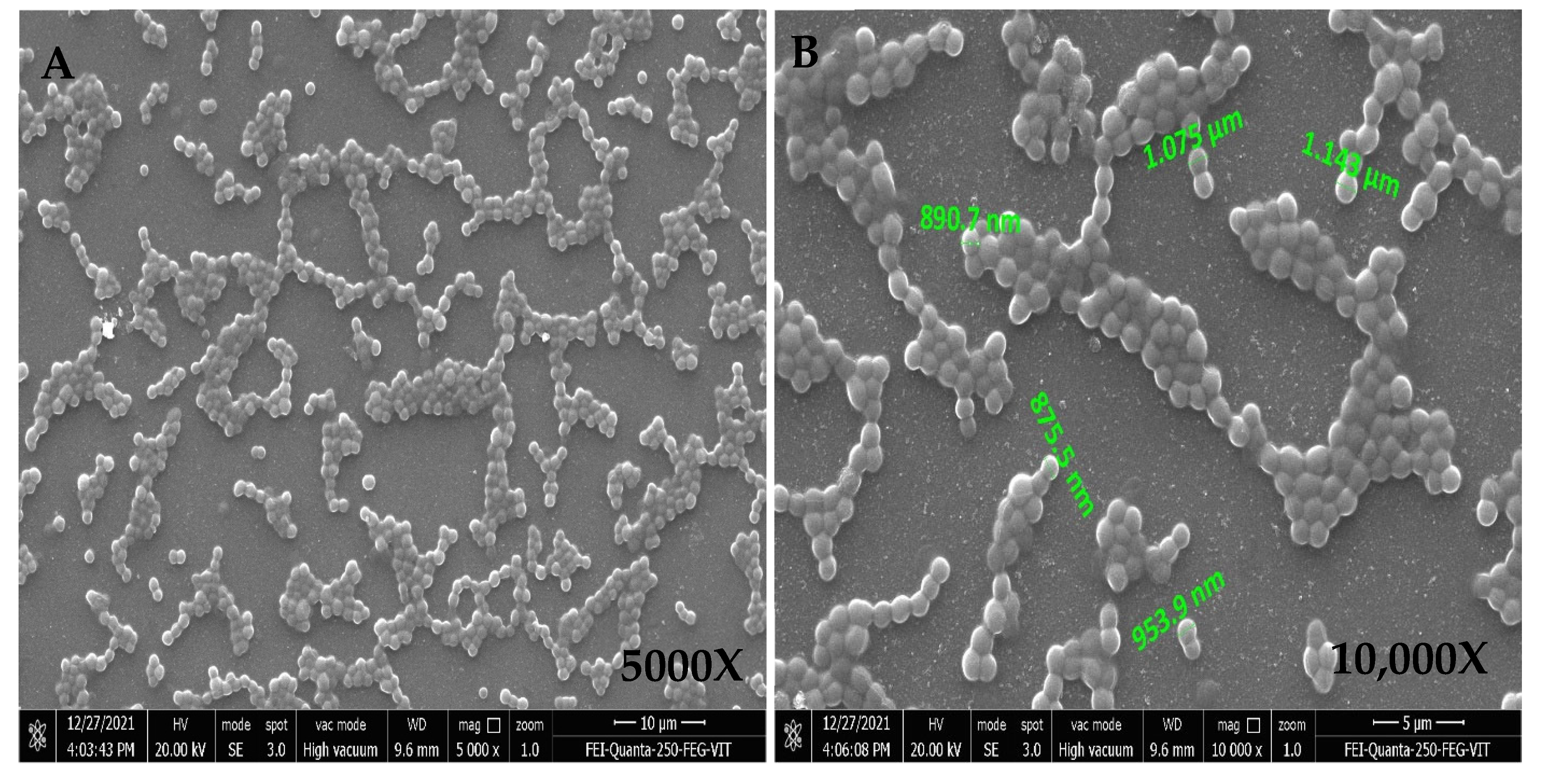
FESEM Analysis
Biochemical Characterization
Physiological Characterization
Molecular Characterization
2.3. FTIR Analysis of Crude Extracts from Actinomycetes
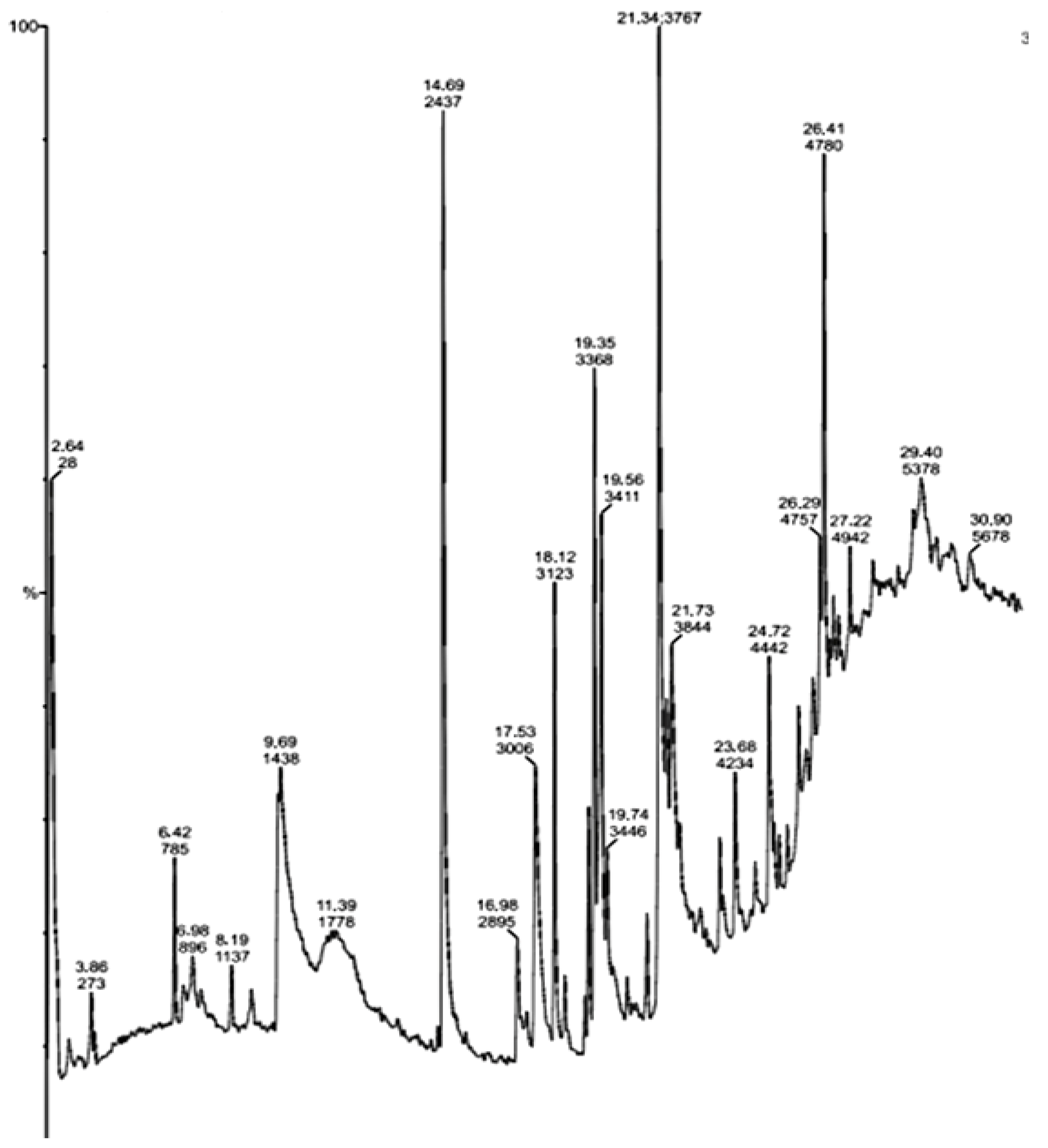
2.4. GC-MS Analysis of Crude Extracts from Actinomycetes
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Media Used
Media
4.3. Pre-Treatment of Sediment Samples
4.4. Isolation of Samples
4.5. Screening of Samples
4.6. Characterization of Isolates
4.6.1. Morphological Characterization
Gram Staining
Motility Test
Capsule Staining
Field Emission Scanning Electron Microscope (FESEM) Analysis
4.6.2. Biochemical Characterization
Biochemical Tests
Starch Utilization Test
Gelatin Hydrolysis Test
Casein Hydrolysis Test
Citrate Utilization Test
4.6.3. Physiological Characterization
4.6.4. Molecular Characterization
4.7. Bacterial Cultures Used
4.8. In Vitro Antibacterial Activity
4.8.1. Primary Screening
4.8.2. Secondary Screening
Agar Well Diffusion Method
Disc Diffusion Method
4.9. Extraction of the Active Compound
4.10. Identification of the Crude Bioactive Compound
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mo, E.; Wa, E.; Gm, D. Marine Actinomycetes the Past, the Present and the Future. Pharm. Res. 2021, 5, 000241. [Google Scholar] [CrossRef]
- Arbat, A.B.; Zodpe, S.N. Biodiversity of Actinomycetes Species Isolated from Saline Belt of Akola District. Indian J. Appl. Res. 2011, 4, 450–452. [Google Scholar] [CrossRef]
- Rashad, F.M.; Fathy, H.M.; El-Zayat, A.S.; Elghonaimy, A.M. Isolation and Characterization of Multifunctional Streptomyces Species with Antimicrobial, Nematicidal and Phytohormone Activities from Marine Environments in Egypt. Microbiol. Res. 2015, 175, 34–47. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Rasool, U.; Hemalatha, S. Diversified Role and Applications of Marine Actinomycetes in the Field of Biology. World Sci. News 2017, 74, 1–14. [Google Scholar]
- Imada, C.; Koseki, N.; Kamata, M.; Kobayashi, T.; Hamada-Sato, N. Isolation and Characterization of Antibacterial Substances Produced by Marine Actinomycetes in the Presence of Seawater. Actinomycetologica 2007, 21, 27–31. [Google Scholar] [CrossRef]
- Valli, S.; Sugasini, S.S.; Aysha, O.S.; Nirmala, P.; Vinoth Kumar, P.; Reena, A. Antimicrobial Potential of Actinomycetes Species Isolated from Marine Environment. Asian Pac. J. Trop. Biomed. 2012, 2, 469–473. [Google Scholar] [CrossRef]
- Helmke, E.; Weyland, H. Rhodococcus Marinonascens. Int. J. Syst. Bacteriol. 1984, 34, 127–138. [Google Scholar] [CrossRef]
- Colquhoun, J.A.; Mexson, J.; Goodfellow, M.; Ward, A.C.; Horikoshi, K.; Bull, A.T. Novel Rhodococci and Other Mycolate Actinomycetes from the Deep Sea. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 1998, 74, 27–40. [Google Scholar] [CrossRef]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and Persistent Populations of a Major New Marine Actinomycete Taxon in Ocean Sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar] [CrossRef]
- Yi, H.; Schumann, P.; Sohn, K.; Chun, J. Serinicoccus Marinus Gen. Nov., Sp. Nov., a Novel Actinomycete with L-Ornithine and L-Serine in the Peptidoglycan. Int. J. Syst. Evol. Microbiol. 2004, 54, 1585–1589. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Lyla, P.S.; Khan, S.A. Distribution and Generic Composition of Culturable Marine Actinomycetes from the Sediments of Indian Continental Slope of Bay of Bengal. Chin. J. Oceanol. Limnol. 2008, 26, 166–177. [Google Scholar] [CrossRef]
- Freel, K.C.; Edlund, A.; Jensen, P.R. Microdiversity and Evidence for High Dispersal Rates in the Marine Actinomycete “Salinispora Pacifica”. Environ. Microbiol. 2012, 14, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, N.B.; Sabry, S.A.; El-Sherif, Z.M.; Abu El-Ela, G.A. Isolation and Enumeration of Marine Actinomycetes from Seawater and Sediments in Alexandria. J. Gen. Appl. Microbiol. 2000, 46, 105–111. [Google Scholar] [CrossRef]
- Sharma, M.; Dangi, P.; Choudhary, M. Actinomycetes: Source, identification, and their applications. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 801–832. [Google Scholar]
- Solanki, R.; Khanna, M.; Lal, R. Bioactive Compounds from Marine Actinomycetes. Indian J. Microbiol. 2008, 48, 410–431. [Google Scholar] [CrossRef]
- Okami, Y.; Okazaki, T. Nocardia and Streptomyces; Gaustav Fischer: Stuttgart, Germany, 1978; pp. 145–151. [Google Scholar]
- Magarvey, N.A.; Keller, J.M.; Bernan, V.; Dworkin, M.; Sherman, D.H. Isolation and Characterization of Novel Marine-Derived Actinomycete Taxa Rich in Bioactive Metabolites. Appl. Environ. Microbiol. 2004, 70, 7520–7529. [Google Scholar] [CrossRef]
- Dilip, C.; Mulaje, S.S.; Mohalkar, R.Y. A Review on Actinomycetes and Their Biotechnological Application. Int. J. Pharm. Sci. Res. 2013, 4, 1730–1742. [Google Scholar] [CrossRef]
- Riedlinger, J.; Reicke, A.; Zähner, H.; Krismer, B.; Bull, A.T.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bister, B.; Bischoff, D.; et al. Abyssomicins, Inhibitors of the Para-Aminobenzoic Acid Pathway Produced by the Marine Verrucosispora Strain AB-18-032. J. Antibiot. 2004, 57, 271–279. [Google Scholar] [CrossRef]
- Soria-Mercado, I.E.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. Antibiotic Terpenoid Chloro-Dihydroquinones from a New Marine Actinomycete. J. Nat. Prod. 2005, 68, 904–910. [Google Scholar] [CrossRef]
- El-Gendy, M.M.A.; Shaaban, M.; Shaaban, K.A.; El-Bondkly, A.M.; Laatsch, H. Essramycin: A First Triazolopyrimidine Antibiotic Isolated from Nature. J. Antibiot. 2008, 61, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically Active Secondary Metabolites of Marine Actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.G. Antibiotics in Aquaculture. Is Responsible Use Possible? 2007. Available online: https://www.globalseafood.org/advocate/antibiotics-in-aquaculture-is-responsible-use-possible/ (accessed on 28 September 2022).
- Duke, J.U.S. Dr. Duke’s Phytochem. Ethanobotanical Databases; Department of Agriculture, Agricultural Research Service: Washington, DC, USA, 1992. [CrossRef]
- Suthindhiran, K.; Kannabiran, K. Cytotoxic and Antimicrobial Potential of Actinomycete Species Saccharopolyspora Salina VITSDK4 Isolated from the Bay of Bengal Coast of India. Am. J. Infect. Dis. 2009, 5, 90–98. [Google Scholar] [CrossRef]
- Gebreyohannes, G.; Moges, F.; Sahile, S.; Raja, N. Isolation and Characterization of Potential Antibiotic Producing Actinomycetes from Water and Sediments of Lake Tana, Ethiopia. Asian Pac. J. Trop. Biomed. 2013, 3, 426–435. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, D.; Ding, L.; Li, X.L.; Liu, X.T.; Li, W.J.; Wei, T.; Yan, S.; Xie, J.H.; Wei, L.; et al. Three-Dimensional Poly-(ε-Caprolactone) Nanofibrous Scaffolds Directly Promote the Cardiomyocyte Differentiation of Murine-Induced Pluripotent Stem Cells through Wnt/β-Catenin Signaling. BMC Cell Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Oggerin, M.; Arahal, D.R.; Rubio, V.; Marín, I. Identification of Beijerinckia Fluminensis Strains CIP 106281T and UQM 1685T as Rhizobium Radiobacter Strains, and Proposal of Beijerinckia Doebereinerae Sp. Nov. to Accommodate Beijerinckia Fluminensis LMG 2819. Int. J. Syst. Evol. Microbiol. 2009, 59, 2323–2328. [Google Scholar] [CrossRef]
- Shwaaiman, H.A.; Shahid, M.; Elgorban, A.M.; Siddique, K.H.M.; Syed, A. Beijerinckia Fluminensis BFC-33, a Novel Multi-Stress-Tolerant Soil Bacterium: Deciphering the Stress Amelioration, Phytopathogenic Inhibition and Growth Promotion in Triticum Aestivum L. (Wheat). Chemosphere 2022, 295, 133843. [Google Scholar] [CrossRef]
- Trakunjae, C.; Boondaeng, A.; Apiwatanapiwat, W.; Kosugi, A.; Arai, T.; Sudesh, K.; Vaithanomsat, P. Enhanced Polyhydroxybutyrate (PHB) Production by Newly Isolated Rare Actinomycetes Rhodococcus Sp. Strain BSRT1-1 Using Response Surface Methodology. Sci. Rep. 2021, 11, 1896. [Google Scholar] [CrossRef]
- Sivanandhini, T.; Subbaiya, R.; Gopinath, M.; Mahavinod Angrasan, J.K.V.; Kabilan, T.; Masilamani Selvam, M. An Investigation on Morphological Characterization of Actinomycetes Isolated from Marine Sediments. Res. J. Pharm. Biol. Chem. Sci. 2015, 6, 1234–1243. [Google Scholar]
- Rajkumar, T.; Manimaran, M.; Taju, G.; Vimal, S.; Majeed, S.A.; Kannabiran, K.; Sivakumar, S.; Kumar, K.M.; Madhan, S.; Hameed, A.S. Antiviral Viral Compound from Streptomyces Ghanaensis like Strain against White Spot 2 Syndrome Virus (WSSV) of Shrimp. bioRxiv 2018. [Google Scholar] [CrossRef]
- Undabarrena, A.; Beltrametti, F.; Claverías, F.P.; González, M.; Moore, E.R.B.; Seeger, M.; Cámara, B. Exploring the Diversity and Antimicrobial Potential of Marine Actinobacteria from the Comau Fjord in Northern Patagonia, Chile. Front. Microbiol. 2016, 7, 1135. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Haque, S.; Singh, H.; Verma, J.; Vibha, K.; Singh, R.; Jawed, A.; Tripathi, C.K.M. Isolation, Screening, and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. Front. Microbiol. 2016, 7, 1921. [Google Scholar] [CrossRef] [PubMed]
- Azhar, N.; Yudiati, E.; Subagiyo; Alghazeer, R. Producing Active Secondary Metabolite Against Pathogenic Vibrio Spp. by Actinobacteria-Sodium Alginate Co-Culture. Ilmu Kelaut. Indones. J. Mar. Sci. 2021, 26, 254–264. [Google Scholar] [CrossRef]
- Norouzi, H.; Danesh, A.; Mohseni, M.; Khorasgani, M.R. Marine Actinomycetes with Probiotic Potential and Bioactivity against Multidrugdrug-Resistant Bacteria. Int. J. Mol. Cell. Med. 2018, 7, 44. [Google Scholar]
- Subramani, R.; Aalbersberg, W. Marine Actinomycetes: An Ongoing Source of Novel Bioactive Metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Dholakiya, R.N.; Kumar, R.; Mishra, A.; Mody, K.H.; Jha, B. Antibacterial and Antioxidant Activities of Novel Actinobacteria Strain Isolated from Gulf of Khambhat, Gujarat. Front. Microbiol. 2017, 8, 2420. [Google Scholar] [CrossRef]
- Ryu, M.J.; Hwang, S.; Kim, S.; Yang, I.; Oh, D.C.; Nam, S.J.; Fenical, W. Meroindenon and Merochlorins e and F, Antibacterial Meroterpenoids from a Marine-Derived Sediment Bacterium of the Genus Streptomyces. Org. Lett. 2019, 21, 5779–5783. [Google Scholar] [CrossRef]
- Wang, C.; Lu, Y.; Cao, S. Antimicrobial Compounds from Marine Actinomycetes. Arch. Pharm. Res. 2020, 43, 677–704. [Google Scholar] [CrossRef]
- Voon, W.W.Y.; Muhialdin, B.J.; Yusof, N.L.; Rukayadi, Y.; Meor Hussin, A.S. Bio-Cellulose Production by Beijerinckia Fluminensis WAUPM53 and Gluconacetobacter Xylinus 0416 in Sago By-Product Medium. Appl. Biochem. Biotechnol. 2019, 187, 211–220. [Google Scholar] [CrossRef]
- Joshi, M.H.; Patil, A.A.; Adivarekar, R.V. Characterization of Brown-Black Pigment Isolated from Soil Bacteria, Beijerinckia Fluminensis. bioRxiv Microbiol. 2021. [Google Scholar] [CrossRef]
- Imada, C.; Masuda, S.; Kobayashi, T.; Hamada-Sato, N.; Nakashima, T. Isolation and Characterization of Marine and Terrestrial Actinomycetes Using a Medium Supplemented with NaCl. Actinomycetologica 2010, 24, 12–17. [Google Scholar] [CrossRef]
- Manimaran, M.; Rajkumar, T.; Vimal, S.; Taju, G.; Majeed, S.A.; Hameed, A.S.S.; Kannabiran, K. Antiviral Activity of 9(10H)-Acridanone Extracted from Marine Streptomyces Fradiae Strain VITMK2 in Litopenaeus Vannamei Infected with White Spot Syndrome Virus. Aquaculture 2018, 488, 66–73. [Google Scholar] [CrossRef]
- Selvan, G.P.; Ravikumar, S.; Ramu, A.; Neelakandan, P. Antagonistic Activity of Marine Sponge Associated Streptomyces Sp. Against Isolated Fish Pathogens. Asian Pac. J. Trop. Dis. 2012, 2, S724–S728. [Google Scholar] [CrossRef]
- Malviya, M.K.; Pandey, A.; Sharma, A.; Tiwari, S.C. Characterization and Identification of Actinomycetes Isolated from “fired Plots” under Shifting Cultivation in Northeast Himalaya, India. Ann. Microbiol. 2013, 63, 561–569. [Google Scholar] [CrossRef]
- El Karkouri, A.; Assou, S.A.; El Hassouni, M. Isolation and Screening of Actinomycetes Producing Antimicrobial Substances from an Extreme Moroccan Biotope. Pan Afr. Med. J. 2019, 33, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cheesbrough, M. Adaptive or Specific Immune Response, District Laboratory in Tropical Countries; Cambridge University Press: Cape Town, South Africa, 2010; p. 11. [Google Scholar]
- Muiru, W.M.; Mutitu, E.W.; Mukunya, D.M. Identification of Selected Actinomycetes Isolates and Characterization of Their Antibiotic Metabolites. J. Biol. Sci. 2008, 8, 1021–1026. [Google Scholar] [CrossRef]
- Thirumurugan, D.; Vijayakumar, R. A Potent Fish Pathogenic Bacterial Killer Streptomyces Sp. Isolated from the Soils of East Coast Region, South India. J. Coast. Life Med. 2013, 1, 175–180. [Google Scholar] [CrossRef]
- Sapkota, A.; Thapa, A.; Budhathoki, A.; Sainju, M.; Shrestha, P.; Aryal, S. Isolation, Characterization, and Screening of Antimicrobial-Producing Actinomycetes from Soil Samples. Int. J. Microbiol. 2020, 2020, 2716584. [Google Scholar] [CrossRef]
- Barakat, K.M.; Beltagy, E.A. Bioactive Phthalate from Marine Streptomyces Ruber EKH2 against Virulent Fish Pathogens. Egypt. J. Aquat. Res. 2015, 41, 49–56. [Google Scholar] [CrossRef]
- Ramalingam, V.; Rajaram, R. Antioxidant Activity of 1-Hydroxy-1-Norresistomycin Derived from Streptomyces Variabilis KP149559 and Evaluation of Its Toxicity against Zebra Fish Danio Rerio. RSC Adv. 2016, 6, 16615–16623. [Google Scholar] [CrossRef]
- Thanigaivel, S.; Vijayakumar, S.; Mukherjee, A.; Chandrasekaran, N.; Thomas, J. Antioxidant and Antibacterial Activity of Chaetomorpha Antennina against Shrimp Pathogen Vibrio Parahaemolyticus. Aquaculture 2014, 433, 467–475. [Google Scholar] [CrossRef]
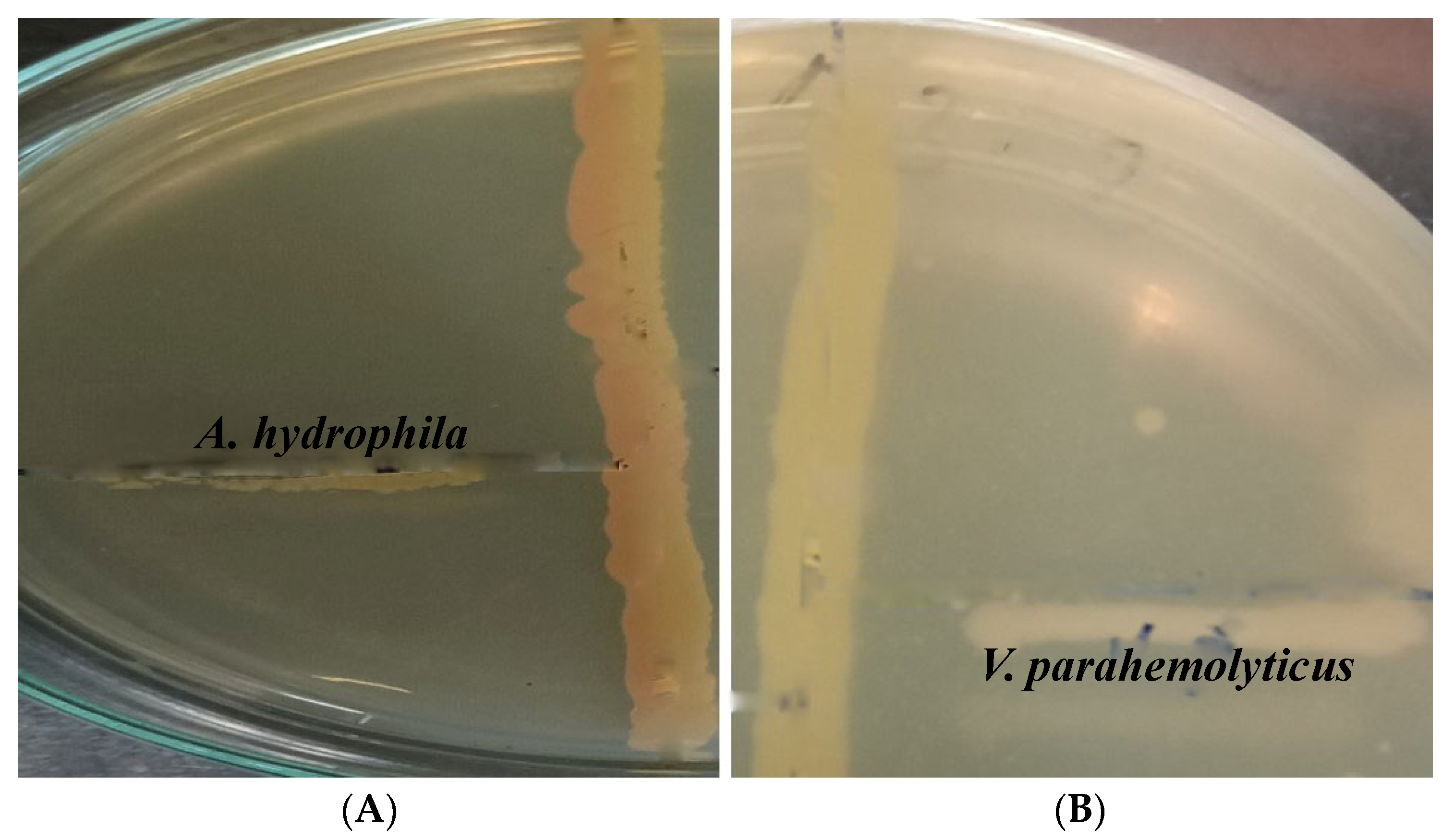

| S.No. | Isolates | Antibacterial Activity/Zone of Inhibition (in mm) | Zone of Inhibition in Gentamicin (in mm) | ||
|---|---|---|---|---|---|
| Aeromonas hydrophila | Vibrio parahemolyticus | Aeromonas hydrophila | Vibrio parahemolyticus | ||
| 1 | Isolate 1 (Beijernickia fluminensis) | 11 | 13 | 12 | 11 |
| 2 | Isolate 2 | 9 | 12 | 12 | 11 |
| Biochemical Characteristics | Beijerinickia fluminensis |
|---|---|
| ONPG | Positive |
| Lysine Utilization | Positive |
| Ornithine Utilization | Positive |
| Urease | Positive |
| Phenylalanine Deamination | Negative |
| Nitrate Reduction | Positive |
| H2S Production | Negative |
| Citrate Utilization | Positive |
| Voges–Proskauer | Negative |
| Methyl Red | Negative |
| Indole | Negative |
| Malonate Utilization | Negative |
| Esculin Hydrolysis | Positive |
| L-Arabinose | Negative |
| Xylose | Negative |
| Adonitol | Negative |
| Rhamnose | Negative |
| Cellobiose | Negative |
| Melibiose | Negative |
| Saccharose | Positive |
| Raffinose | Negative |
| Trehalose | Positive |
| Glucose | Positive |
| Lactose | Positive |
| Oxidase | Negative |
| Casein Utilization | Positive |
| Melezitose | Negative |
| α–Methyl–D-Mannoside | Negative |
| Xylitol | Positive |
| D-Arabinose | Negative |
| Sorbose | Positive |
| Wavelength | Functional Group/Bonds | Band Strength | Compound |
|---|---|---|---|
| Ethyl Acetate Extract of Actinomycetes (Sample) | |||
| 2652.15 | S-H stretch | Weak | Thiol |
| 2573.22 | O-H stretch | Strong, Broad | Carboxylic acid |
| 2041.57 | N=C=S stretch | Strong | Isothiocyanate |
| 2013.71 | N=C=S stretch | Strong | Isothiocyanate |
| 1292.27 | C-F stretch | Strong | Fluoro compound |
| 1167.49 | C-O stretch | Strong | Tertiary alcohol |
| 1097.84 | C-N stretch | Medium | Amine |
| 1042.705 | C-N stretch | Medium | Amine |
| 991.63 | C=C bending | Strong | Alkene |
| 885.41 | C=C bending | Strong | Alkene |
| 751.34 | C-H bending | Strong | 1,2-disubstituted |
| 709.55 | C=C bending | Strong | Alkene |
| 556.909 | C-Cl stretching | Strong | Halo compound |
| 464.62 | C-l stretching | Strong | Halo compound |
| S.No | Compounds | Major Peak Area (%) | Retention Time (RT) | Major Activities * Present in Sample |
|---|---|---|---|---|
| 1 | N, N-Dimethylheptanamide | 28 | 2.64 | Antibacterial, Antitumor |
| 2 | Butanamide,3,N-Dihydroxy | 273 | 3.86 | Antitumor |
| 3 | 3-O-Acetyl-Exo-1,2-O-Ethylidene- Alpha-D-Erythrof | 785 | 6.42 | Anticancer |
| 4 | 5-Aminovaleric acid | 1438 | 9.69 | Anticarcinogenic |
| 5 | Glycine,N-Octyl-,Ethyl Ester | 2437 | 14.69 | Antibacterial, Antitumor |
| 6 | As-Triazine-3,5(2H,4H)-Dione, 6-(Dimethylamino) | 2895 | 16.98 | |
| 7 | Glycyl-L-Proline | 3006 | 17.53 | Antibacterial |
| 8 | Propyl Aldoxime, 2-Methyl-, Syn | 3123 | 18.12 | DNA synthesis inhibitor |
| 9 | Actinomycin C2 | 3368 | 19.35 | Antibacterial, Anticancer |
| 10 | (S)-3,4-Dimethylpentanol | 3411 | 19.56 | Antibacterial, Anticancer, Antidiabetic, DNA synthesis inhibitor |
| 11 | 7-Tetradecene, (E) | 3767 | 21.34 | Antibacterial, Anticancer |
| 12 | 2-Decenioc acid | 3844 | 21.73 | Anticarcinogenic |
| 13 | 1-Decene, 8-Methyl | 4234 | 23.68 | Methyl-guanidine inhibitor |
| 14 | Heneicosane, 11-Phenyl | 4442 | 24.72 | - |
| 15 | Dodecane, 1-ChloroHeneicosane, 11-Phenyl | 4757 | 26.29 | - |
| 16 | Dichloroacetic Acid, 2-Ethylhexyl Ester | 4780 | 26.41 | - |
| 17 | Pterin-6-Carboxylic acid | 4942 | 27.22 | Anticarcinogenic |
| 18 | 2,3-Anhydro-D-Galactosan | 5378 | 29.40 | Anticancer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondal, H.; Thomas, J. Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens. Antibiotics 2022, 11, 1546. https://doi.org/10.3390/antibiotics11111546
Mondal H, Thomas J. Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens. Antibiotics. 2022; 11(11):1546. https://doi.org/10.3390/antibiotics11111546
Chicago/Turabian StyleMondal, Haimanti, and John Thomas. 2022. "Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens" Antibiotics 11, no. 11: 1546. https://doi.org/10.3390/antibiotics11111546
APA StyleMondal, H., & Thomas, J. (2022). Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens. Antibiotics, 11(11), 1546. https://doi.org/10.3390/antibiotics11111546